Abstract
During λ infections, the holin S105 accumulates harmlessly in the membrane until, at an allele-specific time, suddenly triggering to form irregular holes of unprecedented size (>300 nm), releasing the endolysin from the cytoplasm, resulting in lysis within seconds. Here we used a functional S105–GFP chimera and real-time deconvolution fluorescence microscopy to show that the S105–GFP fusion accumulated in a uniformly distributed fashion, until suddenly, within 1 min, it formed aggregates, or rafts, at the time of lethal triggering. Moreover, the isogenic fusion to a nonlethal S105 mutant remained uniformly distributed, whereas a fusion to an early-lysing mutant showed early triggering and early raft formation. Protein accumulation rates of the WT, early, and nonlethal alleles were identical. Fluorescence recovery after photobleaching (FRAP) revealed that the nonlethal mutant and untriggered WT hybrids were highly mobile in the membrane, whereas the WT raft was essentially immobile. Finally, an antiholin allele, S105ΔTMD1–mcherryfp, in the product of which the S105 sequence deleted for the first transmembrane domain was fused to mCherryFP. This hybrid retained full antiholin activity, in that it blocked lethal hole formation by the S105–GFP fusion, accumulated uniformly throughout the host membrane and prevented the S105–GFP protein from forming rafts. These findings suggest that phage lysis occurs when the holin reaches a critical concentration and nucleates to form rafts, analogous to the initiation of purple membrane formation after the induction of bacteriorhodopsin in halobacteria. This model for holin function may be relevant for processes in mammalian cells, including the release of nonenveloped viruses and apoptosis.
Keywords: bacteriophage, latent period, peptide linker
The programmed formation of nonspecific, lethal membrane lesions, or “holes,” is featured in many cytocidal phenotypes, including Bax-mediated apoptosis, virion release in infections of nonenveloped mammalian viruses, and the dissemination of important human toxins from bacteria (1–6). The most genetically tractable hole-formation process is host lysis in double-strand DNA bacteriophage infections (7, 8). In the infection cycle of phage λ, lysis is a precisely timed event controlled by the holin, S105, a 105-aa product of the S gene (Fig. 1). S105 accumulates in the membrane throughout the morphogenesis period of the infection cycle. This accumulation has no effect on membrane integrity or the proton-motive force (PMF), as shown by noninvasive assays measuring the flagellar rotation speed (9), until suddenly triggering to form irregular holes of unprecedented size (>300 nm) (10). These holes allow release of the phage endolysin, R, from the cytoplasm, resulting in destruction of the cell wall within seconds (9). The holin triggering time is allele specific (11, 12), in that it can be advanced or retarded by missense mutations throughout all three transmembrane domains (TMDs) of S105. Holins can be triggered prematurely by energy poisons; for S105, as little as a 40% reduction in the PMF results in instantaneous triggering (9).
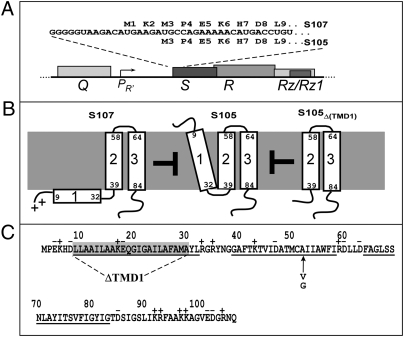
Fig. 1.
Sequence and topology of S gene products. (A) Translational initiation region of λ S, in the context of the λ late gene promoter and lysis cassette. Late promoter pR′ is activated by the product of gene Q. S encodes both S105 holin and S107 antiholin by virtue of dual translational starts at codons Met3 and Met1, respectively (11, 12). The S105 allele used in this study has the start codon for S107 inactivated. (B) Absence of TMD1 in the membrane confers antiholin (negative dominant) character to S gene products (17–22). Topologies of antiholin S107, holin S105, and deletion construct S105ΔTMD1. As long as the membrane is energized, TMD1 of S107 is prevented from entering the membrane because it has two positive charges at its N terminus: the protonated free amino group of Met1 and the Lys2 side chain. The N terminus of S105 is formylated (22). Both S107 and S105ΔTMD1 inhibit the lethal holin function of S105. (C) Sequence of S105, showing extent of ΔTMD1 and positions of A52G and A52V mutations (12, 17).
