Abstract
TRIM5α proteins are restriction factors that protect mammalian cells from retroviral infections by binding incoming viral capsids, accelerating their dissociation, and preventing reverse transcription of the viral genome. Individual TRIM5 isoforms can often protect cells against a broad range of retroviruses, as exemplified by rhesus monkey TRIM5α and its variant, TRIM5-21R, which recognize HIV-1 as well as several distantly related retroviruses. Although capsid recognition is not yet fully understood, previous work has shown that the C-terminal SPRY/B30.2 domain of dimeric TRIM5α binds directly to viral capsids, and that higher-order TRIM5α oligomerization appears to contribute to the efficiency of capsid recognition. Here, we report that recombinant TRIM5-21R spontaneously assembled into two-dimensional paracrystalline hexagonal lattices comprising open, six-sided rings. TRIM5-21R assembly did not require the C-terminal SPRY domain, but did require both protein dimerization and a B-box 2 residue (Arg121) previously implicated in TRIM5α restriction and higher-order assembly. Furthermore, TRIM5-21R assembly was promoted by binding to hexagonal arrays of the HIV-1 CA protein that mimic the surface of the viral capsid. We therefore propose that TRIM5α proteins have evolved to restrict a range of different retroviruses by assembling a deformable hexagonal scaffold that positions the capsid-binding domains to match the symmetry and spacing of the capsid surface lattice. Capsid recognition therefore involves a synergistic combination of direct binding interactions, avidity effects, templated assembly, and lattice complementarity.
Keywords: electron microscopy, HIV-1 capsid, lattice complementarity, retroviral restriction, two-dimensional crystal
The susceptibility of mammals to retroviral infections is restricted by innate immunity factors that protect the host and limit retroviral tropism. One such restriction factor, TRIM5α, can block replication of HIV-1 and other retroviruses at the postentry stage and prevent accumulation of viral reverse transcripts (1–4). As illustrated in Fig. 1, TRIM5α proteins comprise four domains: an N-terminal RING domain that functions as a ubiquitin E3 ligase (5 kDa), a B-box 2 domain required for restriction and higher-order assembly (5 kDa), a dimerization region predicted to form a coiled-coil(s) (13 kDa), and a C-terminal SPRY/B30.2 domain that contacts retroviral capsids (22 kDa). The RING/B-box 2 and coiled-coil/SPRY domains are separated by linker regions, termed L1 and L2, respectively.
Fig. 1.
Schematic representation of the rhesus monkey TRIM5α protein. The four principal domains are illustrated as colored boxes: R, RING domain, yellow; B, B-box 2 domain, red; COIL, predicted coiled-coil domain, blue; SPRY/B30.2 domain, orange. The L1 and L2 linker regions are also labeled. The position of residue Arg121 within the B-box 2 domain is indicated by the arrow. In the TRIM5-21R construct used in this study, the RING domain of rhesus TRIM5α was replaced with the RING domain from human TRIM21.
Retroviral capsids can vary in shape, but in all cases comprise CA protein hexamers and pentamers, with a conserved interhexamer lattice spacing of ∼90 Å [reviewed in (5)]. For example, HIV-1 capsids are closed fullerene cones composed of ∼250 CA hexamers and 12 pentamers (6, 7), whereas other retroviral capsids form cylinders, spheres, or polyhedra (8, 9). In addition to variations in capsid morphology, CA proteins can also vary substantially in primary sequence. Nevertheless, individual TRIM5α proteins can often recognize and restrict a variety of different retroviruses (3, 10–12 and references therein), as exemplified by rhesus monkey TRIM5α, which restricts HIV-1, as well as several distantly related retroviruses. These observations raise the question of how a diverse collection of retroviral capsids can be recognized by a single protein. This issue is of particular importance for understanding viral tropism because the ability of different TRIM5α alleles to restrict specific retroviruses is typically dictated at the level of capsid recognition.
