Abstract
DNA replication fork movement is impeded by collisions with transcription elongation complexes (TEC). We propose that a critical function of transcription termination factors is to prevent TEC from blocking DNA replication and inducing replication fork arrest, one consequence of which is DNA double-strand breaks. We show that inhibition of Rho-dependent transcription termination by bicyclomycin in Escherichia coli induced double-strand breaks. Cells deleted for Rho-cofactors nusA and nusG were hypersensitive to bicyclomycin, and had extensive chromosome fragmentation even in the absence of the drug. An RNA polymerase mutation that destabilizes TEC (rpoB*35) increased bicyclomycin resistance >40-fold. Double-strand break formation depended on DNA replication, and can be explained by replication fork collapse. Deleting recombination genes required for replication fork repair (recB and ruvC) increased sensitivity to bicyclomycin, as did loss of the replication fork reloading helicases rep and priA. We propose that Rho responds to a translocating replisome by releasing obstructing TEC.
Keywords: DNA polymerase/RNA polymerase collisions, pulsed-field gel electrophoresis, SOS
DNA replication forks translocate 20 to 30 times faster (~600 nt/s) than the rate of RNA polymerase (RNAP) elongation (~20 nt/s) (1–3). This finding raises the possibility of repeated collisions between replication forks and transcription elongation complexes (TEC), even when DNA and RNA synthesis are codirectional. Collisions between replication forks and TEC can generate fork collapse and chromosomal double-strand breaks (DSBs) (4, 5). Mechanisms have evolved, therefore, to reduce collisions and to repair collapsed forks. Fork repair is promoted by multiple pathways that involve a variety of DNA repair functions (6). Recombination can restart replisomes blocked by arrested TEC (7, 8). Boubakri et al. (9) have demonstrated that DNA helicases Rep, DinG, and UvrD resolve head-on collisions between TEC in rrn operons and replisomes. A recent report describes the role of transcription elongation factors DksA and GreA in preventing conflicts between replication and transcription (7, 10). In vitro, replication forks that collapse after colliding with a codirectional TEC can restart, using an R-loop (RNA-DNA hybrid) from a transcription bubble as primer (11), although it is not known if this occurs in vivo.
Transcription termination isolates transcription units and prevents inappropriate expression of downstream genes. Termination in Escherichia coli is largely mediated by Rho, an RNA-dependent ATPase that terminates transcription promoter-distal to unstructured and untranslated RNA (12, 13). The RNA/DNA helicase activity of Rho promotes termination by unwinding RNA/DNA hybrid in the RNAP transcription bubble (14). Inhibition of Rho with bicyclomycin (BCM) or deletion of the Rho accessory factors, NusA and NusG, massively disregulate gene expression in E. coli (15). Surprisingly, efficient transcription termination is only required to suppress expression of cryptic prophage toxic genes. A strain lacking these prophages (MDS42) is relatively resistant to BCM, although rho remains an essential gene. Unlike wild-type E. coli, MDS42 can sustain deletions of nusA and nusG, which are required for efficient transcription termination in vivo (15, 16). The deletion strains, however, are hypersensitive to BCM (15), suggesting that Rho has an essential function in addition to terminating transcription at the ends of operons.
Rho associates with TEC immediately after the start of transcription, followed by NusA and NusG (13, 17). Although the majority of TEC thus include Rho, NusA, and NusG, Rho does not act until translation termination, presumably because ribosomes block Rho access to RNA. Competition between ribosomal NusE/S10 and Rho for NusG may also prevent premature transcription termination (18).
Head-on collisions between the replisome and TEC are highly disruptive to replication (3). It is proposed that the genome of E. coli is organized to avoid such collisions; all seven rrn operons and 55% of ORFs transcribe codirectionally with replication (19). Chromosomal inversions that place rrn operons head-on with replisome movement interfere with replication. Thus, the mutant cells are more dependent on the DNA helicases Rep, DinG, and UvrD (9).
We report here that Rho, along with NusA and NusG, plays a major role in preventing replication fork arrest caused by replisome collisions with TEC. We propose that the essential activity of Rho is to remove TEC ahead of the replisome, thus preventing this potentially lethal event.
