Abstract
The transcription factor Krüppel-like factor 2 (KLF2) is critical for normal trafficking of T lymphocytes, but its role in B cells is unclear. We report that B cell-specific KLF2 deficiency leads to decreased expression of the trafficking molecules CD62L and β7-integrin, yet expression of sphingosine-1 phosphate receptor 1 (which is a critical target of KLF2 in T cells) was, unexpectedly, minimally altered. Unexpectedly, Klf2 deletion led to a drastic reduction in the B1 B-cell pool and a substantial increase in transitional and marginal zone B-cell numbers. In addition, we observed that KLF2-deficient B cells showed increased apoptosis and impaired proliferation after B-cell receptor cross-linking. Gene expression analysis indicated that KLF2-deficient follicular B cells display numerous characteristics shared by normal marginal zone B cells, including reduced expression of several signaling molecules that may contribute to defective activation of these cells. Hence, our data indicate that KLF2 plays a critical role in dictating normal subset differentiation and functional reactivity of mature B cells.
The Krüppel-like factor (KLF) family of transcription factors is expressed in many tissues and found to regulate differentiation, trafficking, cellular quiescence, and stem cell pluripotency (1). We and others have shown that KLF2 is critical for T-cell trafficking through regulation of the genes encoding sphingosine-1 phosphate receptor 1 (S1Pr1) and CD62L (l-selectin) (2–4), and additional studies suggest that KLF2 promotes T-cell quiescence (5–7). KLF2 is also expressed in B lymphocytes (8, 9), and both S1Pr1 and CD62L are also important for B-cell trafficking (10, 11). However, the role of KLF2 in B cells is unclear.
Three main B-cell populations have been described in mice: follicular (FO), marginal zone (MZ), and B1 B cells (12). These cells display numerous differences in their tissue distribution, trafficking potential, and functional capacity. The factors regulating differentiation of these populations are still unclear, although B-cell receptor (BCR) signal strength has been proposed to play a key role in dictating B-cell subset development (12–16).
Here, we report that KLF2 is differentially expressed in mature B-cell subsets and that KLF2 deficiency leads to overrepresentation of MZ B cells and a drastic loss of B1 B cells. In addition, we find Klf2−/− FO B cells are functionally compromised and display some gene expression characteristics of MZ B cells. KLF2 loss leads to altered B-cell trafficking and reduced CD62L and β7-integrin expression, but expression of S1Pr1 was only modestly impaired in Klf2 KO B cells. Hence, our data indicated that KLF2 has both overlapping and distinct roles in T and B cells and that KLF2 is critical for normal B-cell tissue localization, subset differentiation, and functional reactivity.
Results
KLF2 Is Differentially Expressed in Mature B-Cell Subsets.
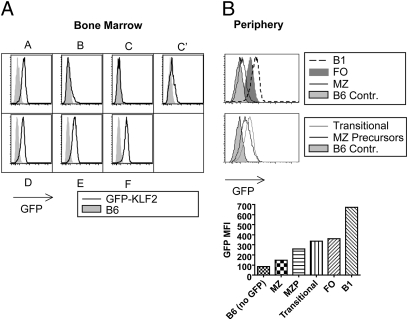
KLF2 is expressed early in B-cell development (8), but its expression pattern in mature B cells has not been defined. We used a GFP-KLF2 fusion protein knock-in mouse (4) to determine KLF2 protein expression in distinct stages of B-cell development. Confirming previous studies (8), we observed that KLF2 expression initiates at the large pre-B stage (fraction C′) of committed B-cell development and is maintained through bone marrow development (Fig. 1A). KLF2 expression was also detected in fraction A cells, but this population includes precursors for other lineages (including natural killer and dendritic cell subsets) besides B cells (17). Unexpectedly, KLF2 expression levels differed substantially between mature B-cell subsets in the periphery, with highest expression detected in B1 B cells, lower expression detected in the FO pool, and minimal KLF2 in MZ cells (Fig. 1B). Furthermore, although splenic transitional B cells expressed KLF2 similar to follicular cells, marginal zone precursor cells (MZP) had reduced KLF2 expression (Fig. 1B) (18).
Fig. 1.