The malleability of the temporal schedule imposed by holins allows rapid evolution of the infection cycle in the face of altered environmental conditions (13–15). Holins are extremely diverse, existing in many unrelated protein families, not only in terms of sequence but also membrane topology, suggesting that holins have evolved independently many times (7). A “death raft” model has been proposed to account for the hole-forming ability, the capacity for precise timing and the structural diversity of holins (16). In this model, the holins accumulate in 2D aggregates, or rafts, characterized by close helical packing of TMDs that excludes lipid. At an allele-specific size, the rafts lose the ability to support the PMF, resulting in a local depolarization, which then triggers the holins to undergo a conformational change in which at least one helical face of a TMD faces the lumen of the hole. In this way, the conceptually difficult problem of how lipids are removed from the lumen of such large holes is avoided, because the lumen is formed from tertiary and quaternary rearrangements of the holin within the lipid-free interior of the raft. We here test this model by following the subcellular distribution of a GFP-tagged holin in real time.
Results
S105–GFP Retains Holin Function.
To determine whether the bacteriophage λ holin S105 accumulates in rafts, we fused the reading frames encoding S105 and GFP. A fusion at the extreme C terminus, with an intervening linker of only 2 aa, was only partially functional in that, although triggering did occur, as judged by the sudden cessation of growth of the induced culture, lysis was more gradual than wild-type S105 (Fig. S1). Progressive lengthening of the linker resulted in better holin function until, with a linker of 30 aa between S105 and GFP, the sharply defined lysis profile characteristic of holin function was obtained, albeit significantly delayed compared with the native allele. Although there were differences in the amount of fusion protein produced, all of the fusion proteins localized to the membrane S105 (Fig. S1). The allele encoding the fusion protein with the 30-aa linker, designated S105–gfp, was used as the wild-type protein in the following experiments, expressed under its normal transcriptional and translational context but without the function of the other λ lysis genes. Cryo-EM imaging confirmed that, under these conditions, S105–GFP also formed the large membrane holes observed for S105 (Fig. S2).
S105–GFP Accumulates Uniformly in the Membrane Before Forming Foci.
To visualize S105–GFP in vivo, deconvolution fluorescence microscopy was used on cells grown and induced on agarose pads. Images captured at 15-min intervals showed that, during the latent period, S105–GFP accumulated throughout the membrane but, after triggering, assumed a punctate distribution (Fig. 2). Analysis of the optical sections from the 60-min and 105-min images confirmed that S105–GFP is evenly distributed in the former but largely localized to foci, which we designate as “rafts,” in the latter (Fig. S3 and Movies S1 and S2). In the triggered cells, there are multiple rafts of varying size, distributed throughout the membrane. On the basis of the approximate number of S105–GFP molecules per cell, estimated from quantitative Western blotting (Fig. S4), and the percentage of total GFP fluorescence in each raft per cell, the largest of the rafts may consist of as many as 1,000 molecules, and the smallest of the rafts may consist of as few as 50 molecules. The number of large rafts (>3 pixels in diameter) per cell averaged 3.3 (for 229 cells counted; Fig. S5).
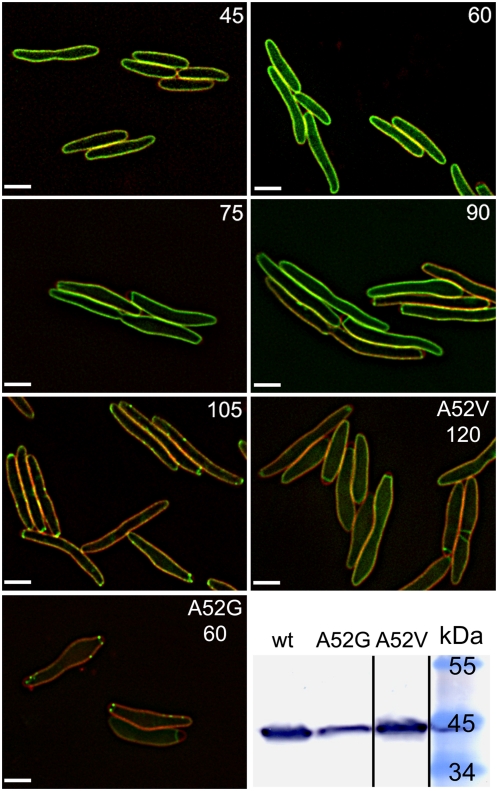
Fig. 2.
Deconvolution fluorescence imaging reveals S105–GFP raft formation. For the first five panels, numbers indicate the time after induction, in minutes, of S105–GFP (green). The last two fluorescence panels are images of inductions of S105A52V–GFP and S105A52G–GFP, taken at 120 min and 60 min, respectively. Membranes (red) are stained with FM 4–64. (Scale bar, 2 μm.) For S105–GFP, some raft formation is observed in a few cells at 90 min after induction, but by 105 min, all cells show rafts. (Lower Right) Immunoblot, using anti-S105 N terminus antibody of whole cell protein samples taken at the time of triggering, for the WT S105–GFP and S105A52G–GFP samples or at 120 min, for S105A52V–GFP.