Results
Mutations That Inhibit Replication Fork Repair Increase Sensitivity to BCM.
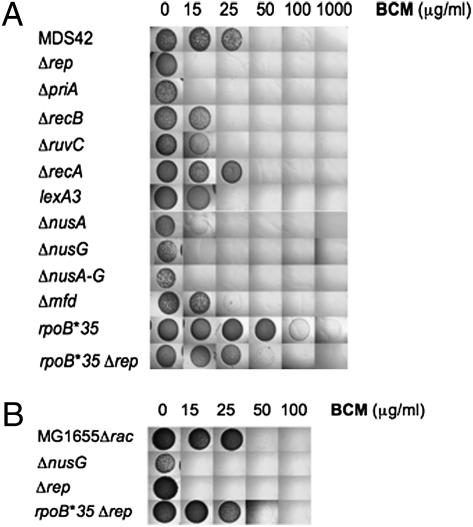
The highly defective rho-15 mutant is UV sensitive and the mutation is synthetic-lethal with mutations in the DNA repair functions, rep and ssb (20). We confirmed and extended this observation by determining the BCM sensitivity of E. coli strain MDS42 mutants defective in repair of damaged replication forks (Figs. 1 and 2). We chose to study BCM sensitivity in MDS42, which is deleted for all cryptic prophages, several of which contain toxic genes and recombination activities that are up-regulated by BCM (15) and might affect the phenotype. Rep and PriA DNA helicases reassemble replisomes at arrested forks to restore DNA synthesis (6, 21, 22). MDS42 carrying rep or priA deletions were exquisitely sensitive to BCM [minimum inhibitory concentration (MIC) <15 μg/mL] (Fig. 2). We propose that inhibition of Rho leads to fork arrest, which can be repaired by Rep and PriA; failure to do so is lethal. Because each mutation alone increased BCM sensitivity, the two functions cannot be completely redundant. Similar results were seen in a wild-type strain deleted for the rac prophage (MG1655Δrac). Deletion of rep resulted in hypersensitivity to BCM (MIC <15 μg/mL BCM) (Fig. 2B). RecB processes regressed replication forks and prevents subsequent DSB formation (8) (Fig. 1). We found that introduction of a recB deletion into MDS42 enhanced BCM sensitivity (MIC <25 μg/mL) (Fig. 2). Deletion of the Holliday junction resolvase, ruvC, which processes regressed replication forks (Fig. 1), similarly sensitized cells to BCM (MIC <25 μg/mL). In contrast, deletion of recA had no significant effect on BCM sensitivity (Fig. 2). However, RecA is proposed to both promote fork regression and to process regressed forks (8). Consistent with this model, we found that MDS42 lexA3 is hypersensitive to BCM (MIC <25 μg/mL BCM) (Fig. 2A). MDS42 lexA3 expresses RecA but is not inducible for SOS DNA repair. The strain is predicted to promote fork regression, but to be defective in repair (23). MDS42 ΔrecA and MDS42 lexA3 were UV sensitive (Fig. 3C), confirming the loss of SOS repair functions.
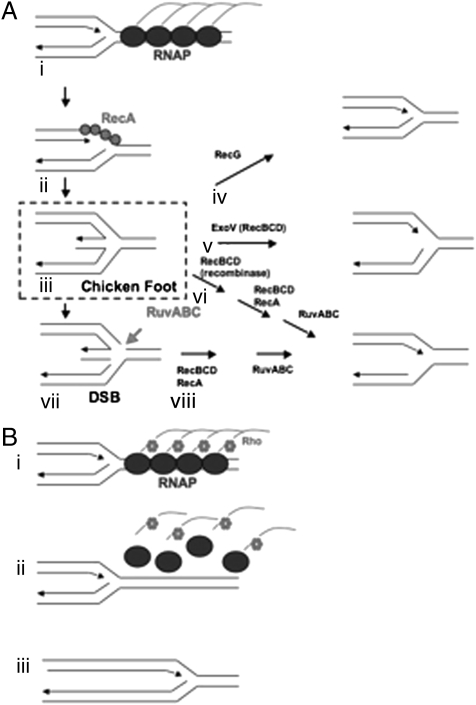
Fig. 1.
