Abstract
Two bombsin peptides, GRPR agonist [Aca-QWAVGHLM-NH2] and antagonist [fQWAVGHL-NHEthyl] were evaluated. We employed the highly sensitive Waters Q-Tof Premier MS coupled with a UPLC system to identify the metabolites produced by rat hepatocytes or PC-3 human prostate cancer cells; and we utilized the AB/MDS 4000 Q-Trap LC/MS/MS system with highly sensitive quantitative and qualitative performance, to quantitatively analyze the internalization of GRPR agonist and antagonist in PC-3 cells. The major metabolites of both GRPR agonist and antagonist were the result of peptide bond hydrolysis between W and A which was demonstrated by observation of the N-terminal fragment m/z 446 (Aca-QW-OH) for agonist and m/z 480 (fQW-OH) for antagonist. Both peptides were also hydrolyzed between A and V which formed peaks m/z 517 [Aca-QWA-OH] and m/z 555 (VGHLM-NH2) for the agonist and m/z 551 [fQWA-OH] and m/z 452 (VGHL-NHEthyl) for the antagonist. The peptide agonist also formed a unique metabolite that resulted from hydrolysis of the C-terminal amide. The antagonist showed significantly slower metabolism as compared to the agonist in both rat hepatocytes and PC-3 cells. The antagonist also showed significantly lower PC-3 cell internalization rate than that of the agonist. In conclusion, the metabolism profiles of both GRPR agonist and antagonist peptides were identified by LC/MS. The antagonist peptide was more stable than the agonist peptide in rat hepatocyte incubation. One major factor could be the hydrolysis-resistant C-terminal L-NHEthyl group compared with the unsubstituted amide of the agonist. Another factor could be different amino acid sequences of the agonist and antagonist that may also influence the enzymatic hydrolysis. The antagonist ligand is potentially more useful for receptor-targeted imaging due primarily to its higher metabolic stability.
Keywords: LC/MS/MS, Gastrin-releasing peptide receptor (GRPR), Bombesin (BBN), Agonist, Antagonist
Introduction
Gastrin-releasing peptide receptor (GRPR) is expressed on a variety of human cancers, including breast, lung, pancreatic and prostate, making it a viable target toward site-directed localization or therapy of these human diseases (Smith et al. 2005; Markwalder and Reubi 1999; Gugger and Reubi 1999; Cornelio et al. 2007a, b). This receptor also represents a viable molecular target for radiolabeled bombesin (BBN) analogs for diagnostic or radiotherapeutic applications in these tumors. We and others have applied a number of radiolabeled BBN peptide derivatives for positron emission tomography imaging of tumor GRPR expression (Liu et al. 2009a, b; Shi et al. 2008; Yang et al. 2006; Zhang et al. 2006; Chen et al. 2004; Linder et al. 2009; Nock et al. 2003) targeted for GRPR. An optimal BBN-like radiotracer needs to meet several requirements: high affinity for GRPR, with rapid and specific tumor uptake; high hydrophilicity, with excretion profile that can be tailored to the desired pathway; high metabolic stability; and relatively rapid clearance from blood. Previous studies indicated that these ligands were stable over time in human plasma, but degraded rapidly in the kidneys and liver homogenates (Linder et al. 2009; Shipp et al. 1991; Zhang et al. 2004). The goal of this study was to identify the metabolites of GRPR agonist and antagonist (Fig. 1) using LC/MS and to use those results to provide guidance for future ligand development that may lead to clinical translation.
Fig. 1.