KLF2 expression in B-cell precursors and mature subsets. (A) Bone marrow B-cell developmental stages A–F were distinguished (SI Methods) and assessed for KLF2 expression using the GFP-KLF2 reporter mouse (4). (B) Expression of GFP-KLF2 reporter expression levels in peripheral B cells was determined by examination of splenic MZ, splenic FO, peritoneal B1, transitional, and marginal zone precursor (MZP) cells. C57BL/6 B cells were used as GFP-negative control cells. The GFP mean fluorescent intensity (MFI) for all peripheral populations is put into one graph for direct comparison. Bone marrow and peripheral subsets are representative of two and four experiments, respectively.
KLF2 Deficiency Leads to Altered B-Cell Tissue Distribution and Expression of Trafficking Molecules.
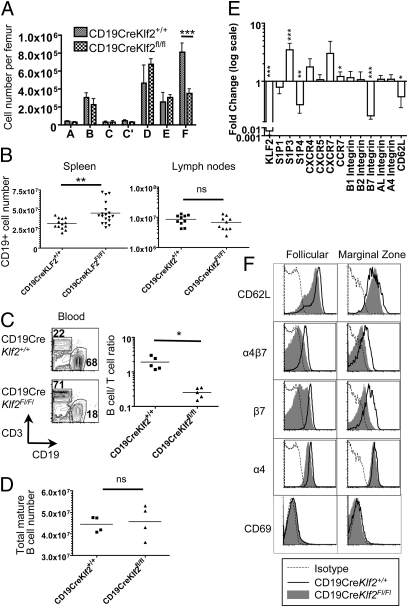
To investigate the requirement of KLF2 in B cells, we generated mice carrying floxed Klf2 alleles (4) and CD19-Cre (19) to yield conditional Klf2 deletion in committed B cells (CD19CreKlf2fl/fl). We previously showed that T cell-specific KLF2 deficiency had little effect on early thymocyte differentiation but dramatically altered thymic emigration and peripheral migration because of loss of key trafficking molecules, including S1Pr1 and CD62L (2, 4). Analysis of CD19CreKlf2fl/fl bone marrow suggested no substantial changes in B-cell development, although we noted a reduction in recirculating mature B cells (fraction F) in this pool (Fig. 2A). In the periphery, loss of KLF2 led to changes in B-cell tissue distribution: B-cell numbers were increased in the spleen (Fig. 2B), whereas the frequency of Klf2−/− B cells in the blood was significantly reduced (Fig. 2C). The number of Klf2−/− B cells was modestly (but not significantly) reduced in lymph nodes (Fig. 2B). However, total B-cell numbers (obtained by summing the numbers of B cells found in spleen, lymph nodes, and the mature B-cell pool in the bone marrow) were unchanged by KLF2 deficiency (Fig. 2D).
Fig. 2.
KLF2 deficiency alters tissue distribution and expression of trafficking molecules by B cells. (A) Assessment of B-cell developmental stages in bone marrow from CD19CreKlf2+/+ (control) and CD19CreKlf2Fl/Fl (KLF2-deficient) mice delineated as in Fig. 1A. (B) Numbers of CD19+ B cells in spleen and lymph nodes from CD19CreKlf2+/+ and CD19CreKlf2Fl/Fl mice. C shows the ratio of CD3+ T-cell to CD19+ B-cell frequencies in peripheral blood from CD19CreKlf2+/+ and CD19CreKlf2Fl/Fl mice. In D, total B-cell numbers were calculated by summing mature B numbers in spleen, peripheral, and mesenteric lymph node and bone marrow. (E) Sorted follicular cells from <6-mo-old C57BL/6 and CD19CreKlf2Fl/Fl mice were compared for gene expression of KLF2 and trafficking genes by real-time PCR (n = 3). (F) Two-month-old CD19CreKlf2+/+ and CD19CreKlf2Fl/Fl mice were compared for surface expression of CD62L, α4β7 integrin pair, β7 integrin alone, α4 alone, and CD69 for FO and MZ cells. Data are representative of three experiments.