(A) Model of replication fork collapse and DSB formation. Adapted from Mahdi et al. (8) (i) Replication fork encounters TEC. The TEC arrest and form arrays that induce fork regression. (ii) RecA binds to exposed ssDNA and promotes recombination between leading and lagging strand forming “chicken foot” structures. (iii) “Chicken foot” structures are processed through multiple redundant pathways to allow reloading of replication forks. (iv) RecG separates leading and lagging strands and restores the fork. (v) RecBCD exonuclease activity degrades the hybrid, reforming the fork. (vi) RecBCD, RecA and RuvABC can restore the fork in a three step process without cleaving the Holliday junction. (vii) RuvABC cleaves the “chicken foot” structure across the Holliday junction. Failure to repair the “chicken foot” structure is proposed to lead to DSB formation. (viii) RecBCD, RecA and RuvABC restore replication forks in a two-step process. (B) Model of Rho-dependent release of TEC array. (i) Replication fork encounters an array of TEC promoter-proximal to an arrested TEC (7). (ii) Before the replication fork can regress, Rho factor removes the arrayed RNAP. (iii) Replication proceeds.
Fig. 2.
BCM sensitivity. MICs of the indicated strains were determined by spotting 104-fold dilutions of fresh overnight cultures onto LB-agar plates containing the indicated concentration of BCM. Plates were incubated at 37 °C for 24 h. (A) MDS42, RSW586, RSW585, RSW464, RSW554, RSW455, RSW919, RSW421, RSW422, RSW427, RSW738, RSW712, RSW828. (B) RSW472, RSW481, RSW921, RSW915.
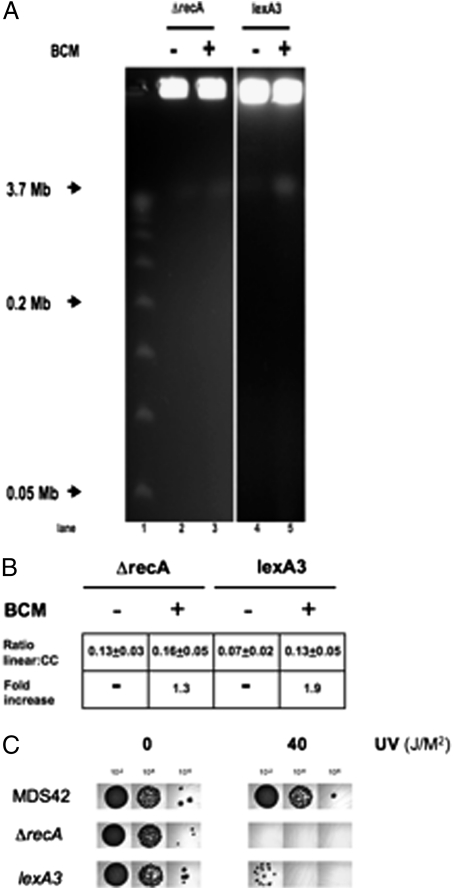
Fig. 3.
Pulsed-field gel analysis of BCM-induced DSBs. Cells were treated with 100 μg/mL BCM for 12 h where indicated. DNA fragments were separated on a 1% agarose gel in 0.5× TBE at 14 °C for 26.4 h at 6 V/cm (200 V) using a 120° included angle with a 2.8- to 26.3-s linear switch time ramp. Intact chromosomes remain in the well, and linearized DNA runs at 3.7 Mb. (A) PFGE. lane1: λ concateners from 0.05 to 1.0 Mb; lane 2: RSW464; lane 3: BCM-treated RSW464; lane 4: RSW919; lane 5: BCM-treated RSW919. (B) Ratios of linearized to intact chromosomes. Ratio linear:cc = amount of linearized chromosomal DNA divided by amount of DNA retained in well. Fold-increase = fold-increase in linear DNA after BCM treatment. (C) UV sensitivity. Dilutions (102-, 104-, and 106-fold) of fresh overnight cultures of MDS42, RSW455, and RSW919 were exposed to 0 or 40 J/M2 UV irradiation, and incubated at 37 °C for 24 h.
Mutations That Inhibit Transcription Termination Increase Dependence on Rho.