Raft Formation Reflects Hole-Forming Phenotypes of S105 Mutant Alleles.
These results falsified the original death raft model and suggest an alternative that triggering time could be correlated with the sudden formation of such rafts. To explore this notion further, we tested the effect of representative S105 missense alleles with opposing lysis phenotypes on inducible triggering and raft formation in the context of the GFP fusion (Fig. 2). The A52V substitution (Fig. 1), which abolishes triggering and, as judged by chemical cross-linking, causes S105 to accumulate as inactive dimers in the membrane, also blocks triggering and raft formation in the context of the fusion protein (11). In contrast, the A52G change, which causes catastrophically early (i.e., before the assembly of the first virion) lysis in S105 (17) and, like all lytic alleles, can be cross-linked into a multimeric ladder (11), greatly advances the timing of triggering and raft formation of S105–GFP. As a consequence of the early triggering, S105A52G–GFP triggered at a lower level of fusion protein and formed fewer smaller rafts than the parental, although the average number of large rafts is similar (3.7 for 57 cells counted; Fig. S5).
Antiholin Fusion Accumulates Uniformly and Blocks S105–GFP Raft Formation.
Multiple lines of evidence indicate that TMD1 of S105 is essential for triggering and hole formation. Indeed, λ S encodes a second gene product, S107 (Fig. 1), which has been designated as an antiholin because it specifically dimerizes with and inhibits S105 (12, 18). Antiholins are produced by many phages, in some cases as alternate products of the holin gene but also, in some cases, as products of separate genes (8). The antiholin character of S107 is due to its extra N-terminal positive charge, which prevents TMD1 from entering the energized (positive outside) bilayer (12, 19–21). S105ΔTMD1, in which the entire first TMD has been deleted, was shown to inhibit S105 even more robustly, in the sense that, unlike S107, its antiholin function cannot be subverted by collapsing the membrane potential (22). When we examined the effect of producing the TMD1 deletion protein, tagged with mCherryFP, in trans to S105–GFP, we observed that it blocked triggering in liquid culture (Fig. S6) and raft formation in individual cells (Fig. 3). These results suggest that antiholins block lysis by preventing the formation of rafts.
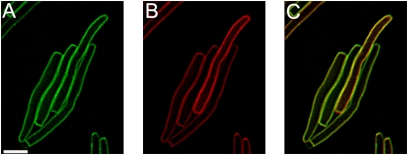
Fig. 3.
Dominant-negative antiholin allele blocks S105–GFP from raft formation. Cells coexpressing S105–gfp and S105ΔTMD1–mcherryfp at 150 min. (A) S105–gfp, (B) S105ΔTMD1–mcherryfp, and (C) overlay. (Scale bar, 2 μm.)
Raft Formation Occurs Within a 1-Min Interval at Triggering Time.
To further refine the triggering event at the molecular level, images were captured every minute from 75 to 105 min after induction. These images showed that although some small, transient rafts are present before the appearance of large rafts, formation of the large, stable rafts occurs in less than a minute (Fig. 4 and Movies S3 and S4). The timing of the appearance of the first large rafts coincides with the triggering time of S105–GFP in liquid culture (Fig. S7).
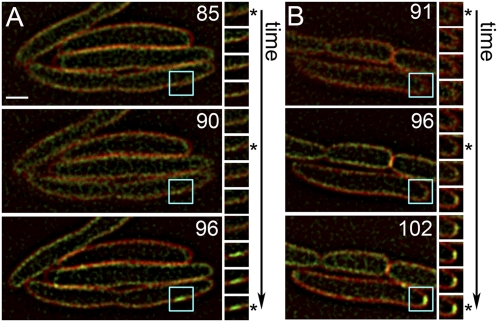
Fig. 4.
Sudden formation of S105–GFP rafts. Two representative time-lapse images (A and B) of cells induced for S105–GFP are shown, with the time (in minutes) after induction indicated on the large panels. The small panels show the area circumscribed in blue in the large panels, imaged at sequential 1-min intervals from Top to Bottom. Asterisks indicate the same time shown on the large panels on the Left. (Scale bar, 1 μm.)
FRAP Analysis Contrasts the Pre- and Posttriggered States of the Holin–GFP Fusion.