RT-PCR analysis on sorted FO B cells revealed small but significant reductions in expression of Sell (encoding CD62L) and Itgb7 (encoding β7-integrin) in Klf2−/− B cells, although expression of CXCR7 was elevated (Fig. 2E). Given that KLF2 is required for expression of S1Pr1 by T cells, it was surprising that expression of Edg1 (encoding S1Pr1) was only modestly reduced in KLF2-deficient FO B cells (Fig. 2E). However, the expression of mRNA encoding S1Pr3 was elevated, and the expression of mRNA encoding S1Pr4 was decreased. Although the role of S1Pr4 is obscure, S1Pr3 has been implicated in positioning of MZ B cells (20), but its function in FO B cells is less clear.
Reduced CD62L and β7-integrin expression on Klf2−/− FO B cells could be confirmed at the level of cell surface expression (Fig. 2F). Reduction of β7-integrin but not CD62L was also observed on MZ B cells (Fig. 2F). Cell surface staining for S1Pr1 on B cells has proven technically difficult (21), but elevated CD69 expression acts as a surrogate marker for reduced S1Pr1 expression (10, 20); it is noticeable that CD69 levels were unchanged in CD19CreKlf2fl/fl B cells (Fig. 2F). We also saw reduced cell surface staining for α4-integrin and the α4/β7-heterodimer on CD19CreKlf2fl/fl FO B cells (Fig. 2F); this was likely secondary to reduced β7-integrin expression, because mRNA levels for Itga4 gene were not noticeably altered (Fig. 2E).
Other trafficking molecules important for B-cell positioning, including CXCR4, CXCR5, LFA-1, and β1-integrin, were found to be unchanged at the level of cell surface expression in KLF2-deficient B cells (Fig. S1).
Together, these data indicate that, similar to its function in T cells, KLF2 influences expression of trafficking molecules and tissue distribution of B cells; however, the role of KLF2 is clearly distinct, which is exemplified by the substantial loss of Edg1 expression in KLF2-deficient T cells (2, 3) but not B cells (Fig. 2E).
KLF2 Deficiency Leads to an Increase in the MZ Pool and Decrease in the B1 B-Cell Population.
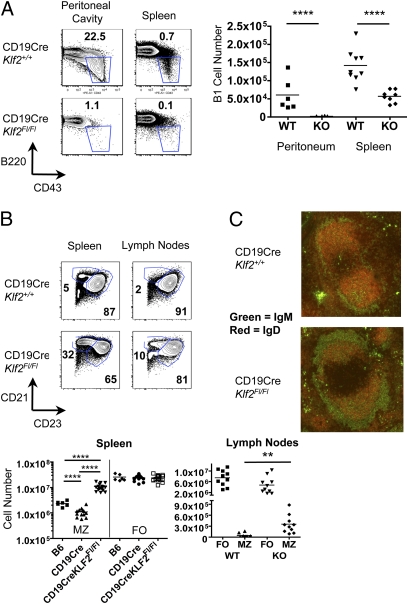
Given the differences in KLF2 expression by mature B-cell subsets (Fig. 1A), we next explored the impact of KLF2 deficiency on these populations. Strikingly, the B1 B-cell pool of CD19CreKlf2fl/fl mice was substantially reduced, this pool being virtually absent from the peritoneal cavity and significantly diminished in the spleen (Fig. 3A), with the residual splenic B1 population being predominantly B1b rather than B1a cells (Fig. S2A).
Fig. 3.
CD19CreKlf2Fl/Fl mice have significantly decreased B1 cells and increased splenic MZ B cells, with aberrant localization of MZ cells in the lymph nodes. (A) Frequencies and numbers of B1 phenotype B cells were determined in the spleen and peritoneal cavity of control and CD19CreKlf2Fl/Fl mice. (B) Frequencies and numbers of MZ phenotype B cells were determined in the spleen and lymph nodes of control and CD19CreKlf2Fl/Fl mice. (C) Immunohistochemistry of the spleen sections were stained for IgM (green) and IgD (red). MZ B cells (IgM hi and IgD lo) are localized in the MZ in both control and CD19CreKlf2Fl/Fl mice. Representative of three experiments.
In contrast, splenic MZ cells were more than fivefold increased in CD19CreKlf2fl/fl animals (Fig. 3B). We also observed elevated expression of CD21 and CD23 on CD19CreKlf2fl/fl FO B cells (Fig. 3B), presenting a potential complication for demarcation of FO and MZ populations using these markers. However, staining for the MZ markers CD9 and CD1d as well as differential IgD and IgM (12) expression confirmed the increase in MZ numbers in Klf2−/− splenocytes (Fig. S2 B and C). It is important to note that the increased Klf2−/− MZ pool occurred, despite the reduction in the MZ population observed in CD19-Cre heterozygous animals (Fig. 3B) (22).