MDS42 nusA or nusG deletion mutants are hypersensitive to BCM (MIC <25 μg/mL BCM and <15 μg/mL BCM, respectively) (Fig. 2A) (12). MDS42 deleted for both nusA and nusG was as sensitive to BCM as MDS42 deleted for nusG alone (<15 μg/mL). Deleting nusG in MG1655Δrac increased sensitivity to BCM (MIC <15 μg/mL BCM) (Fig. 2B). These results indicate that Rho retains some activity in the absence of NusA and NusG, and NusA and NusG contribute to cell viability when Rho activity is compromised. We propose that Rho, NusA, and NusG act in concert to release TEC ahead of moving replisomes.
Mfd removes TEC stalled at UV-induced thymine dimers, and recruits repair enzymes UvrA and UvrB to the damaged DNA (24). Mfd also terminates TEC arrested by protein roadblocks or by phage HK022 Nun protein (25, 26). As shown in Fig. 2, MDS42 deleted for mfd was more sensitive to BCM than wild-type cells (MIC <25 μg/mL). We suggest that TEC forms arrays at sites of DNA damage in mfd mutants. Rho releases these arrays, reducing the likelihood of replication fork collapse.
rpoB Mutation Confers Resistance to BCM.
The RNAP mutation, rpoB*35, destabilizes TEC in vitro (27). We thought that rpoB*35 might allow release of arrested TEC and permit replisome movement, effectively replacing Rho. We predicted that rpoB*35 would increase resistance to BCM. Fig. 2 shows that MDS42 rpoB*35 formed colonies even at 1,000 μg/mL BCM, the highest concentration tested, although colony size was reduced. We were, however, unable to delete rho in MDS42 rpoB*35, indicating that rho is still essential. The rpoB*35 mutation also partially suppressed the BCM sensitivity of MDS42 Δrep and MG1655 Δrac Δrep, increasing the MIC from <15 to >50 μg/mL BCM (Fig. 2).
Taken together, these results support the idea that Rho releases TEC ahead of replisomes in E. coli, which may be the essential role of Rho in MDS42.
BCM Induces DSBs.
DSBs indicate replication fork collapse. They are generated when arrested forks form “chicken foot” structures, which include a Holliday junction (8) (Fig. 1). Holliday junctions are a substrate for RuvC cleavage, which, if not repaired, will yield a DSB. We used pulsed-field gel electrophoresis (PFGE) to detect chromosomal DSBs in MDS42 treated with BCM (Fig. 4). Circular E. coli chromosomes remain in the agarose plug and do not migrate into the gel (28). Linearized chromosomes containing a single DSB migrate as a 3.7-Mb species. Two breaks at specific sites will generate two visible bands, whereas two breaks, at least one of which is random, cannot be detected as discrete bands. Replication and recombination intermediates of linear fragments also remain in the well and do not migrate into the gel (29).
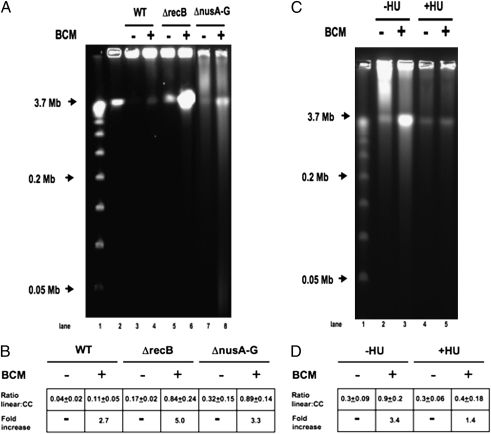
Fig. 4.