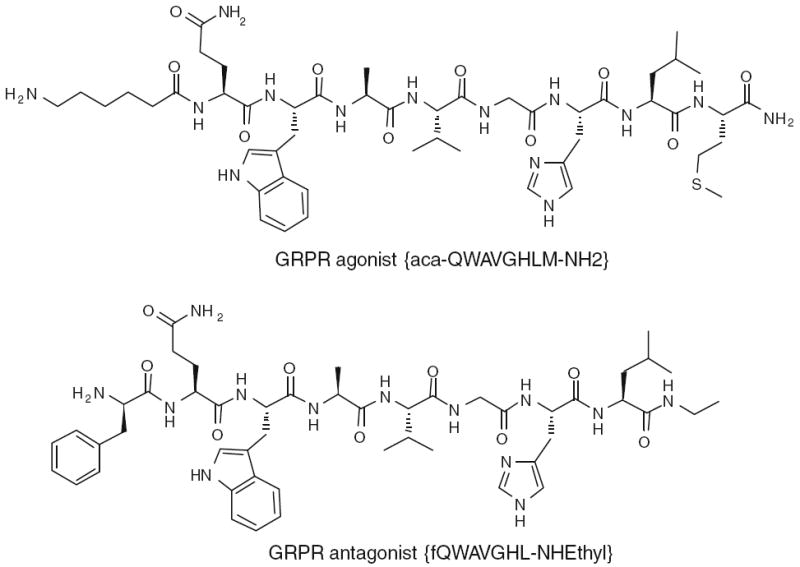
Structures of GRPR agonist [Aca-QWAVGHLM-NH2] and antagonist [fQWAVGHL-NHEthyl] peptides
It is well known from molecular–pharmacologic investigations that efficient internalization was predominantly observed for agonists (Wagner 1979). However, high-affinity receptor antagonists that poorly internalize into tumor cells can perform equally or even better in terms of in vivo uptake in animal tumor models than the corresponding agonists, which internalize efficiently (Cescato et al. 2008b). In this study, we explored the metabolism of a potent BBN agonist (Aca-QWAVGHLM-NH2) and a comparably potent BBN antagonist (fQWAVGHL-NHEthyl) (Fig. 1) in rat hepatocytes and PC-3 human prostate cancer cells using LC/MS. In addition, the cell binding and cell internalization of the two peptides were investigated in the same assays with the same batch of PC-3 cells under identical conditions.
Methods
Chemicals, reagents and solutions
Acetonitrile (CH3CN, HPLC grade) was purchased from Fisher Scientific (Pittsburgh, PA). All other reagents for synthesis and analysis were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise indicated in the context. Aca-QWAVGHLM-NH2 was prepared according to the published procedure using solid-phase Fmoc chemistry by American Peptide Inc. fQWAVGHL-NHEthyl was synthesized in our laboratory using solid-phase Fmoc chemistry and purified by semi-preparative reversed-phase HPLC. Purity and identity were established by LC/MS; Aca-QWAVGHLM-NH2 (m/z 1,054 [M+1], 95% purity) and fQWAVGHL-NHEthyl (m/z 985 [M+1], 97% purity). Stock solutions of the two peptides were prepared in water at a concentration of 1 mg/mL.
LC/MS
For qualitative analysis of metabolites, LC/MS analysis employed a Waters LC-MS system (Waters, Milford, MA) that included an Acquity UPLC system coupled to the Waters Q-Tof Premier high resolution mass spectrometer. An Acquity BEH Shield RP18 column (150 × 2.1 mm) was employed for chromatography. Elution was achieved with a binary mixture of two components. Solution A was composed of 2 mM ammonium formate, 0.1% formic acid and 5% CH3CN; solution B was composed of 2 mM ammonium formate and 0.1% formic acid in CH3CN. The elution profile, at 0.2 mL/min, had the following components: initial condition at 100% (v:v) A and 0% B; gradient 0–40% B over 15 min; isocratic elution at 40% B for an additional 3 min; 40–80% B over 2 min; and re-equilibrated with A for an additional 4 min. The retention time for Aca-QWAVGHLM-NH2 and fQWAVGHL-NHEthyl were 10.47 and 11.47 min, respectively. The injection volume was 10 μL. The entire column elute was introduced into the Q–Tof mass spectrometer. Ion detection was achieved in ESI mode using a source capillary voltage of 3.5 kV, source temperature of 110°C, desolvation temperature of 200°C, cone gas flow of 50 L/h (N2) and desolvation gas flow of 700 L/h (N2).
For quantitative analysis of peptides for cell internalization studies, an LC/MS system consisting of an Agilent 1200 autosampler, Agilent 1200 LC pump and an AB/MDS Sciex 4000 Q-TRAP (AB Sciex, Foster City, CA, USA). Standards were prepared for each peptide covering the concentration range from 0.001 to 10 μM in 1:1 CH3CN–water. Three replicate injections (10 μL) were made for each concentration level.