We also assayed serum Ig of control and CD19CreKlf2fl/fl animals. Overall levels of all Ig isotypes were slightly reduced in KLF2-deficient animals, and this reached statistical significance for IgA, IgG1, and IgE (Fig. S3). Given the near absence of detectable B1 B cells, the fact that IgM levels were unaltered was initially surprising, because B1 cells are thought to contribute significantly to the natural IgM pool (23, 24). However, this finding might be explained by the compensatory expansion of the MZ pool and/or the residual splenic B1b pool found in CD19CreKlf2fl/fl animals.
We also observed a small but reproducible population in lymph nodes of CD19CreKlf2fl/fl mice that displayed a MZ B-cell phenotype, being CD23low, CD21high, IgMbright, and IgDdim (Fig. 3B and Fig. S2C). Because MZ phenotype B cells are normally absent from lymph nodes, these data suggest altered distribution/trafficking by Klf2−/− MZ B cells. Manipulation of several trafficking molecules (including S1Pr1 and LFA-1) leads to MZ displacement into follicles (20, 25), and although we did not find expression changes for these particular receptors (Fig. 2 and Fig. S1), we assessed the distribution of splenic MZ cells by immunohistology. As shown in Fig. 3C, the CD19CreKlf2fl/fl IgMhi IgDlo (MZ phenotype) population was positioned appropriately in the MZ, with few cells of this phenotype in the follicles. However, the marginal zone was generally thicker in CD19CreKlf2fl/fl animals, presumably reflecting the increased MZ pool.
Alterations in CD19CreKlf2fl/fl B Cells Are Cell-Autonomous.
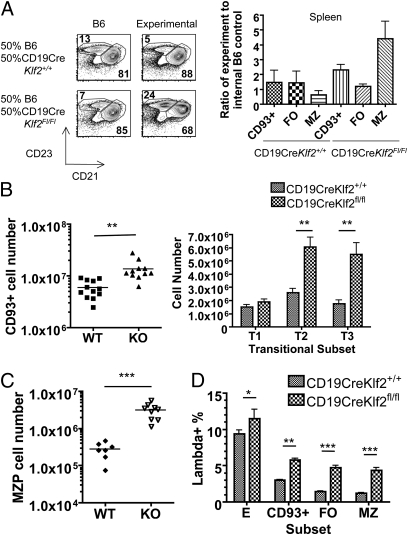
We recently reported nonautonomous effects of KFL2 deficiency in T cells (4, 26); hence, it was important to assess the intrinsic effects of KLF2 in B cells. Accordingly, we examined mixed bone marrow chimeras involving CD19CreKlf2fl/fl (or control CD19CreKlf2+/+) bone marrow mixed 1:1 with congenic B6 bone marrow. The enhancement in MZ B-cell frequency, increased expression of CD23 and CD21, and reduced expression of CD62L and β7-integrin on FO B cells were selective to the CD19CreKlf2fl/fl donor B cells, suggesting that these effects were autonomous (Fig. 4A and Fig. S4A). It was possible that the MZ B-cell phenotype of the CD19CreKlf2fl/fl was secondary to effects of CD19 heterozygosity (because of use of the CD19-Cre system). However, we also generated Klf2fl/fl mice carrying a Vav-Cre transgene, in which Cre is expressed early in hematopoietic differentiation (27), and bone marrow chimeras derived from these cells also showed an autonomous increase in the MZ pool (Fig. S4B). B1 cells are not efficiently generated from adult bone marrow, limiting use of chimeras to assess whether loss of this pool was autonomous to KLF2-deficiency. However, the B1 pool was reduced in VavCreKlf2fl/fl animals (Fig. S4C), suggesting that this feature is common to distinct Cre systems. Hence, our data revealed dramatic changes in representation of B-cell subsets after Klf2 deletion.
Fig. 4.