Pulsed-field gel analysis of BCM-induced DSBs. Cells were treated with 100 μg/mL BCM for 12 h where indicated. DNA fragments were separated on a 1% agarose gel in 0.5× TBE at 14 °C for 26.4 h at 6 V/cm (200 V) using a 120° included angle with a 2.8- to 26.3-s linear switch time ramp. Intact chromosomes remain in the well, and linearized DNA runs at 3.7 Mb. (A) PFGE. lane1: λ concateners from 0.05 to 1.0 Mb; lane 2: 3.7-Mb linearized E. coli chromosomes (MDS42 I-SceI); lane 3: MDS42; lane 4: BCM-treated MDS42; lane 5: MDS42ΔrecB; lane 6: BCM-treated MDS42ΔrecB; lane 7: MDS42 ΔnusA-G; lane 8: BCM-treated MDS42 ΔnusA-G. (B) Ratios of linearized to intact chromosomes. Ratio linear:cc = amount of linearized chromosomal DNA divided by amount of DNA retained in well. Fold-increase = fold-increase in linear DNA after BCM treatment. (C) PFGE of HU-treated Cells. MDS42 ΔrecB cells were prepared as above, except that cells were treated with 100 μg/mL BCM for 6 h, where indicated, in the presence or absence of 150 mM hydroxyurea (HU) to inhibit DNA replication. (D) Ratios of linearized to intact chromosomes. All values are the average of three or more independent experiments.
As expected, DNA extracted from untreated MDS42 remained almost entirely in the well. Exposure of MDS42 to 100 μg/mL BCM induced DSBs, as shown by a 2.7-fold increase in linear, full-length chromosomes (Fig. 4A). Linear DNA was readily evident in untreated MDS42 ΔnusA ΔnusG, as predicted for mutations that inhibit Rho-dependent termination. Exposure of MDS42 ΔnusA ΔnusG to BCM increased the yield of linear DNA by 3.3-fold, confirming that some Rho activity remains in the absence of NusA and NusG. We could also detect DSBs in untreated MDS42 carrying a recB deletion, presumably because of unrepaired replication errors (30) (Fig. 4A). BCM treatment of MDS42 ΔrecB dramatically increased the frequency of DSBs (fivefold).
The background levels of DSB were increased in MDS42 ΔrecA and MDS42 lexA3 relative to MDS42. Treatment with BCM enhanced DSB formation in both strains (1.3- and 1.9-fold, respectively) (Fig. 3). The approximately twofold increase in DSB formation in MDS42 lexA3 exposed to BCM illustrates the two roles of RecA. RecA promotes fork regression, which sensitizes cells to BCM, but also induces SOS repair of DSBs, which increases BCM-resistance (8). Linear DNA in the Δrep and ΔpriA strains with or without BCM treatment was below the limit of detection. This finding is consistent with their proposed role in reloading replication forks, rather than in recombination and repair of regressed forks; it is the regressed forks that generate DSBs.
BCM Induction of DSBs Requires DNA Replication.
To confirm our hypothesis that BCM-induced DSBs are generated by replication fork collisions with TEC, stationary phase MDS42 ΔrecB cells were treated with hydroxyurea (HU) to inhibit DNA replication (31). The cells were then diluted into media with or without BCM. As shown in Fig. 4C, HU inhibited DSB formation in BCM-treated cells. BCM induced a 3.4-fold increase in linear DNA in control cells vs. a 1.4-fold increase in cells blocked in DNA synthesis. Cultures exposed to HU continued to increase in OD600, indicating that DNA replication was preferentially inhibited over RNA and protein synthesis. We conclude that DNA replication is required for induction of DSBs by BCM.
Discussion
Transcription termination factor Rho is essential in E. coli, even though efficient transcription termination at the ends of operons is not required for cell viability (15). Recall that MDS42 ΔnusA or MDS42 ΔnusG mutants, despite extensive operon read-through, form colonies (15). That Rho is still required in these mutant strains suggests another role for the enzyme. We propose that this role is to release TEC from DNA ahead of moving replisomes. In this study we show that: (i) Mutants with impaired ability to restore collapsed replication forks are more dependent on Rho activity, and (ii) Inhibition of Rho leads to chromosomal DNA DSBs.
Given that transcription and replication occur simultaneously in bacteria, and that replication proceeds at least 20-fold faster than transcription (1–3), encounters between the TEC and replisomes must occur frequently, in particular within genes. However, they can only rarely result in fork collapses, which are highly damaging to cells (3, 7–9, 11). Our data suggest that Rho removes TEC ahead of replisomes, preventing collision, fork collapse, and DSB formation.