Separation was achieved on an Agilent RP18 column (1.8 μm, 100 × 4.6 mm, with 2 mM ammonium acetate and CH3CN) with gradient system at flow rate of 1.0 mL/min with the same solvent system. A gradient of 0% B for 3 min ramped to 40% B was utilized and isocratic elution at 40% B for an additional 4 min; 80% B for 2 min; and re-equilibrated with A for additional 1 min. Five different combinations of multiple reaction monitoring (MRM) [Aca-QWAVGHLM-NH2 (527.3/114.1, 446.3/114.1, 517.3/114.1, 630.3/114.1, 527.8/110.1) and fQWAVGHL-NHEthyl (492.8/120.1, 480.3/120.1, 551.3/120.1, 650.3/120.1)] and full-scan MS/MS experiments were performed. The specific comparisons made for quantitation used a single MRM transition per analyte.
Incubation with hepatocytes
Cryopreserved hepatocytes from male Sprague-Dawley rats (Celsis In Vitro Technologies, Inc., Baltimore, MD) were used in the in vitro metabolism studies. The cells, which were stored in liquid nitrogen (6–12 months), were thawed rapidly at 37°C in a water bath and gradually diluted with cell culture medium (Celsis In Vitro Technologies, Inc.). After washing the cells with the medium and adjusting the viable cell concentration to 1.0 million/mL, the resulting cell suspension was incubated at 37°C for 15 min prior to the introduction of the test compound. The Aca-QWAVGHLM-NH2 and fQWAVGHL-NHEthyl (10 μL from a stock solution of 1.0 mg/mL in water) was added to 1.0 mL suspension of cells. The final concentration of test compound was 10 μg/mL. The suspension was maintained at 37°C; 100 μL of cell suspension was removed and added to 100 μL acetonitrile at 10, 30, 60, 120 and 240 min. Each aliquot was centrifuged at 5,000 rpm for 5 min. The supernatants (10 μL) were analyzed by LC–MS.
Internalization studies in PC-3 cell line
The PC-3 human prostate carcinoma cell line was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). PC-3 cells were grown in DMEM (Mediatech Inc., Manassas, VA) supplemented with 10% (v/v) fetal bovine serum (Mediatech), 100 IU/mL penicillin and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA), at 37°C in a humidified atmosphere containing 5% CO2.
For the uptake assay, PC-3 cells were seeded into 6-well plates at a density of 1 × 106 cells/well and incubated for 24 h. Cells were rinsed three times with phosphate-buffered saline, followed by the addition of Aca-QWAVGHLM-NH2 or fQWAVGHL-NHEthyl to the cultured wells in triplicate (1 nmol/well). After incubation at 37°C for 15, 30, 60 and 120 min, the samples were processed in two different manners at each time point. For the first set, the media was collected for quantitative analysis of the unbound ligand. The cells were washed twice with normal saline (1 mL) and the washings discarded. Finally, the cells were lysed by the addition of 500 μL 0.1 N NaOH and then neutralized with 500 μL 0.1 N HCl. This final solution was collected for analysis of total cellular binding. A second set was first separated from the medium; the medium was analyzed for unbound ligand. The cells were washed twice (0.5 mL) with 50 mM glycine and 0.1 M NaCl (pH 2.8). These acid washes contained the cell surface binding component. The cells were then lysed by adding 1 N NaOH and 1 N HCl and the resulting mixture was analyzed for internalized ligand. Thus, two fractions from the first processing method and three fractions from the second processing method were analyzed by LC/MS to determine the amount in the four pools: unbound ligand, total cellular binding, cell surface binding and internalized ligand.
Results
Metabolism in rat hepatocytes
We incubated cultured rat hepatocytes with both agonist and antagonist peptides. Because we harvested the metabolites by treating an aliquot of the cell suspension with CH3CN, we presume that all metabolites are sampled regardless of site of generation. The metabolites in the supernatant solution were analyzed by LC/MS (Fig. 2 for agonist and Fig. 3 for antagonist). The major metabolites of both GRPR agonist [Aca-QWAVGHLM-NH2] and antagonist [fQWAVGHL-NHEthyl] were the result of peptide bond hydrolysis between W and A, which was demonstrated by observation of the N-terminal fragment m/z 446 (Aca-QW-OH) for agonist and m/z 480 (fQW-OH) for antagonist. Both peptides were also hydrolyzed between A and V which formed peaks at m/z 517 [Aca-QWA-OH] and m/z 555 (VGHLM-NH2) for the agonist and m/z 551 [fQWA-OH] and m/z 452 (VGHL-NHEthyl) for the antagonist. The agonist peptide also formed a unique metabolite from hydrolysis of the C-terminal amide. The parent amide (Aca-QWAVGHLM-NH2) and acid metabolite (Aca-QWAVGHLM-OH) both exhibit a doubly charged molecular ion ([M+1]/2). The ion observed for the acid metabolite was m/z 0.503 larger than the amide. The ability to conduct high resolution mass measurements was important to allow the characterization of this metabolite.