(A) Mixed chimeras comprising CD19CreKlf2+/+ or CD19CreKlf2fl/fl bone marrow mixed (at a 1:1 ratio) with WT congenic bone marrow into irradiated B6.SJL CD45.1 hosts. The actual chimerism achieved was used to normalize chimerism to a ratio of 1, and representation of mature B-cell subsets was assessed through ratios of experimental to WT control. (B) CD19CreKlf2+/+ and CD19CreKlf2Fl/Fl splenic bulk transitional and T1, T2, and T3 cell numbers from mice ages 2–6 mo. (C) MZ precursor numbers from the spleen. CD19CreKlf2fl/fl MZP number significantly increased. (D) Two-month-old CD19CreKlf2+/+ and CD19CreKlf2Fl/Fl mice were compared for λ-light chain expression. CD19CreKlf2Fl/Fl transitional, MZ, and FO cells had significantly increased λ-chain expression compared with control mice.
KLF2 Deficiency Leads to Increased Frequency of Transitional and MZ B Cells.
Although bone marrow B-cell precursors were normally represented in CD19CreKlf2fl/fl animals (Fig. 2B), we observed a significant increase in the numbers of splenic transitional B cells, which are precursors to the mature B2 B-cell subsets and are identified by expression of CD93 (12). Both T2 and T3 (but not T1) transitional cells were increased in number (Fig. 4B). However, some caution is required in this interpretation; CD23 expression is typically used to discriminate T1 and T2 cells, and this marker is increased on KLF2-deficient FO B cells (Fig. 3B). In addition, we found increased numbers of MZP cells (18) in CD19CreKlf2fl/fl animals (Fig. 4C), consistent with the increased representation of mature MZ cells. A candidate mechanism for enhancing MZ B-cell survival would be elevated B-cell activating factor (BAFF) sensitivity, because this factor is limiting for MZ cell maintenance (28, 29). However, expression levels for the BAFF receptors were not significantly altered by KLF2 loss on either FO or MZ populations (Fig. S5).
Increased numbers of immature FO and MZ B cells might imply a partial block in late B-cell maturation or improved survival of developing B cells through this stage. Previous studies have suggested that B-cell maturation accompanies progressive loss of λ-light chains-expressing B cells, correlating with editing and subsequent elimination of autoreactive B cells (30, 31). Interestingly, we observed a consistent increase in the frequency of λ expressing cells among transitional and mature B-cell populations from CD19CreKlf2fl/fl mice (Fig. 4D), consistent with enhanced survival of developing B cells in KLF2-deficient mice. A potential consequence of altered selection would be enhanced survival of autoreactive B cells, but anti-dsDNA antibodies were not elevated in the serum of young or old CD19CreKlf2fl/fl mice (Fig. S6).
KLF2-Deficient FO B Cells Display Impaired Functional Reactivity.
Gene expression microarrays showed that a number of genes with a potential role in B-cell signaling were altered in KLF2-deficient FO B cells (Figs. S7 and S8). This included several genes encoding proteins proposed to influence calcium and potassium mobilization or signaling (Fig. S7) but also genes for other molecules with potential roles in B-cell signaling, including Smad-1, Lilrb3 (Pirb), Pira2, and Dusp3 (down-regulated in Klf2−/− B cells) as well as the phosphatases Ptpn22 and Ptpn14 (up-regulated in Klf2−/− B cells) (Figs. S7 and S8).