Rho termination requires untranslated RNA and NusG. Within genes, ribosomes block Rho access to RNA. In addition, the first ribosome sequesters NusG via contact with S10 (NusE) (18). It is possible that contact with the replisomes obviates both of these requirements and allows Rho to terminate within a gene. Alternatively, Rho may be required to remove backtracked TEC within nontranslated regions. The almost complete resistance of MDS42 rpoB*35 (H1244Q) to BCM suggests that the latter is the essential role of Rho. RpoB*35 appears to be less prone to form stalled, backtracked TEC (7, 27). In this model, the function responsible for removing TEC within genes ahead of a moving replisome remains to be identified. One candidate is Mfd. Trautinger et al. (7) proposed a role for Mfd similar to that of Rho in removing arrested TEC. The modest increase in BCM sensitivity in a Δmfd strain (Fig. 2) is consistent with this notion.
Our model provides a rationale for the observation that Rho is found associated with nearly all TEC (32), even though not all operons are subject to Rho-dependent termination (15). Rho is thus positioned to remove arrested TEC and prevent clashes with the replisome.
Regressed replication forks can result in DSB formation. BCM induced a DNA synthesis-dependent increase in chromosomal DSBs, which we attribute to DNA polymerase regression after collision with stalled TEC (Fig. 4). We suggest that the primary mechanism of killing by BCM in MDS42 is, however, the generation of arrested and unrestored replication forks rather than unrepaired DSBs. Thus, lethal concentrations of BCM result in <1 DSB per chromosome (Fig. 4). In addition, although lethal concentrations of BCM induced a dramatic increase in DSBs in wild-type cells, untreated ΔrecB mutants are viable despite a substantially higher percentage of DSBs (Fig. 4). The notion that arrested and unrestored forks are the primary mechanism of killing by BCM is consistent with the extreme sensitivity of Δrep and ΔpriA to the drug.
Rho may also play this role in other Gram-negative bacteria, most of which, including Salmonella sp., Shigella sp., and Neisseria sp., are sensitive to BCM (33). Rho is not essential in the Gram-positive bacteria Bacillus subtilis and Staphylococcus aureus (34, 35), and the level of Rho in B. subtilis is <5% that of E. coli (36). Perhaps Gram-positive bacteria maintain Rho to reduce conflicts with replication, but express a redundant TEC release activity not found in E. coli. NusA is essential in B. subtilis; it might play this hypothetical role. Alternatively, Rho may not be essential in Gram-positive organisms because TEC are less stable or because the replisome has the capacity to remove TEC by itself.
Experimental Methods
Strain Construction.
Standard bacteriological techniques used in strain construction (e.g., transformation, transduction and media preparation) are as described in Silhavy et al. (37). Standard molecular biology techniques were as described in Sambrook and Russell (38). RSW454 was constructed by P1 transduction of Δrep::CamR from STL7371 into MDS42. RSW455 was constructed by P1 transduction of ΔrecA::TetR from N8095 into MDS42. RSW464 was constructed by P1 transduction of ΔrecB::TetR from N8096 into MDS42. RSW554 was constructed by P1 transduction of ΔruvC::CamR from STL7505 into MDS42. RSW585 was constructed by P1 transduction of ΔpriA::KanR from N95A7 into MDS42. RSW680 was constructed by P1 transduction of codA21::miniTn7kan(I-Sce I) from N95A7 into MDS42 to introduce a unique restriction site. RSW712 was constructed by P1 transduction of rpoB*35 argE86::Tn10 from N5530 into MDS42. RSW738 was constructed by P1 transduction of Δmfd::KanR from UNCNOMFD into MDS42. RSW828 was constructed by P1 transduction of Δrep::CamR from STL7371 into RSW712. RSW876 was constructed by P1 transduction of Δrac::CamR from RSW472 into N4849. RSW915 was constructed by P1 transduction of Δrep::KanR from JW5604 into RSW876. RSW919 was constructed by P1 transduction of lexA3:CamR from DE407 into MDS42. RSW921 was constructed by P1 transduction of Δrep::KanR from JW5604 into RSW472.
See Table 1 for a list of bacterial strains used in this study.
Table 1.