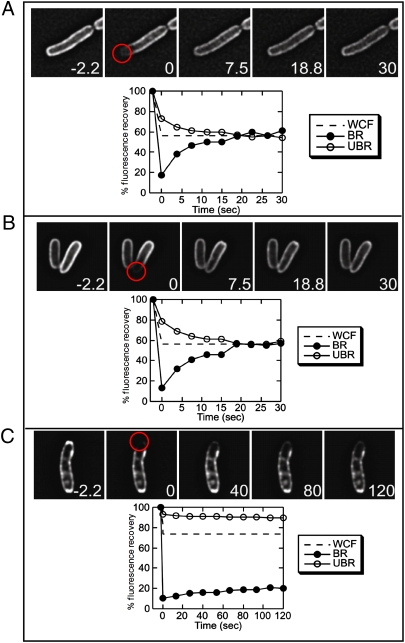
The mobility of S105–GFP in the two states was probed with fluorescence recovery after photobleaching (FRAP) experiments (Fig. 5). The results indicated that before triggering, S105–GFP is mobile in the membrane, with a half time of recovery of 5–7 s. The control, the nonlytic mutant S105A52V–GFP, showed similar high mobility, indicating that both molecules freely diffuse within the membrane. After triggering, however, the large rafts formed by S105–GFP are immobile and stable, showing very little recovery over a 2-min period, suggesting that triggering involves the sudden aggregation of freely diffusing molecules into a higher molecular weight complex of limited mobility. These results are consistent with results of cross-linking analysis of S105 in membranes, in which the A52V and other nonlethal mutant S105 proteins were blocked at the level of dimer formation, whereas the WT and timing variants exhibited oligomer formation. In addition, purified S105 and S105A52G, but not S105A52V, can form high-order oligomers in the detergent dodecylmaltoside (23).
Fig. 5.
Triggered S105–GFP forms immobile rafts. Cells expressing S105–gfp constructs were laser bleached in the region indicated by the red circle at t = 0. Whole cell fluorescence (WCF), and the relative fluorescence intensity of the bleached region (BR; black circles) and an unbleached region (UBR; open circles) are shown versus time after bleaching. The dashed line indicates the calculated full recovery level at which fluorescence is equilibrated across the cell. Mobility is indicated by the equilibration of fluorescence intensity in the bleached and unbleached regions as the proteins diffuse throughout the membrane, leading to a decreased fluorescence in the unbleached region and increased fluorescence in the bleached region, as observed before raft formation (A and B). Little recovery is observed for S105–GFP after triggering (C), and the bleached and unbleached regions show only minor changes in recovery. (A) S105A52V–GFP, taken 140 min after induction. (B and C) S105–GFP, taken before (45 min) and after (140 min) triggering.
Discussion
Here we show that triggering reflects the sudden redistribution of holins from a state of uniform dispersal and high mobility in the membrane into a punctate pattern dominated by a few large and immobile aggregates, or holin rafts. These observations falsify the previous model, which predicted that holins would accumulate in such rafts, which would then, in an allele-specific manner, eventually collapse into holes to initiate lysis (16). Instead, these results suggest a revised model for the ability of the λ holin, and by extension, all holins, to impose a temporally precise schedule on the lytic termination of the phage infective cycle (Fig. 6). During the late gene expression period, in parallel to virion assembly, the holin accumulates throughout the cytoplasmic membrane in a high-mobility and biologically innocuous form (Fig. 6). This form is likely to be a dimer, on several grounds. First, both the wild-type S107 antiholin and the ΔTMD1 antiholin block S105 triggering by heterodimerizing with it (11, 18, 22). A homodimer interface must exist in the C-terminal domain of S105, presumably in TMD2 and/or TMD3. Second, most lysis-defective mutants of S105 are blocked at a dimer stage, as judged by cross-linking studies (11). Moreover, the homodimer interface involves one face of TMD2, because a thiol at position 51, but not at any other nearby positions, can be quantitatively driven into a homodisulfide by Cu++-phenanthroline treatment. Finally, S105 can be purified in a dimer form in the zwitterionic detergent Empigen BB (EBB), as judged by cross-linking and gel filtration analysis (23).
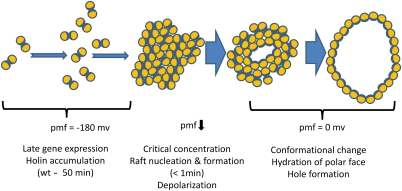
Fig. 6.
Model for holin triggering. In this cartoon, a top-down view of the cytoplasmic membrane, each holin is represented by an orange circle, with one face shaded blue, representing a relatively hydrophilic face (Fig. S8). From the onset of late gene expression, the holin protein accumulates as mobile homodimers, without perturbation of membrane integrity or the PMF. When a critical concentration is reached, which for WT λ would be at ∼50 min after infection, the 2D holin aggregates (“rafts”) rapidly form, within a 1-min window (Fig. 4). These rafts cause the collapse of membrane energization, which in turn leads to a concerted tertiary and/or quaternary conformational change in the holins, resulting in the hydration of the more hydrophilic surfaces of the holin and hole formation. These holes may rapidly grow until all or most of the holins are oriented with the hydrophilic face toward the lumen and the hydrophobic face toward the lipid.