Perhaps surprisingly, TRIM5α proteins bind individual CA subunits very weakly, if at all (4). Instead, robust binding is only observed when CA is assembled into hexagonal lattices that mimic the surfaces of retroviral capsids (2, 13–16), indicating that TRIM5α recognizes epitopes that are formed when CA assembles and/or that TRIM5α recognizes a repeating pattern of epitopes on the capsid surface. Consistent with the latter model, there are several reports that oligomerization of TRIM5α contributes to the efficiency of capsid recognition, implying that avidity effects probably play an important role in TRIM5α/capsid interactions. In cells, both TRIM5α dimerization and higher-order oligomerization are important for retroviral capsid binding and restriction (14, 15, 17–21). Dimerization of TRIM5α enhances binding to capsid mimics in vitro (15) and requires the putative coiled-coil domain (18, 22–24). Higher-order oligomerization is dependent on a hydrophobic patch on the surface of the B-box 2 domain (21, 25) and a region located immediately downstream of the predicted coiled-coil domain (residues 263–278) (26). How these elements contribute to capsid binding has not yet been examined in vitro.
To learn how TRIM5 restriction factors form higher-order assemblies, we examined a recombinant TRIM5α construct, designated TRIM5-21R, in which the N-terminal RING domain of rhesus TRIM5α was replaced by the homologous RING domain from the related human TRIM21 protein. This construct was selected for study because it potently restricts HIV-1 replication in culture (10, 11) and can be expressed and purified more readily than wild-type TRIM5α (14, 15). Electron microscopy (EM) and biochemical studies revealed that recombinant TRIM5-21R spontaneously forms two-dimensional hexagonal arrays that are complementary in symmetry and dimensions to retroviral capsids. Moreover, assembly of TRIM5-21R was promoted by preformed two-dimensional crystals of the HIV-1 CA protein, suggesting that TRIM5α proteins employ “pattern recognition” to bind the hexagonal CA arrays found in all retroviral capsids.
Results
TRIM5-21R Spontaneously Assembles into Two-Dimensional Paracrystalline Hexagonal Lattices.
Negative-stain EM was used to examine TRIM5-21R assemblies formed under a range of different conditions. These experiments revealed that purified TRIM5-21R dimers spontaneously formed two-dimensional paracrystalline hexagonal arrays when incubated at low protein concentrations (∼10 μM), and under moderate ionic strength conditions [20 mM Tris, pH 8, 25 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP)] (Fig. 2A). This hexagonal TRIM5-21R lattice was also observed in unstained samples preserved in vitreous ice (Fig. 2B). Although the TRIM5-21R crystals did not exhibit long-range order, image processing without imposed symmetry confirmed that the protein formed a lattice composed of six-sided rings, with inter-ring spacings of 315–355 Å (Fig. 2A, inset) and p3 or p6 plane-group symmetry (Table 1). Within each ring, regions of high density at the threefold and twofold symmetry axes were connected by thinner regions of weaker density (Fig. 2C).
Fig. 2.
Hexagonal assemblies of TRIM5-21R. (A) Representative negatively-stained EM image of a hexagonal array formed spontaneously by dimeric TRIM5-21R upon incubation on a carbon-coated grid. (Scale bar,100 nm). The red circle demarcates a region with clear hexagonal order, as shown in the computed Fourier transform (inset). (B) Fourier-filtered image of a TRIM5-21R array preserved in vitreous ice. (C) Projection density map of (B) shown in gray scale (with no imposed symmetry), illustrating that the hexagonal lattice is composed of six-sided rings. The unit cell parameters for the crystals are: (A) a = 347 Å, b = 335 Å, γ = 119.3°; (B) and (C) a = 348 Å, b = 346 Å, and γ = 120.5°.
Table 1.
Analyses of the TRIM5-21R lattices
| I. Unit cell parameters from images of 10 negatively-stained crystals | ||
| a = b, Å (average/range) | 334 ± 12/310 - 353 | |
| γ, ° (average) | 119.0 ± 1.4 | |
| II. ALLSPACE (50) analysis of the TRIM5-21R lattice shown in Fig. 2B and C | ||
| Candidate plane group | Residual | Target residual |
| p1 | 14.9 | |
| p2 | 14.9* | 21.2 |
| p3 | 9.4* | 14.9 |
| p312 | 10.1* | 15.2 |
| p321 | 9.4* | 15.1 |
| p6 | 10.5* | 16.2 |
| p622 | 10.3* | 15.5 |
*Most probable.
TRIM5-21R Lattice Formation Does Not Require the Capsid-Binding SPRY Domain.