Bacterial strains
| Strain | Genotype | Reference |
| DE407 | lexA3(Ind−) malB::Tn9 | (23) |
| JW3906 | BW25113 ΔpriA::KanR | National BioResource Project (National Institute of Genetics, Japan) |
| JW5604 | BW25113 Δrep::KanR | National BioResource Project (National Institute of Genetics, Japan) |
| MDS42 | MG1655 deleted for ~14% of genome | (40) |
| N4849 | MG1655 rpoB*35 | (7) |
| N5530 | MG1655 rpoB*35 argE86::Tn10 | (7) |
| N8095 | N99 ΔrecA::TetR | Laboratory collection |
| N8096 | N99 ΔrecB::TetR | Laboratory collection |
| RSW421 | MDS42 nusA::CamR | (15) |
| RSW422 | MDS42 nusG::KanR | (15) |
| RSW427 | RSW422 ΔnusA::CamR | Present study |
| RSW454 | MDS42 Δrep::CamR | Present study |
| RSW455 | MDS42 ΔrecA::TetR | Present study |
| RSW472 | MG1655 Δrac::CamR | (15) |
| RSW481 | RSW472 nusG::KanR | (15) |
| RSW554 | MDS42 ΔruvC::CamR | Present study |
| RSW585 | MDS42 ΔpriA::KanR | Present study |
| RSW464 | MDS42 ΔrecB::TetR | Present study |
| RSW680 | MDS42 codA21::miniTn7kan(I-Sce I) | Present study |
| RSW712 | MDS42 rpoB*35 argE86::Tn10 | Present study |
| RSW738 | MDS42 mfd::KanR | Present study |
| RSW828 | RSW712 Δrep::CamR | Present study |
| RSW876 | N4849 Δrac::CamR | Present study |
| RSW915 | RSW876 Δrep::KanR | Present study |
| RSW919 | MDS42 lexA3 malB::CamR | Present study |
| RSW921 | MG1655 Δrep::KanR | Present study |
| STL7371 | MG1655 Δrep::CamR | S. Lovett |
| STL7505 | MG1655 ΔruvC::CamR | S. Lovett |
| SMR8476 | MG1655 codA21::miniTn7kan(I-Sce I) | (41) |
| UNCNOMFD | mfd::KanR | (23) |
BCM Sensitivity.
Strains were grown to saturation in liquid LB medium at 37 °C with shaking. BCM sensitivity was determined by plating serial dilutions of the bacteria onto LB-agar plates containing 0, 15, 25, 50, 100, or 1,000 μg/mL BCM (Fujisawa Pharmacueticals). Plates were then incubated at 37 °C for 24 h.
UV Sensitivity.
Strains were grown to saturation in liquid LB medium at 37 °C with shaking. UV sensitivity was determined by plating serial dilutions of the bacteria onto LB-agar plates exposed to 0, or 40 J/M2 UV-radiation in a Stratalinker 2400 (Stratagene). Plates were then incubated at 37 °C for 24 h.
Pulsed-Field Gel Electrophoresis.
Fresh overnight cultures were grown in LB media and diluted to OD600 0.02 to 0.04 in M9 minimal medium supplemented with 0.2% glucose and 0.2% casamino acids with or without 100 μg/mL BCM. Cells were then incubated at 37 °C with shaking for 12 h and harvested. Agarose plugs were prepared for PFGE analysis using ca. 3 × 108 cells/plug as estimated by OD600, following the manufacturer's instructions (CHEF Bacterial Genomic DNA Plug Kit, Cat.#170–3592; BioRad). Samples of RSW464 for the HU experiments were prepared identically except that, where indicated, 150 mM HU from a freshly prepared stock was added to inhibit DNA replication, and all samples were harvested after 6 h incubation at 37 °C with shaking. Lambda ladder PFG marker (Cat. #N0340S) was purchased from New England Biolabs. Linearized RSW680 chromosomal DNA was prepared by digestion of a genomic plug of RSW680 (prepared as above) with I-SceI to cleave at the unique site, generating a size standard of 3.7 Mb.
DNA fragments were separated on a 1% agarose gel in 0.5× TBE at 14 °C for 26.4 h at 6 V/cm (200 V) using a 120° included angle with a 2.8- to 26.3-s linear switch time ramp using a BioRad CHEF-DR II System. Gels were then stained with ethidium bromide and visualized with UV-transillumination. DNA was quantified using ImageJ software (39).