Whatever the state of the mobile, pretriggered holin, a dramatic transition occurs at time of lethal triggering, with large, immobile aggregates forming within the time span of ∼1 min. We propose that this marks the time when the holin reaches a critical concentration (Cr) and nucleates the formation of holin rafts. Critical-concentration behavior for membrane proteins has a well-established precedent in the formation of the purple membrane of halobacteria (24, 25). After induction by light exposure, bacteriorhodopsin (BR) accumulates in the bilayer as a monomer. After reaching a critical concentration, BR accumulates exclusively in the pseudocrystalline 2D array of the purple membrane. Mutations in residues important for the intermolecular helical interactions result in up to 10-fold changes in Cr and thus in advancement or retardation of the onset of purple membrane formation.
One important difference between purple membrane formation by BR and raft formation by S105 and other holins are the properties of the 2D aggregate. Unlike S105, the purple membrane protein BR does not compromise the integrity of the membrane and, after appropriate maturation by retinal binding, promotes energy transduction. The original death raft model posited that allelic differences in triggering times reflected the differential stabilities of the growing rafts with respect to spontaneous local ion leakage and depolarization, which would then spread throughout the raft and throughout the cytoplasmic membrane and confer the requisite “all or nothing” feature to holin timing (16). Our revised model suggests instead that the allelic differences in lysis timing, both advancing and retarding the triggering time, observed for many missense S alleles would thus simply reflect changes in the Cr for holin raft formation.
A principal attraction of both versions of the death raft model is that lipid is excluded from the site of hole formation by helical packing of the three TMDs (16). Thus, because the hole can form within the raft, the raft concept solves the problem of how to remove the lipids from the hole, a problem made even more significant with the discovery of the enormous μm-scale sizes of the holin lesions. In the revised model, the simplest notion is that the holin rafts are inherently incapable of supporting the membrane potential, likely because the raft interior is either lipid-free or lipid depleted. The collapse of the membrane potential would then drive the holin into a new conformation in which at least one face of a TMD becomes hydrated and ultimately faces the lumen of the hole (Fig. 6). Helical projections suggest that all three TMDs of S105 have relatively hydrophilic and/or hydroxylated faces and could thus be candidates for facing the lumen (Fig. S8).
The model also implies that, if the PMF is collapsed artificially, e.g., by an energy poison or sudden anaerobiosis, the holin molecules in the high-mobility form can rapidly reorganize into holes. The fact that mutant proteins blocked at the dimer stage cannot be triggered exogenously by energy poisons suggests that premature hole formation of the functional holins via exogenous energy poison still requires a transitory raft step.
Chief among the unresolved questions is the relationship between the rafts and the S105-dependent membrane holes observed by cryoelectron microscopy in cells that have experienced S105 triggering (10). Identical lesions are seen with S105−GFP (Fig. S2), but it remains unclear if they form at the same time as the rafts or if they represent a later stage in assembly. The rapid development of correlative microscopy (26) may allow direct interrogation of these holes to ascertain whether, as the model predicts, the holin protein lines the lumen.
Materials and Methods
Materials, Strains, Bacteriophage, Plasmids, and Growth Media.
The Escherichia coli K-12 host strain, RY17303, the phages λΔ(SR) and λSam7, and the plasmid pS105 (22, 27), as well as the media, growth conditions, and thermal induction of the λ lysis genes from a prophage and/or plasmid (11, 21, 28), have been described previously. Bacterial cultures were grown in standard LB medium supplemented with ampicillin (100 μg/mL) and chloramphenicol (10 μg/mL) for the maintenance of plasmids and prophage, respectively. FM 4–64 was purchased from Invitrogen.
Standard DNA Manipulations.
Isolation of plasmid DNA, DNA amplification by PCR, DNA transformation, and DNA sequencing were performed as previously described. Primers were obtained from Integrated DNA Technologies and were used without further purification. Restriction and DNA-modifying enzymes were purchased from New England Biolabs; all reactions using these enzymes were performed according to the manufacturer's instructions. Standard recombinant DNA and site-directed mutagenesis techniques, described previously (11), were used in the construction of plasmids. The DNA sequence of all constructs was verified by automated fluorescence sequencing performed at the Laboratory for Plant Genome Technology at Texas AgriLife.
Structures of Plasmids and Phages.