Although the C-terminal SPRY/B30.2 domain of TRIM5α is required for capsid binding (2), this domain was not necessary for TRIM5-21R assembly in vitro because a truncated construct that lacked the SPRY domain (TRIM5-21R1-276) could still form hexagonal arrays (Fig. S1). These crystals closely resembled those formed by wild-type TRIM5-21R, although the efficiency of TRIM5-21R1-276 assembly was somewhat lower (see Materials and Methods for details). Thus, the RING, B-box 2, and coiled-coil domains are sufficient to form a hexagonal scaffold that displays different capsid-binding domains, including the SPRY/B30.2 of TRIM5α, as well as the cyclophilin A domains of the related TRIMCyp family of restriction factors (27–34). Notably, these three N-terminal domains are conserved across the entire TRIM/RBCC (Tripartite Motif/RING, B-box, Coiled-Coil) family of proteins. It is therefore possible that analogous scaffolds display the different recognition domains found in other TRIM family members. Indeed, higher-order assemblies have even been reported for other RING domain proteins outside of the TRIM family (35–38).
TRIM5-21R Lattice Formation and Capsid Recognition Exhibit Similar Requirements for Coiled-Coil and B-box 2 Oligomerization.
To evaluate the biological relevance of the hexagonal TRIM5-21R assemblies, we tested whether there were common requirements for efficient TRIM5α restriction in vivo, retroviral capsid binding within cells, binding to pure recombinant HIV-1 CA, and hexagonal lattice formation. These experiments built on the observations that efficient retroviral restriction in cells requires both: (i) TRIM5α dimerization mediated by the putative coiled-coil motif (14, 15, 17–20), and (ii) higher-order assembly mediated by a hydrophobic patch in the B-box 2 domain and a key exposed arginine residue (Arg121) (21, 25). We previously used cosedimentation binding assays to show that TRIM5-21R dimerization enhances TRIM5-21R binding to helical tubes of HIV-1 CA hexamers (15). Here, we employed a similar binding assay to analyze the effect of the B-box 2 R121E mutation on TRIM5-21R binding to CA tubes in vitro. As shown in Fig. 3, pure dimeric TRIM5-21R alone did not pellet through a 70% sucrose cushion, but the protein did copellet through the cushion when bound to assembled HIV-1 CA tubes (positive control, compare lanes 1 and 4 or lanes 7 and 10). In contrast, a TRIM5-21R construct that lacked the SPRY domain failed to copellet with CA tubes, consistent with the known requirement for the SPRY domain in capsid binding (negative control, compare lanes 4 and 6 or lanes 10 and 12). In the actual experiment, the TRIM5-21RR121E mutant did not bind detectably to CA tubes when the binding reactions were performed at low protein concentrations (0.5 μM TRIM5-21R proteins and 3 μM CA subunits, compare lanes 4 and 5). However, attenuated capsid binding by TRIM5-21RR121E was detected when the binding reaction was performed at higher protein concentrations (1 μM TRIM5-21R proteins and 6 μM CA subunits, compare lanes 10 and 11). Thus, TRIM5-21RR121E retained some CA binding activity, presumably because the B-box 2 mutation did not affect the integrity of the SPRY binding domain, but the mutation reduced binding efficiency. These observations are consistent with the idea that higher-order TRIM5-21R assembly contributes to the avidity of capsid binding.
Fig. 3.
TRIM5-21R binding to helical tubes of cysteine-crosslinked HIV-1 CA hexamers requires the SPRY domain and is enhanced by B-box 2 domain interactions. TRIM5-21R proteins were incubated in the absence of CA (lanes 1–3 and 7–9) or in the presence of a 6-fold molar excess of CA subunits assembled into helical tubes that mimic the hexagonal capsid lattice (lanes 4–6 and 10–12). Binding experiments were performed at two different TRIM5-21R protein concentrations (0.5 μM, lanes 1–6 or 1 μM, lanes 7–12) using either wild-type, full-length TRIM5-21R proteins (WT, lanes 1, 4, 7, and 10), TRIM5-21R proteins with the R121E mutation (R121E, lanes 2, 5, 8, and 11), or truncated TRIM5-21R proteins that lacked the SPRY domain (ΔSPRY, lanes 3, 6, 9, and 12). Pelletable CA and associated TRIM5-21R proteins (Pellet, 4% of total), were separated from unassembled and soluble CA proteins and unbound TRIM5-21R (Supernatant, 1% of total) by centrifugation, and analyzed by Western blotting, with the input levels of both proteins shown for reference (Input, 1% of total).