Fig. 2.
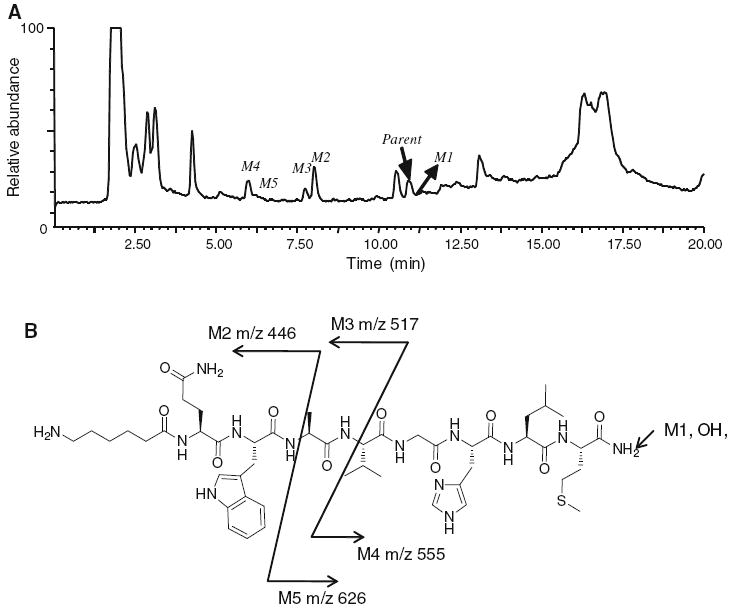
a The total ion chromatography (TIC) of agonist [Aca-QWAVGHLM-NH2] metabolites after 30 min incubation with rat hepatocytes. The other peaks were from the incubation media which as confirmed by the control experiments without adding the peptide. b Hydrolysis sites of possible metabolites and their molecular weights [m/z] observed in positive ESI mass spectrometry
Fig. 3.
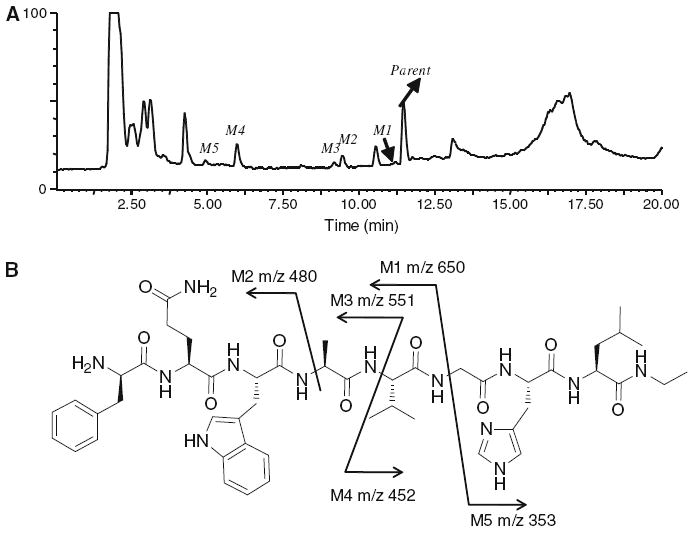
a The total ion chromatography (TIC) of antagonist [fQWAVGHL-NHEthyl] metabolites after 30 min of incubation in rat hepatocytes. The other peaks were from the incubation media which as confirmed by the control experiments without adding the peptide. b Hydrolysis sites of possible metabolites and their molecular weights [m/z] observed in positive ESI mass spectrometry
The metabolic decomposition rate of the two peptides was examined in the same preparation of rat hepatocytes (Fig. 4a). Within 2 h, the parent agonist peptide had nearly disappeared from the assay solution while the antagonist parent compound persisted. The half life of parent from the medium was estimated to be 4 and 32 min for the agonist and antagonist, respectively.