All fluorescence experiments were done using lysogens of RY17303 carrying a thermo-inducible prophage in single copy and carrying a derivative of the plasmid pS105, a medium copy plasmid carrying an ampicillin-resistance gene and the λ-lysis cassette in its native late promoter context (11). In each chimeric plasmid, the RRzRz1 genes encoding the endolysin and spanin have been replaced by the DNA encoding the fluorescent protein, inserted in frame at the end of the S105 reading frame (Fig. 1) with the following linkers: 2 aa = PG; 9 aa = PGASSGAGG; 13 aa = PGSAGAASGSAGG; 20 aa = PGASSGAGGSAGAASGSAGG; and 30 aa = PGSASGAAGAGSASSGAGGSAGAASGSAGG. In all GFP constructs, the gfp gene carries the monomerization mutation A206K (29). The plasmid-borne lysis cassette carrying genes encoding holin–GFP chimeras are expressed by transactivation after thermal induction of the prophage, which provides the λ late gene activator, Q, at the time of late gene induction. For most experiments, the prophage was λΔ(SR), in which the holin and endolysin genes have been deleted to observe holin localization without endolysin-mediated destruction of the murein. For testing complementation with the various S105–gfp constructs, the prophage was λSam7 (27). For Fig. 3, the bacterium carried an additional plasmid with the S105ΔTMD1–mcherryfp allele. This plasmid has the same structure and transcriptional control as the pS105 derivatives, except that it has the compatible pACYC177 backbone and carries a kanamycin-resistance gene. The plasmid pR′–GFP was constructed by cloning the BamHI/HindII GFPmut2 fragment from pDS439 (30) into pRE (31) and introducing the monomerization mutation, A206K, by site-directed mutagenesis. For cryo-EM experiments, the chimeric plasmid was pS105–GFPRamRzamRz1am, in which the three other λ-lysis genes, each inactivated by amber codons, have been reinserted downstream of S105–gfp, as described previously (10).
Protein Sample Preparation, SDS/PAGE, and Western Blotting.
Preparation of soluble and membrane fractions was done as previously described (21). Briefly, cultures were induced at an A550 of 0.3–0.4. After lysis was complete or 100 min after induction in cases where lysis does not occur, 10-mL aliquots of the induced cultures were disrupted by passage through a French pressure cell (Spectronic Instruments) at 16,000 lb/in2 (1 lb/in2 = 6.89 kPa). Following a brief clearing spin, the membrane fraction was collected by centrifugation at 100,000 × g for 60 min at 18 °C. The membrane pellet was directly resuspended in SDS/PAGE buffer. The soluble fraction was subjected to trichloroacetic acid (TCA) precipitation (see below) and resuspended in SDS/PAGE buffer. Membrane and soluble samples were balanced to reflect equal volume of culture. Proteins were separated on 16.5% SDS/PAGE with a 4% stacking gel. Western blotting and immunodetection with anti-S antibodies were performed as described previously (32), with the exception that the GFP fusion proteins were detected with either the α-S105 N terminus (Bethyl Laboratories) or α-GFP antibodies (Stressgen).
TCA Precipitation.
One-milliliter or 5-mL culture aliquots were added to 111 μL or 555 μL, respectively, of cold, 6.1 N of TCA, then placed on ice for 30 min. The precipitate was collected by centrifugation (15,000 rpm in a tabletop microcentrifuge or 3,000 rpm in a clinical centrifuge, respectively) and washed once with acetone. Pellets were air dried and resuspended in SDS/PAGE loading buffer and run on gels as stated above.
Fluorescence Microscopy.
Fluorescence microscopy was performed as previously described (33, 34), with the exception that the cells were thermally induced. Briefly, 100 μL of an overnight culture was harvested and resuspended in 1 mL PBS; 10 μL of the cell suspension was then placed on an agarose pad (1.2% agarose, 0.25× LB, 0.5 μg/mL of FM 4–64; for cells producing mCherryFP fusions, the FM 4–64 was omitted). The agarose pad was heated to 30 °C on the stage of the microscope via the WeatherStation environmental chamber; the pads were incubated for 2–2.5 h and then placed between two modular heating blocks in a 42 °C incubator (the blocks were equilibrated to 42 °C in the incubator overnight before the experiment) for 15 min to thermally induce the λ-prophage. Following thermal induction, the pads were placed back on the stage (the temperature of the stage and WeatherStation was increased to 37 °C during the thermal induction period) and images of cells were captured at the times indicated. For time lapse experiments, cells were grown on agarose pads as described and midplane images were captured at 1-min intervals from 75 to 105 min after induction. Images were captured with an Applied Precision Spectris optical sectioning microscope system equipped with an Olympus IX70 microscope, an Olympus Plan Apo 100× oil immersion objective (NA 1.4), a Photometrics Cool SNAP HQ digital camera, and Delta Vision standard fluorescence filters: FITC for GFP visualization (excitation, blue 490/20 nm; emission, green 528/38 nm) and RD-TR-PE for FM 4–64 and mCherryFP visualization (excitation, green 550/28 nm; emission, orange 617/73 nm). Using softWoRx software (Applied Precision, Inc.), 12 optical sections, 0.2 μm apart, were collected for each sample and deconvolved using the constrained iterative deconvolution algorithm. Following deconvolution, the brightness and contrast of each fluorochrome were adjusted with softWoRx, setting the area outside of cells to be background; images were saved as TIFF files.