We also tested whether TRIM5-21R dimerization and higher-order assembly were required for hexagonal lattice formation in vitro. We previously reported that recombinant TRIM5-21R expressed in insect cells can be isolated in both monomeric and dimeric forms that do not interconvert rapidly (15). Two-dimensional crystallization trials with the kinetically trapped TRIM5-21R monomer revealed that this species was impaired in lattice formation (see Materials and Methods for details). This result suggested that the TRIM5-21R dimer, which is the predominant species in mammalian cells (15), is the likely building block of the hexagonal lattice. Furthermore, a TRIM5-21R construct that contained the R121E mutation failed to form hexagonal crystals, even at concentrations that were 30-fold higher than those required for assembly of the wild-type protein. Sedimentation equilibrium experiments showed that this mutant protein remained dimeric in solution (Fig. S2), implying that the R121E mutation disrupted an interface that mediates lattice formation. Thus, the hydrophobic surface patch on the B-box 2 domain that is required for restriction and for efficient retroviral capsid binding in cells was also required for efficient CA binding and for assembly of the TRIM5-21R hexagonal lattice in vitro.
Hexagonal Arrays of HIV-1 CA That Mimic the Capsid Surface Act as a Template for Assembly of TRIM5-21R Arrays.
Given that TRIM5 protein assembly enhances capsid binding, we reasoned that two-dimensional hexagonal crystals of HIV-1 CA that mimic the capsid lattice might also promote TRIM5-21R assembly. To test this idea, we first had to develop a stable HIV-1 CA lattice that could withstand extended incubation with TRIM5-21R, which was necessary because the TRIM5-21R and CA crystals are stabilized by different buffer conditions and because TRIM5-21R binding destabilizes CA lattices (2, 15–17). The successful construct was an HIV-1 CA fusion protein, termed CA-NCA14C/E45C/W184A, that: (i) spanned the CA and NC domains of HIV-1 Gag, (ii) contained A14C and E45C mutations within the CA N-terminal domain that allowed disulfide crosslinking to stabilize the CA hexamers (39, 40), and (iii) contained a W184A mutation in the C-terminal domain of CA that promoted planar sheet formation (40). The two-dimensional hexagonal crystals formed by this protein were stabilized by both the engineered disulfide bonds (39, 40) and by interactions between the NC domains and a 25-mer repeating TG oligodeoxynucleotide (25-TG) (41). It was also necessary to identify suitable incubation conditions that minimized spontaneous assembly of TRIM5-21R, which was achieved by performing the incubations under more basic conditions than those used in the untemplated assembly reactions (pH 9.0 vs. 8.0).
TRIM5-21R dimers were incubated in solution with the preassembled CA-NC crystals, and aliquots of the reaction mixtures were applied to carbon-coated grids and examined by negative-stain EM (Fig. 4A). At incubation times > 6 h, the surfaces of the CA-NC crystals were decorated with TRIM5-21R (compare Fig. 4B with Fig. 4C). In contrast, spontaneous assembly of TRIM5-21R crystals was rarely observed in the absence of added CA-NC crystals. Fourier transforms of the decorated crystals revealed well defined reflections that corresponded to the CA lattice, as well as more diffuse peaks corresponding to the first-order reflections of the TRIM5-21R lattice (Fig. 4D). The boundaries of ordered TRIM5-21R and CA lattices typically coincided almost exactly (Fig. 4A), and regions directly adjacent to the crystals lacked both CA and TRIM5-21R diffraction. Furthermore, the CA-NC crystals did not serve as a template for assembly of TRIM5-21R constructs that carried the R121E mutation or lacked the SPRY domain. Thus, the hexagonal CA lattice promoted assembly of the TRIM5-21R lattice, and templated assembly required both the capsid-binding activity of the SPRY domain, as well as higher-order interactions mediated by the B-box 2 domain.
Fig. 4.