Fig. 4.
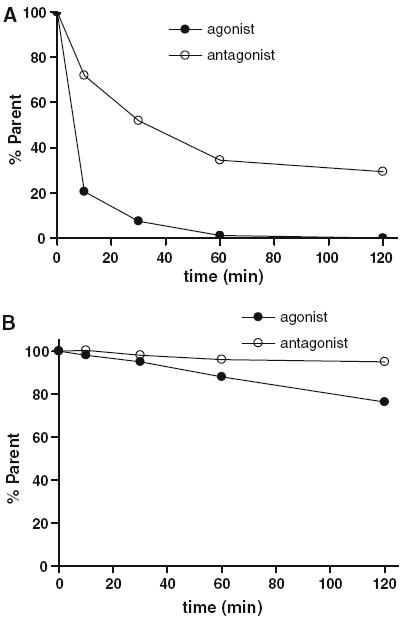
a The metabolic rate of agonist [Aca-QWAVGHLM-NH2] and antagonist [fQWAVGHL-NHEthyl] presented as the percent of parent ligands at various time points during 120 min of incubation with rat hepatocytes. b The percent of parent ligands of agonist [Aca-QWAVGHLM-NH2] and antagonist [fQWAVGHL-NHEthyl] during 120 min of incubation with PC-3 cells
Cell internalization
The highly sensitive 4000 Qtrap MS system was used to quantitate ligand binding to the PC-3 cells. Standard curves were established with various concentrations of agonist [Aca-QWAVGHLM-NH2] and antagonist [fQWAVGHL-NHEthyl] in CH3CN:water (1:1); a linear response from 0.001 to 10.0 μM (0.01–100 pmol on column) was observed. The detection limit (s/n > 3) for both ligands was approximately 0.5 nM.
The direct binding study was performed by incubating 1 nmol agonist or antagonist with GRPR expressing PC-3 prostate cancer cells (Markwalder and Reubi 1999). At specified time points, aliquots were taken and the cells were separated from the media. The media contained unbound ligand. The cells were washed with acid to recover surface bound ligand and then lysed to recover the internalized ligand. A second aliquot at each time point omitted the acid wash to give, after cell lysis, the total binding and internalization. Each of the fractions was quantitatively analyzed for parent ligand by LC/MS (Fig. 5). The cell uptake was expressed as the percent of total ligand added to the cell. Aca-QWAVGHLM-NH2 showed relatively high uptake inside the cell and lower uptake on the surface of cell, while fQWAVGHL-NHEthyl showed very low uptake inside the cell and similar level of cell surface binding when compared with Aca-QWAVGHLM-NH2.
Fig. 5.
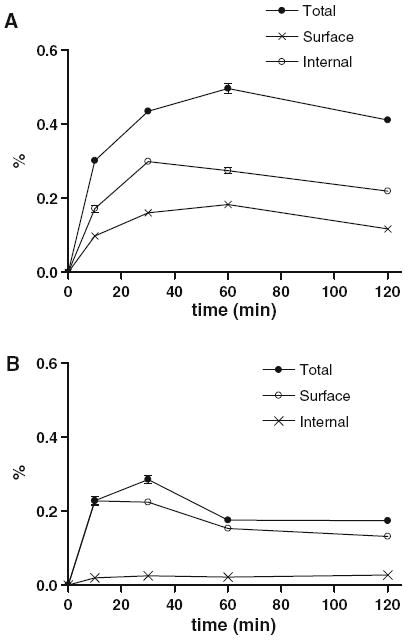
a The percent of parent ligands of agonist [Aca-QWAVGHLM-NH2] presented by total cell binding, surface binding and internalization during 120 min of incubation in PC-3 cells. b The percent of parent ligands of antagonist [fQWAVGHL-NHEthyl] presented by total cell binding, surface binding and internalization during 120 min of incubation in PC-3 cells
Both ligands were relatively stable in PC-3 cell culture solution when compared with hepatocyte incubation (Fig. 4b). The half-lives of parent ligands in all the pools in PC-3 cell were 3.5 and 8 h for the agonist and antagonist, respectively. The acid metabolite (Aca-QWAVGHLM-OH) from Aca-QWAVGHLM-NH2 was found in the unbound pool in the incubation media, and on the surface of PC-3 cells.