Acknowledgments
We thank Lorraine Symington, Vanessa Marrero, Jue Wang, Christophe Herman, and Jeffrey H. Miller for helpful discussions and technical advice, and Susan Lovett (Brandeis University, Waltham, MA), Robert Lloyd (University of Nottingham, Nottingham, UK), Roger Woodgate (National Institute of Child Health and Human Development, Rockville, MD), and Susan Rosenberg (Baylor College of Medicine, Houston) for bacterial strains. This work was supported by Grant GM37219 from the National Institutes of Health (to M.E.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair (Amst) 2007;6:981–993. doi: 10.1016/j.dnarep.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vilette D, Ehrlich SD, Michel B. Transcription-induced deletions in Escherichia coli plasmids. Mol Microbiol. 1995;17:493–504. doi: 10.1111/j.1365-2958.1995.mmi_17030493.x. [DOI] [PubMed] [Google Scholar]
- 5.Tourrière H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 2007;6:900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Marians KJ. Replication and recombination intersect. Curr Opin Genet Dev. 2000;10:151–156. doi: 10.1016/s0959-437x(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 7.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Mahdi AA, Buckman C, Harris L, Lloyd RG. Rep and PriA helicase activities prevent RecA from provoking unnecessary recombination during replication fork repair. Genes Dev. 2006;20:2135–2147. doi: 10.1101/gad.382306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tehranchi AK, et al. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson JP. Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- 13.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan CA, Dombroski AJ, Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987;48:945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- 15.Cardinale CJ, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–938. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan SL, Gottesman ME. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- 17.Mooney RA, et al. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burmann BM, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 19.Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- 20.Fassler JS, Tessman I, Tessman ES. Lethality of the double mutations rho rep and rho ssb in Escherichia coli. J Bacteriol. 1985;161:609–614. doi: 10.1128/jb.161.2.609-614.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Marians KJ. PriA-directed assembly of a primosome on D loop DNA. J Biol Chem. 1999;274:25033–25041. doi: 10.1074/jbc.274.35.25033. [DOI] [PubMed] [Google Scholar]
- 22.Heller RC, Marians KJ. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J Biol Chem. 2005;280:34143–34151. doi: 10.1074/jbc.M507224200. [DOI] [PubMed] [Google Scholar]
- 23.Ennis DG, Fisher B, Edmiston S, Mount DW. Dual role for Escherichia coli RecA protein in SOS mutagenesis. Proc Natl Acad Sci USA. 1985;82:3325–3329. doi: 10.1073/pnas.82.10.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 25.Park JS, Marr MT, Roberts JW. E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 26.Washburn RS, Wang Y, Gottesman ME. Role of E. coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J Mol Biol. 2003;329:655–662. doi: 10.1016/s0022-2836(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 27.Trautinger BW, Lloyd RG. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 2002;21:6944–6953. doi: 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birren B, Lai E. Pulse Field Gel Electrophoresis, A Practical Guide. New York: Academic Press; 1993. [Google Scholar]
- 29.Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006;20:3104–3116. doi: 10.1101/gad.1478906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinha NK, Snustad DP. Mechanism of inhibition of deoxyribonucleic acid synthesis in Escherichia coli by hydroxyurea. J Bacteriol. 1972;112:1321–1324. doi: 10.1128/jb.112.3.1321-1334.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters JM, et al. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci USA. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams RM, Durham CA. Bicyclomycin: Synthetic, mechanistic, and biological studies. Chem Rev. 1988;88:511–540. [Google Scholar]
- 34.Quirk PG, Dunkley EA, Jr, Lee P, Krulwich TA. Identification of a putative Bacillus subtilis rho gene. J Bacteriol. 1993;175:647–654. doi: 10.1128/jb.175.3.647-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Washburn RS, Marra A, Bryant AP, Rosenberg M, Gentry DR. rho is not essential for viability or virulence in Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1099–1103. doi: 10.1128/AAC.45.4.1099-1103.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingham CJ, Dennis J, Furneaux PA. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- 37.Silhavy T, Berman M, Enquist L. Experiments with Gene Fusions. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 38.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 39.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 40.Pósfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 41.Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nat Genet. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]