FRAP.
FRAP experiments were conducted as previously described (35, 36). Briefly, photobleaching experiments to assess the mobility of S105–GFP (or derivatives) used cells grown in liquid culture (0.25× LB, 0.5 μg/mL of FM 4–64); 10 μL of cells were removed at the indicated times and applied to coverslips. After collecting a prebleach image, photobleaching was achieved using a 0.05-s pulse of a 488-nm argon laser at 50% power, and subsequent GFP images were collected at 3.5-s intervals for up to 10 min. Exposure times were limited to 1.5–2.5 s. Images were quantified as previously described (35). Briefly, the “edit polygon” function of the Delta Vision version 2.10 software was used to define individual polygons to represent the background fluorescence, the entire membrane region, and completely unbleached or bleached regions of the cell. Average mean fluorescence from the background region was subtracted from all time points before data processing. Because there is an overall loss of fluorescence caused by photobleaching during image acquisition, mean fluorescence for each data region was adjusted for the losses by correcting the values with the application of a ratio created by dividing the first whole-cell fluorescence postbleach value by the last whole-cell fluorescence postbleach value. Data were plotted as time versus mean fluorescence for the unbleached region, the bleached region, and the whole cell fluorescence. To calculate the pixel intensity expected for the theoretical equilibration point, each whole-cell fluorescence ratio was applied to its corresponding mean whole-cell fluorescence value, and it is represented on the graphs as a dotted line.
Cryoelectron Microscopy.
A 500-μL aliquot of an overnight culture of RY17303(λΔ(SR)) pS105–GFPRamRzamRz1am grown in LB, supplemented with ampicillin and chloramphenicol, was used to inoculate 100 mL of the medium. The culture was aerated at 30 °C, thermally induced at 42 °C by aeration for 15 min at A550 = 0.3–0.4, and then aerated at 37 °C. At t = 105 min, the culture was concentrated by filtration with a 0.45-μm filter, and the cells were resuspended in fresh LB. Five microliters of the resuspended culture were applied onto freshly glow discharged C-Flat holey carbon grids and plunge frozen in liquid ethane using an FEI Vitrobot. Specimens were observed using an FEI Tecnai F20 transmission electron microscope equipped with a GATAN Tridiem energy filter under zero-loss and low-dose imaging conditions, with typical doses of 3–5 e−/Å2 per image, as previously described (10).
Supplementary Material
Acknowledgments
This work was supported by Public Health Service Grants GM27099 (to R.Y.) and GM57045 (to K.P.) and by funds from the Program for Membrane Structure and Function from Texas A&M University (to R.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011921108/-/DCSupplemental.
References
- 1.Gonzalez ME, Carrasco L. Viroporins. FEBS Lett. 2003;552:28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 2.Daniels R, et al. Simian virus 40 late proteins possess lytic properties that render them capable of permeabilizing cellular membranes. J Virol. 2006;80:6575–6587. doi: 10.1128/JVI.00347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniels R, Sadowicz D, Hebert DN. A very late viral protein triggers the lytic release of SV40. PLoS Pathog. 2007;3:e98. doi: 10.1371/journal.ppat.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madan V, Castelló A, Carrasco L. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell Microbiol. 2008;10:437–451. doi: 10.1111/j.1462-5822.2007.01057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayles KW. The biological role of death and lysis in biofilm development. Nat Rev Microbiol. 2007;5:721–726. doi: 10.1038/nrmicro1743. [DOI] [PubMed] [Google Scholar]
- 6.Waldor MK, Friedman DI, Adhya SL. Phages: Their Role in Bacterial Pathogenesis and Biotechnology. Washington, DC: ASM; 2005. p. v-450. [Google Scholar]
- 7.Wang IN, Smith DL, Young R. Holins: The protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 8.Young R. Bacteriophage holins: Deadly diversity. J Mol Microbiol Biotechnol. 2002;4:21–36. [PubMed] [Google Scholar]