Discussion
The inherent inability of radionuclide imaging to differentiate between a parent compound and its radiolabeled metabolites confounds the interpretation of images and may impact the identification of the pathologically induced biochemical changes under investigation. Cytochrome P450 isoforms play a major role in mammalian xenobiotic biotransformation. The utility of liver hepatocytes to generate phase 1 and 2 metabolites of proposed radioligands followed by the analysis of the metabolites has been exploited to determine metabolic pathways (Lahoz et al. 2008a; Lahoz et al. 2008b; Ma et al. 2010). Once these cells have been used to generate a metabolism profile, LC/MS can be applied to identify the possible structures and quantitate the metabolism rate of the parent radioligand.
LC/MS has the unique combination of sensitivity and mass selectivity to provide sensitive detection and mass to charge ratio (m/z) data that can be used to propose parent and metabolite structures. In addition, the ability to conduct LC/MS/MS studies provides even greater power for assignment of possible structure. The various m/z components from collision-induced fragmentation of the parent ion can provide information on the structure of the parent ion. MRM is an LC/MS/MS technique that allows scanning for several daughter ions from a single parent ion and can be used to provide optimal sensitivity for quantitative analysis.
In our rat hepatocyte incubations, we found that both agonist and antagonist were metabolized in a similar pathway to that which had previously been elucidated (Linder et al. 2009). The rapid proteolysis between the tryptophan and alanine was expected for the BBN-like peptides and this cleavage was observed for both agonist and antagonist peptides. In addition, the agonist showed hydrolysis of the C-terminal amide. Unfortunately, the MS results cannot be used to determine the relative amounts of each of the metabolites because of the compound dependence on the ionization response. However, the half-life of the parent peptide could be determined for each compound. The half-lives in rat hepatocytes were 4 and 32 min for the agonist and antagonist, respectively. We attribute the shorter half-life of the agonist to the relatively rapid formation of the amide hydrolysis product and the difference in peptide sequence when compared with the antagonist. In our case, a methionine amino acid residue was present in the agonist instead of leucine in the antagonist, which may influence the enzymatic hydrolysis.
PC-3 cells are derived from human prostate cancer. Since GRPR is related to neoplastic transformation and is expressed at high levels in the intraepithelial neoplasia and primary carcinoma of prostate, GRPR can be used as a marker for imaging and staging prostate cancer (Linder et al. 2009).
The agonist and antagonist activities had been discerned from previous studies (Nock et al. 2003). By far, most BBN derivatives are agonists. Agonists are internalized into and accumulated within cells and exhibit higher uptake than antagonists. The expectation that the agonist would internalize while the antagonist would not was tested using our quantitative assay of the various components of cells following cellular uptake. We found both agonist and antagonist have similar cell surface binding, but the agonist had much higher total cell uptake due to the higher internalization. The antagonist displayed almost no internalization.
The only metabolite observed in the PC-3 cell incubation was the amide hydrolysis product derived from the agonist. The major reason that both BBN agonist and antagonist are more stable in PC-3 cells than in rat hepatocytes is that hepatocytes contain higher expression of P450 enzymes and probably other important enzymes involved in protein degradation than PC-3 tumor cells.
These results suggest that the superior in vivo imaging properties (Cescato et al. 2008a, b) of the antagonist were not due to superior tumor cell binding, but may relate to the much slower metabolism compared with agonist. The successful imaging of BBN-like ligands (both agonist and antagonist) is directly related to the presence of parent ligand in the targeted tissue.
Conclusion
The metabolism profiles of both GRPR agonist and antagonist peptides were identified by LC/MS. The major metabolites of both GRPR agonist and antagonist were the result of peptide bond hydrolysis between W and A in rat hepatocyte incubation. The antagonist peptide was observed to be more stable than the agonist peptide. This difference was believed to be due to the added stability of the N-ethyl amide when compared with an un-substituted amide, the differences in enzymatic hydrolysis rates because of the differing peptide sequences, or a combination of these two properties. Both agonist and antagonist have similar cell surface binding, but the agonist had much higher total cell uptake due to the higher internalization. The antagonist displayed almost no internalization.