TRIM5-21R assembly is promoted by preformed, stabilized two-dimensional CA-NCA14C/E45C/W184A crystals that mimic the surface of the HIV-1 capsid. (A) Representative EM image of a CA-NC/TRIM5-21R cocrystal (1∶1 molar protein ratio, 24 h incubation). (B) Expanded view of the region boxed in red in (A). The contrast was stretched using Adobe Photoshop to enhance the clarity of the TRIM5-21R hexagons. (C) EM image of CA-NC alone at the same magnification as (B), shown for comparison. Scale bars,100 nm. (D) and (E) Computed Fourier transforms of the crystals shown in (B) and (C), respectively. In (D), diffraction from the TRIM5-21R lattice is evident (circled in green). Note that in this case, there are at least two stacked TRIM lattices (the first-order reflections of each correspond to a hexagonal lattice with ∼350 Å spacing), and at least three stacked CA lattices (a = 93.1 Å, b = 93.1 Å, and γ = 119.0°; a = 92.5 Å, b = 94.2 Å, and γ = 118.9°; a = 93.1 Å, b = 92.6 Å, and γ = 118.8°). The TRIM and CA diffraction patterns do not overlap in spatial frequency owing to the size of the TRIM lattice and its lack of long-range order. As expected, TRIM diffraction is absent in the transform of CA-NC alone, which shows two CA lattices (E).
Discussion
We have shown that TRIM5-21R, a variant of TRIM5α that potently restricts HIV-1 replication in culture, assembles in vitro into two-dimensional arrays comprising hexagonal rings. Hexagonal lattice formation is an intrinsic property of the protein, because it occurred spontaneously under standard buffer conditions, in the absence of added precipitant. We note that TRIM5α and its variants can also spontaneously assemble in cells, particularly when the protein is expressed at high levels or when proteasome function is inhibited (10, 22, 42–45). We speculate that these “TRIM bodies” may contain hexagonal arrays of TRIM5 proteins, although other cellular proteins are clearly also associated with the bodies (10, 46). More importantly, we found that a preassembled mimic of the HIV-1 capsid surface promoted formation of the hexagonal TRIM5-21R lattice in vitro, suggesting that incoming retroviral capsids can serve as templates for TRIM5 assembly, even under conditions where assembly would otherwise be disfavored.
Similarities between the requirements for retroviral restriction and TRIM5-21R assembly in vitro suggest that the hexagonal TRIM5-21R arrays described here are functionally relevant. Specifically, we observed that kinetically trapped, monomeric TRIM5-21R proteins were deficient in assembly, suggesting that the basic assembly unit is the TRIM5-21R dimer. TRIM5-21R dimerization is also essential for retroviral restriction in cells (17, 18). Similarly, the R121E mutation inhibited TRIM5-21R assembly in vitro, and this mutation also inhibits retroviral restriction and blocks formation of capsid-associated, higher-order TRIM5 assemblies in cells (21, 25).
Our results support a model in which TRIM5α proteins recognize retroviral capsids through a number of cooperative interactions that include: (i) direct, but weak binding of the SPRY domain to the capsid surface, (ii) TRIM5α dimerization, (iii) assembly of a hexagonal lattice of TRIM5α dimers, and (iv) complementarity between the symmetries and spacings of the TRIM5α and capsid lattices, which would reinforce TRIM5α assembly and create powerful avidity effects. In essence, we suggest that TRIM5α proteins employ “pattern recognition” to bind the hexagonal CA lattices found in all retroviral capsids (4). Although this cooperative mode of binding does not alleviate the requirement for direct capsid binding, it does reduce the affinity required for isolated TRIM5/CA interactions, thereby making it easier for individual TRIM5α proteins to restrict a variety of highly divergent retroviruses and also buffering the system against CA mutations that diminish binding affinity. Additional mechanisms that could regulate cooperative TRIM5α binding and restriction include auto-inhibition of unassembled TRIM5α subunits, TRIM5α phosphorylation (15, 47), TRIM5α ubiquitylation (10, 15, 48, 49), and the coupling of lattice formation to TRIM5α ubiquitin E3 ligase activity via conformational changes and/or proximity effects.