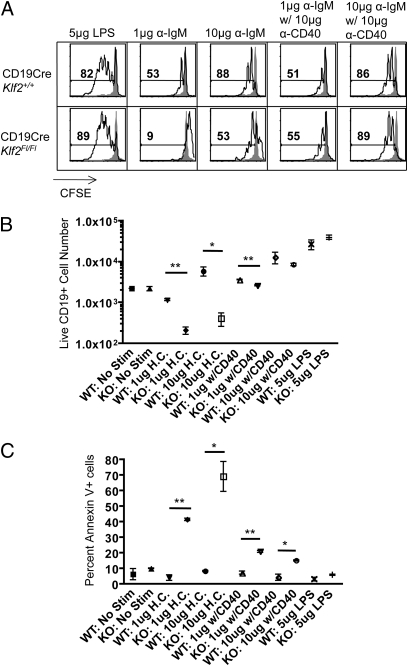
This raised the possibility that KLF2-deficient FO B cells may exhibit altered activation properties. Thus, we next tested the impact of KLF2 loss on enriched FO B-cell proliferation and expansion after LPS, anti-IgM, or anti-IgM/anti-CD40 stimulation in vitro (Fig. 5 A and B). Proliferation [as measured by carboxyfluorescein diacetate succinimidyl ester (CFSE) dye dilution] and accumulation (as measured by viable cell numbers) were similar for both control and CD19CreKlf2fl/fl B cells in their response to LPS (Fig. 5 A and B). In contrast, KLF2-deficient B cells underwent less CFSE dye dilution after anti-IgM activation, although this response was substantially restored by stimulation with high-dose anti-IgM (Fig. 5A). Even more striking, accumulation of CD19CreKlf2fl/fl B cells was impaired after stimulation with high or low doses of anti-IgM stimulation (Fig. 5B). Furthermore, analysis at earlier time points indicated that Klf2−/− B cells had increased susceptibility to apoptosis induced by anti-IgM (but not LPS) (Fig. 5C). Interestingly, stimulation with anti-IgM plus anti-CD40 substantially enhanced the proliferation, expansion, and survival of the Klf2−/− pool (Fig. 5 A–C). Together, these data suggest that KLF2 deficiency leads to impaired survival after BCR stimulation and that engagement of CD40 can, at least partially, protect against this increased cell death.
Fig. 5.
Impaired expansion and survival of KLF2-deficient B cells after BCR stimulation. FO B cells were enriched from CD19CreKlf2Fl/Fl (KO) or control CD19CreKlf2+/+ (WT) animals, labeled with CFSE, and stimulated in vitro with 5 μg LPS or with anti-IgM F(ab’)2 (1 or 10 μg) with or without 10 μg anti-CD40. After culture for 3 (A and B) or 2 (C) d, cells were assayed by flow cytometry. B cells (gated as CD19+ve and DAPI-ve cells) were assessed for (A) CFSE dye dilution, (B) accumulation of live cells, and (C) apoptosis (measured as Annexin-V+ve DAPI-ve cells). Data are representative of triplicate samples analyzed from two mice per group, and similar results were obtained in three separate experiments.
KLF2-Deficient FO B Cells Display Some Gene Expression Features of MZ B Cells.
On further examination of gene microarray data, we noticed that many genes that were substantially up- or down-regulated when comparing KLF2-deficient and control FO B cells were also changed in published (32) and public (Gene Expression Omnibus databases) comparisons between normal MZ and FO B cells. This feature is illustrated in a cluster map (Fig. S8). Genes altered in expression include those for molecules associated with trafficking (including CD62L, β7- and α6-integrins, S1Pr3, and CXCR7), signaling (including Kctd-14, Ahnak-1, S100a6, Fkbp11, PirA2, Nedd4, Lilrb3, Ptpn14, Ptpn22, and Kcnn4), and transcriptional regulation (KLF3 and Pdlim-1) as well as several other molecules with less obvious potential functional roles. Overall, these data are consistent with the concept that KLF2 is required to impart an element of the FO B-cell identity, which distinguishes such cells from MZ B cells.
Discussion
Our data indicate a critical role for KLF2 in trafficking, function, and terminal differentiation of B cells. KLF2 loss in T cells leads to impaired S1Pr1 and CD62L expression as well as nonautonomous effects on CXCR3 and β7-integrin expression (2, 4). In contrast, although B cell-specific KLF2 loss caused reduced blood recirculation and impaired CD62L and β7-integrin expression, S1Pr1 expression was only modestly reduced. This lack of correlation between S1Pr1 and KLF2 expression is consistent with the fact that KLF2 expression is very low in MZ B cells (Fig. 1), but these cells express higher levels of S1Pr1 than FO B cells (20). The basis for reduced Klf2−/− B-cell frequencies in the blood is, therefore, not resolved, but it is interesting to speculate that this may be related to increased expression of S1Pr3 or decreased expression of S1Pr4 in CD19CreKlf2fl/fl FO B cells. In addition, the up-regulation of CXCR7 in Klf2−/− FO B cells is noteworthy. This receptor (which is highly expressed by normal MZ B cells) is thought to lack intrinsic signaling capacity but may cooperate with CXCR4 in reactivity to CXCL12 (33); hence, its overexpression may contribute to altered Klf2−/− B-cell trafficking.