LC/MS data can be used to probe future different labeling ligands in both the identity of metabolites and the internalization of cell binding. This study provides an efficient and sensitive method for screening radiotracer candidates without the need for radiolabeling. The combination of LC and MS/MS can provide both structural information for identification of metabolites and high selectivity for accurate quantitation of ligands in cell binding.
Acknowledgments
The research was supported by Intramural Research Program of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH).
Contributor Information
Dongyu Gu, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), 31 Center Dr, Suite 1C14, Bethesda, MD 20892-2281, USA, Xinjiang Technical Institute of Physics and Chemistry, Chinese Academy of Sciences, Urumqi, China, Graduate University of the Chinese Academy of Sciences, Beijing, China.
Ying Ma, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), 31 Center Dr, Suite 1C14, Bethesda, MD 20892-2281, USA.
Gang Niu, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), 31 Center Dr, Suite 1C14, Bethesda, MD 20892-2281, USA.
Yongjun Yan, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), 31 Center Dr, Suite 1C14, Bethesda, MD 20892-2281, USA.
Lixin Lang, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), 31 Center Dr, Suite 1C14, Bethesda, MD 20892-2281, USA.
Haji Akber Aisaand, Xinjiang Technical Institute of Physics and Chemistry, Chinese Academy of Sciences, Urumqi, China.
Haokao Gao, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), 31 Center Dr, Suite 1C14, Bethesda, MD 20892-2281, USA.
Dale O. Kiesewetter, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), 31 Center Dr, Suite 1C14, Bethesda, MD 20892-2281, USA, NIBIB/NIH, 10 Center Drive MSC 1180, Bethesda, MD 20892, USA, dk7k@nih.gov
Xiaoyuan Chen, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), 31 Center Dr, Suite 1C14, Bethesda, MD 20892-2281, USA, shawn.chen@nih.gov.
References
- Cescato R, Erchegyi J, Waser B, Piccand V, Maecke HR, Rivier JE, Reubi JC. Design and in vitro characterization of highly SST2-selective somatostatin antagonists suitable for radiotargeting. J Med Chem. 2008a;51:4030–4037. doi: 10.1021/jm701618q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cescato R, Maina T, Nock B, Nikolopoulou A, Charalambidis D, Piccand V, Reubi JC. Bombesin receptor antagonists may be preferable to agonists for tumor targeting. J Nucl Med. 2008b;49:318–326. doi: 10.2967/jnumed.107.045054. [DOI] [PubMed] [Google Scholar]
- Chen X, Park R, Hou Y, Tohme M, Shahinian AH, Bading JR, Conti PS. Micropet and autoradiographic imaging of grp receptor expression with 64Cu-DOTA-[Lys3]bombesin in human prostate adenocarcinoma xenografts. J Nucl Med. 2004;45:1390–1397. [PubMed] [Google Scholar]
- Cornelio DB, Meurer L, Roesler R, Schwartsmann G. Gastrin-releasing peptide receptor expression in cervical cancer. Oncology. 2007a;73:340–345. doi: 10.1159/000134478. [DOI] [PubMed] [Google Scholar]
- Cornelio DB, Roesler R, Schwartsmann G. Gastrin-releasing peptide receptor as a molecular target in experimental anticancer therapy. Ann Oncol. 2007b;18:1457–1466. doi: 10.1093/annonc/mdm058. [DOI] [PubMed] [Google Scholar]
- Gugger M, Reubi JC. Gastrin-releasing peptide receptors in non-neoplastic and neoplastic human breast. Am J Pathol. 1999;155:2067–2076. doi: 10.1016/S0002-9440(10)65525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahoz A, Donato MT, Castell JV, Gomez-Lechon MJ. Strategies to in vitro assessment of major human CYP enzyme activities by using liquid chromatography tandem mass spectrometry. Curr Drug Metab. 2008a;9:12–19. doi: 10.2174/138920008783331112. [DOI] [PubMed] [Google Scholar]