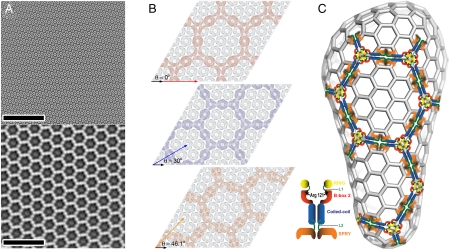
As illustrated in Fig. 5, idealized TRIM5-21R arrays with unit cell spacings of 315–355 Å could overlay on retroviral capsid lattices with unit cell spacings of 90–100 Å in at least three ways, which differ slightly in the matching of spacings and symmetry elements. Unfortunately, the EM images of the TRIM5-21R/CA cocrystals typically revealed multiple, stacked CA and TRIM layers (e.g., at least three distinct CA lattices and two TRIM lattices can be observed in the power spectrum shown in Fig. 4D). This overlap precluded us from identifying the interacting TRIM and CA lattices and determining their exact spatial relationship. In any case, the TRIM lattice must presumably be distorted from ideal geometries in order to accommodate the irregularly curved surfaces of retroviral capsids. This requirement may explain why the TRIM5-21R lattice appears to be mosaic and flexible, and may also limit the range over which the two lattices can interact. Indeed, slight mismatches between the interacting lattices will tend to create discontinuities in the extended CA lattice that could contribute to the accelerated capsid dissociation that accompanies restriction (2, 15–17).
Fig. 5.
Possible modes of interaction between the TRIM5-21R and CA lattices. (A) Fourier-filtered images of CA-NCA14C/E45C/W184A crystals alone (top) and TRIM5-21R crystals alone (bottom) illustrate the dramatic difference in unit cell size of the two lattices. (Scale bars,100 nm). (B) Assuming rigid, planar lattices, we can envision at least three different binding modes between the larger TRIM5-21R hexagonal lattice (colored) and the smaller CA lattice (gray). In these different binding modes, the TRIM5-21R lattice vectors (colored arrows) are rotated by either 0° (top), 30° (center), or 46.1° (bottom) relative to the CA lattice vectors (black arrows), and the interactions require TRIM5-21R lattice spacings of 360–400 Å, 312–346 Å, or 324–361 Å, respectively (given CA lattice spacings of 90–100 Å). The experimentally observed range of lattice spacings in our TRIM5-21R arrays is shown in Table 1. (C) Schematic model of an HIV-1 fullerene cone illustrating how TRIM5α could recognize the capsid lattice. Notice that only a small number of TRIM5α hexameric rings are required to create high avidity and that the lattice-matching geometry must be able to accommodate a greater radius of curvature in the outer TRIM5α lattice as compared to the inner CA lattice. Although the reconstructions do not allow us to position the different domains unambiguously, we modeled the coiled-coil domains within the narrowest density along each edge of the TRIM5α hexagon because the coiled-coil is expected to be the thinnest of the TRIM5α domains. The length of each hexagon edge (∼17 nm), together with the apparent twofold symmetry, suggests that each edge may be composed of two TRIM5α dimers, with coiled-coil elements linking L2/SPRY and RING/B-box 2 domains that occupy the regions of high local density at the midpoint and ends of each edge. We tentatively positioned six RING/B-box 2 domains at each threefold symmetry axis and four SPRY domains about the midpoint of each ring edge. Other models are possible, however, and higher resolution reconstructions will be required to define the TRIM lattice architecture unambiguously.
In summary, we propose that retroviral capsid binding and higher-order TRIM5 assembly are coupled because the hexagonal TRIM5 lattice can align multiple SPRY domains over repeating binding sites on the hexagonal CA lattice. Implicit in our model is the idea that retroviruses could evade TRIM5α recognition either by evolving surface mutations that eliminate SPRY domain recognition or by altering the regularity of capsid subunit conformations and lattice spacings. These selective pressures will tend to favor plasticity in both the TRIM5α and capsid lattices, consistent with the extensive shape polymorphism exhibited by retroviral capsids.
Materials and Methods
Construction, Expression, and Purification of Recombinant Proteins.
TRIM5-21R and HIV-1 CA proteins were expressed and purified as described in SI Text.
Binding Assays.
TRIM5-21R binding to disulfide-stabilized HIV-1 CA tubes was measured by using a published cosedimentation assay (2). Experimental details are described in SI Text.
TRIM5-21R Assembly.