- 9.Gründling A, Manson MD, Young R. Holins kill without warning. Proc Natl Acad Sci USA. 2001;98:9348–9352. doi: 10.1073/pnas.151247598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dewey JS, et al. Micron-scale holes terminate the phage infection cycle. Proc Natl Acad Sci USA. 2010;107:2219–2223. doi: 10.1073/pnas.0914030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gründling A, Bläsi U, Young R. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J Bacteriol. 2000;182:6082–6090. doi: 10.1128/jb.182.21.6082-6090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raab R, Neal G, Sohaskey C, Smith J, Young R. Dominance in lambda S mutations and evidence for translational control. J Mol Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- 13.Bull JJ, Pfennig DW, Wang IN. Genetic details, optimization and phage life histories. Trends Ecol Evol. 2004;19:76–82. doi: 10.1016/j.tree.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Wang IN. Lysis timing and bacteriophage fitness. Genetics. 2006;172:17–26. doi: 10.1534/genetics.105.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, Struck DK, Dankenbring CA, Young R. Evolutionary dominance of holin lysis systems derives from superior genetic malleability. Microbiology. 2008;154:1710–1718. doi: 10.1099/mic.0.2008/016956-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang IN, Deaton JF, Young R. Sizing the holin lesion with an endolysin-β-galactosidase fusion. J Bacteriol. 2003;185:779–787. doi: 10.1128/JB.185.3.779-787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson-Boaz R, Chang CY, Young R. A dominant mutation in the bacteriophage lambda S gene causes premature lysis and an absolute defective plating phenotype. Mol Microbiol. 1994;13:495–504. doi: 10.1111/j.1365-2958.1994.tb00444.x. [DOI] [PubMed] [Google Scholar]
- 18.Gründling A, Smith DL, Bläsi U, Young R. Dimerization between the holin and holin inhibitor of phage lambda. J Bacteriol. 2000;182:6075–6081. doi: 10.1128/jb.182.21.6075-6081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bläsi U, Chang CY, Zagotta MT, Nam KB, Young R. The lethal λ S gene encodes its own inhibitor. EMBO J. 1990;9:981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steiner M, Bläsi U. Charged amino-terminal amino acids affect the lethal capacity of lambda lysis proteins S107 and S105. Mol Microbiol. 1993;8:525–533. doi: 10.1111/j.1365-2958.1993.tb01597.x. [DOI] [PubMed] [Google Scholar]
- 21.Chang CY, Nam K, Young R. S gene expression and the timing of lysis by bacteriophage λ. J Bacteriol. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White R, Tran TA, Dankenbring CA, Deaton J, Young R. The N-terminal transmembrane domain of λ S is required for holin but not antiholin function. J Bacteriol. 2010;192:725–733. doi: 10.1128/JB.01263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savva CG, et al. The holin of bacteriophage lambda forms rings with large diameter. Mol Microbiol. 2008;69:784–793. doi: 10.1111/j.1365-2958.2008.06298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krebs MP, Reznikoff WS. Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. J Mol Biol. 1986;192:781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- 25.Krebs MP, Khorana HG. Mechanism of light-dependent proton translocation by bacteriorhodopsin. J Bacteriol. 1993;175:1555–1560. doi: 10.1128/jb.175.6.1555-1560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sartori A, et al. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Smith DL, Young R. Oligohistidine tag mutagenesis of the lambda holin gene. J Bacteriol. 1998;180:4199–4211. doi: 10.1128/jb.180.16.4199-4211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DL, Struck DK, Scholtz JM, Young R. Purification and biochemical characterization of the lambda holin. J Bacteriol. 1998;180:2531–2540. doi: 10.1128/jb.180.9.2531-2540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 30.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 31.Park T, Struck DK, Deaton JF, Young R. Topological dynamics of holins in programmed bacterial lysis. Proc Natl Acad Sci USA. 2006;103:19713–19718. doi: 10.1073/pnas.0600943103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gründling A, Bläsi U, Young R. Biochemical and genetic evidence for three transmembrane domains in the class I holin, λ S. J Biol Chem. 2000;275:769–776. doi: 10.1074/jbc.275.2.769. [DOI] [PubMed] [Google Scholar]
- 33.Pogliano J, Ho TQ, Zhong Z, Helinski DR. Multicopy plasmids are clustered and localized in Escherichia coli. Proc Natl Acad Sci USA. 2001;98:4486–4491. doi: 10.1073/pnas.081075798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu NJ, Dutton RJ, Pogliano K. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol Microbiol. 2006;59:1097–1113. doi: 10.1111/j.1365-2958.2005.05004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broder DH, Pogliano K. Forespore engulfment mediated by a ratchet-like mechanism. Cell. 2006;126:917–928. doi: 10.1016/j.cell.2006.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiba S, Coleman K, Pogliano K. Impact of membrane fusion and proteolysis on SpoIIQ dynamics and interaction with SpoIIIAH. J Biol Chem. 2007;282:2576–2586. doi: 10.1074/jbc.M606056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.