- Lahoz A, Donato MT, Montero S, Castell JV, Gomez-Lechon MJ. A new in vitro approach for the simultaneous determination of phase I and phase II enzymatic activities of human hepatocyte preparations. Rapid Commun Mass Spectrom. 2008b;22:240–244. doi: 10.1002/rcm.3359. [DOI] [PubMed] [Google Scholar]
- Linder KE, Metcalfe E, Arunachalam T, Chen J, Eaton SM, Feng W, Fan H, Raju N, Cagnolini A, Lantry LE, Nunn AD, Swenson RE. In vitro and in vivo metabolism of Lu-AMBA, a GRP-receptor binding compound, and the synthesis and characterization of its metabolites. Bioconjug Chem. 2009;20:1171–1178. doi: 10.1021/bc9000189. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li ZB, Cao Q, Liu S, Wang F, Chen X. Small-animal pet of tumors with 64Cu-labeled RGD-bombesin heterodimer. J Nucl Med. 2009a;50:1168–1177. doi: 10.2967/jnumed.108.061739. [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan Y, Chin FT, Wang F, Chen X. Dual integrin and gastrin-releasing peptide receptor targeted tumor imaging using 18F-labeled PEGylated RGD-bombesin heterodimer 18F-FB-PEG3-Glu-RGD-BBN. J Med Chem. 2009b;52:425–432. doi: 10.1021/jm801285t. [DOI] [PubMed] [Google Scholar]
- Ma Y, Kiesewetter DO, Lang L, Gu D, Chen X. Applications of LC–MS in PET radioligand development and metabolic elucidation. Curr Drug Metab. 2010;11:483–493. doi: 10.2174/138920010791636167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwalder R, Reubi JC. Gastrin-releasing peptide receptors in the human prostate: relation to neoplastic transformation. Cancer Res. 1999;59:1152–1159. [PubMed] [Google Scholar]
- Nock B, Nikolopoulou A, Chiotellis E, Loudos G, Maintas D, Reubi JC, Maina T. [99mTc]demobesin 1, a novel potent bombesin analogue for GRP receptor-targeted tumour imaging. Eur J Nucl Med Mol Imaging. 2003;30:247–258. doi: 10.1007/s00259-002-1040-x. [DOI] [PubMed] [Google Scholar]
- Shi J, Jia B, Liu Z, Yang Z, Yu Z, Chen K, Chen X, Liu S, Wang F. 99mTc-labeled bombesin(7-14)NH2 with favorable properties for SPECT imaging of colon cancer. Bioconjug Chem. 2008;19:1170–1178. doi: 10.1021/bc700471z. [DOI] [PubMed] [Google Scholar]
- Shipp MA, Tarr GE, Chen CY, Switzer SN, Hersh LB, Stein H, Sunday ME, Reinherz EL. CD10/neutral endopeptidase 24.11 hydrolyzes bombesin-like peptides and regulates the growth of small cell carcinomas of the lung. Proc Natl Acad Sci USA. 1991;88:10662–10666. doi: 10.1073/pnas.88.23.10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Volkert WA, Hoffman TJ. Radiolabeled peptide conjugates for targeting of the bombesin receptor superfamily subtypes. Nucl Med Biol. 2005;32:733–740. doi: 10.1016/j.nucmedbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Wagner JG. Fundamentals of clinical pharmacokinetics. Drug Intelligence Publications, Hamilton; 1979. [Google Scholar]
- Yang YS, Zhang X, Xiong Z, Chen X. Comparative in vitro and in vivo evaluation of two 64Cu-labeled bombesin analogs in a mouse model of human prostate adenocarcinoma. Nucl Med Biol. 2006;33:371–380. doi: 10.1016/j.nucmedbio.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Zhang H, Chen J, Waldherr C, Hinni K, Waser B, Reubi JC, Maecke HR. Synthesis and evaluation of bombesin derivatives on the basis of pan-bombesin peptides labeled with indium-111, lutetium-177, and yttrium-90 for targeting bombesin receptor-expressing tumors. Cancer Res. 2004;64:6707–6715. doi: 10.1158/0008-5472.CAN-03-3845. [DOI] [PubMed] [Google Scholar]
- Zhang X, Cai W, Cao F, Schreibmann E, Wu Y, Wu JC, Xing L, Chen X. 18F-labeled bombesin analogs for targeting GRP receptor-expressing prostate cancer. J Nucl Med. 2006;47:492–501. [PubMed] [Google Scholar]