Aliquots (∼5 μL) of freshly concentrated TRIM5-21R protein solutions (0.7–1.2 mg/mL) were incubated on carbon-coated EM grids for 5–30 min. Grids were then moved directly onto a 40-μL drop of 0.1 M KCl for 3 min, blotted, placed on a 20-μL drop of 2% uranyl acetate for 2 min, blotted, and air dried. Alternatively, TRIM5-21R (> 0.7 mg/mL) was stored for 3–5 d at 4 °C in 20 mM Tris, pH 8, 25 mM NaCl, 1 mM TCEP, whereupon the protein assembled into large hexagonal sheets that precipitated from solution and could be analyzed by EM. Thus, hexagonal sheet assembly is an intrinsic property of TRIM5-21R.
Spontaneous assembly of TRIM5-21R dimers was observed in 20/22 independent protein preparations, but there was substantial variability in assembly efficiencies across the different preparations. This variability precluded quantitative assessment of the relative assembly efficiencies of wild-type and mutant proteins. However, clear trends were observed. Specifically, the R121E mutant failed to assemble under all conditions tested. The SPRY domain deletion mutant, TRIM5-21R1-276, spontaneously assembled in 2/2 independent preparations. However, these crystals were sparse, and were generally smaller and less well ordered than assemblies formed by wild-type TRIM5-21R dimers. To assess the assembly efficiencies of monomeric and dimeric TRIM5-21R, we compared matched samples that were separated by ion exchange chromatography. In three independent experiments, hexagonal arrays were observed for the dimeric TRIM5-21R fraction but not for the monomeric fraction. In a fourth trial, assemblies were observed in the monomeric TRIM5-21R incubation, but were ∼4-fold less frequent than in the dimer incubation.
Templated Assembly of TRIM5-21R on Hexagonal Arrays of HIV-1 CA-NC.
To form stable two-dimensional crystals that mimicked the surface of the viral capsid, HIV-1 CA-NCA14C/E45C/W184A (232 μM) was incubated with 25-TG (143 μM) in 50 mM Tris, pH 8, 250 mM NaCl, 50 mM β-mercaptoethanol (βME) for 90 min at 37 °C. EM analyses confirmed that the CA-NC arrays assembled into the same hexagonal arrays as wild-type HIV-1 CA, with unit cell spacings of ∼90 Å in both cases (Fig. 5A). Following assembly reactions, the CA-NC crystals were diluted 25-fold into 50 mM Tris, pH 8, 300 mM NaCl, and incubated for an additional 10 min at room temperature to promote disulfide bond formation. TRIM5-21R proteins were then added in 1- to 10-fold molar excess, and the pH was immediately adjusted to 9.0 by addition of Tris buffer to a final concentration of 100 mM. Samples were incubated for 1–60 h, applied to carbon-coated EM grids, washed and stained as described above, and visualized by EM.
Electron Microscopy and Image Analysis.
Sample and EM grid preparation are described above. For cryoEM, samples were applied to carbon-coated molybdenum grids, washed with 0.1 M KCl, blotted to near dryness, and plunged into a slurry of liquid ethane. Images were recorded at a magnification of 11,000–30,000× under low electron-dose conditions (∼20 e-/Å2) using a 2k × 2k CCD camera (Gatan) fitted to a Tecnai T12 transmission electron microscope (Phillips/FEI) or a 4k × 4k camera fitted to a Tecnai F20. For processing, images were converted to MRC format (50). Manual indexing, unbending (to correct for lattice distortions), and corrections for the contrast transfer function were performed with the program 2dx (51). Fourier-filtered images were created after one or two rounds of unbending, and a 1-pixel hole was used to mask diffraction spots.
Supplementary Material
Acknowledgments.
We thank Debbie Eckert for assistance with analytical ultracentrifugation experiments. This work was funded by the National Institutes of Health (NIH) through Grants R01-AI63987 (J.G.S), R37 AI-45405-06 (W.I.S.), P50-GM082545 (to W.I.S. and M.Y.), and R01-GM066087 (M.Y.). Electron microscopy experiments were conducted at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through the National Center for Research Resources’ P41 program (RR17573) and at the University of Virginia Molecular Electron Microscopy Core facility (1S10RR025067 and 1G20RR031199).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013426108/-/DCSupplemental.
References
- 1.Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 2.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huthoff H, Towers GJ. Restriction of retroviral replication by APOBEC3G/F and TRIM5α. Trends Microbiol. 2008;16:612–619. doi: 10.1016/j.tim.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luban J. Cyclophilin A TRIM5, and resistance to human immunodeficiency virus type 1 infection. J Virol. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 7.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 8.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heymann JB, Butan C, Winkler DC, Craven RC, Steven AC. Irregular and semi-regular polyhedral models for Rous sarcoma virus cores. Comput Math Method M. 2008;9:197–210. doi: 10.1080/17486700802168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Griffero F, et al. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 11.Li X, et al. Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5α (TRIM5α) by heterologous TRIM domains. J Virol. 2006;80:6198–6206. doi: 10.1128/JVI.00283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 2003;22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian S, Luban J. TRIM5α selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kar AK, Diaz-Griffero F, Li Y, Li X, Sodroski J. Biochemical and biophysical characterization of a chimeric TRIM21- TRIM5α protein. J Virol. 2008;82:11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langelier CR, et al. Biochemical characterization of a recombinant TRIM5α protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black LR, Aiken C. TRIM5α disrupts the structure of assembled HIV-1 capsid complexes in vitro. J Virol. 2010;84:6564–6569. doi: 10.1128/JVI.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perron MJ, et al. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol. 2007;81:2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javanbakht H, et al. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Yap MW, Mortuza GB, Taylor IA, Stoye JP. The design of artificial retroviral restriction factors. Virology. 2007;365:302–314. doi: 10.1016/j.virol.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Javanbakht H, et al. The ability of multimerized cyclophilin A to restrict retrovirus infection. Virology. 2007;367:19–29. doi: 10.1016/j.virol.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Griffero F, et al. A B-box 2 surface patch important for TRIM5α self-association, capsid binding avidity, and retrovirus restriction. J Virol. 2009;83:10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mische CC, et al. Retroviral restriction factor TRIM5α is a trimer. J Virol. 2005;79:14446–14450. doi: 10.1128/JVI.79.22.14446-14450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Sodroski J. The TRIM5α B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82:11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sastri J, et al. Identification of residues within the L2 region of rhesus TRIM5α that are required for retroviral restriction and cytoplasmic body localization. Virology. 2010;405:259–266. doi: 10.1016/j.virol.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 28.Gamble TR, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 29.Brennan G, Kozyrev Y, Hu SL. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci USA. 2008;105:3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci USA. 2008;105:3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson SJ, et al. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci USA. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman RM, et al. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yap MW, Dodding MP, Stoye JP. Trim-cyclophilin A fusion proteins can restrict human immunodeficiency virus type 1 infection at two distinct phases in the viral life cycle. J Virol. 2006;80:4061–4067. doi: 10.1128/JVI.80.8.4061-4067.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 36.Kentsis A, Gordon RE, Borden KL. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc Natl Acad Sci USA. 2002;99:15404–15409. doi: 10.1073/pnas.202608799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mace PD, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 39.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pornillos O, Ganser-Pornillos BK, Banumathi S, Hua Y, Yeager M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J Mol Biol. 2010;481:985–995. doi: 10.1016/j.jmb.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song B, et al. TRIM5α association with cytoplasmic bodies is not required for antiretroviral activity. Virology. 2005;343:201–211. doi: 10.1016/j.virol.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Caballero D, Hatziioannou T, Zhang F, Cowan S, Bieniasz PD. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J Virol. 2005;79:15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell EM, Perez O, Anderson JL, Hope TJ. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5α. J Cell Biol. 2008;180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Connor C, et al. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5α. J Virol. 2010;84:5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daub H, et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi K, Wada K, Tanji K, Tanaka M, Kamitani T. Ubiquitination of E3 ubiquitin ligase TRIM5α and its potential role. FEBS J. 2008;275:1540–1555. doi: 10.1111/j.1742-4658.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- 49.Javanbakht H, Diaz-Griffero F, Stremlau M, Si Z, Sodroski J. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5α. J Biol Chem. 2005;280:26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- 50.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 51.Gipson B, Zeng X, Zhang ZY, Stahlberg H. 2dx—user-friendly image processing for 2D crystals. J Struct Biol. 2007;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.