Unexpectedly, our studies indicated a role for KLF2 in differentiation and maintenance of mature peripheral B-cell subsets. Loss of KLF2 led to a profound reduction in the B1 pool, whereas the MZ population was substantially increased. Current models suggest that B-cell subset differentiation is driven, at least in part, by the BCR signal strength: genetic manipulations that amplify or diminish BCR signal strength alter B-cell subset frequencies (12–16), leading to models in which differentiation of MZ, FO, and B1 subsets is favored by low-, medium-, and high-intensity BCR signals, respectively (13, 14). Given that KLF2 expression levels follow the pattern MZ < FO < B1 and that BCR-induced responses were impaired in KLF2-deficient FO B cells, it is tempting to speculate that KLF2 may be responsible for regulating and/or interpreting BCR signal strength during B-cell subset selection. Indeed, examination of the gene expression profiles for KLF2-KO FO B cells (Figs. S7 and S8) indicates several changes that may impair B-cell activation. Little is known about the function for most of these genes in lymphocytes, making it difficult to assess their significance in B-cell activation/differentiation. The fact that expression of multiple molecules with potential roles in B-cell signaling is altered by KLF2 loss makes it less likely that single targets will account for changes in response or differentiation. At the same time, the overall patterns of differential gene expression reveal notable similarities between KLF2-deficient FO B cells and conventional MZ B cells (Fig. S8). This included many genes that do or could influence trafficking, signaling and transcription. It should be stressed that several MZ B cell markers (including CD1d and CD9) were not expressed by KLF2-deficient FO B cells (Figs. S2B and S8), arguing against overt contamination of analyzed FO B cells by contaminating MZ B cells. However, the fact that KLF2 loss leads to merging of the FO and MZ B cell identities suggests that this transcription factor may play a role in directing and/or interpreting the cues that define B-cell subset differentiation.
In addition, we observed that enriched Klf2−/− FO B cells showed reduced capacity to respond to anti-IgM stimulation, exhibiting increased susceptibility to apoptosis compared with controls. In contrast, stimulation with anti-IgM/anti-CD40 largely restored the Klf2−/− B-cell proliferative response, and their proliferative responses to LPS were similar to controls. These features are reminiscent of those reported for normal MZ B cells (34) and reinforce the potential overlap between KLF2−/− FO and WT MZ B cells. The basis for the enhanced apoptosis by Klf2−/− cells is not clear, and further studies will be required to understand the basis of defective Klf2−/− B-cell responses.
During preparation of this manuscript, a report from Hoek et al. (35) was published that also described KLF2-deficient B cells. Although features of this analysis, such as enhanced MZ and reduced B1 B-cell numbers coincide with our findings, Hoek et al. (35) had a substantially different interpretation of the role played by S1Pr1 and CXCR5. Hoek et al. (35) proposed that KLF2 deficiency caused S1Pr1 and CXCR5 gene expression to be elevated in FO B cells and reduced in MZ B cells, leading to a model in which Klf2−/− FO B cells encroach on the MZ. In contrast, we found that S1Pr1 expression was modestly reduced in KLF2-deficient FO B cells and saw no evidence of changes in CXCR5 levels at the mRNA or protein level. The basis for these divergent findings is unclear; however, our results are further reinforced by the paper from Winkelmann et al. (36) that reached similar conclusions using a distinct KLF2 deletion system. In addition, our study shows that KLF2-deficient B cells exhibit impaired expansion after BCR stimulation and homology between gene expression patterns by CD19CreKlf2fl/fl FO and normal MZ B cells, suggesting that the altered differentiation and trafficking of KLF2-deficient FO and MZ B cells reflects numerous changes but not altered S1Pr1 or CXCR5 expression.
In summary, our data suggest that KLF2 is crucial for B-cell subset differentiation as well as regulation of B-cell activation and trafficking. These aspects may be interlinked with KLF2, contributing to the identity of mature B-cell subsets.
Methods
Please see SI Methods for further information.
Mice.
C57BL/6 (B6) and B6.SJL-Ptprca Pep3b (B6.SJL) were purchased from the National Cancer Institute. Klf2fl and Klf2GFP reporter mice were described earlier (4). CD19-Cre mice were purchased from the Jackson Laboratory. Vav-Cre mice (27) were generated by Dimitris Kioussis (National Institute of Medical Research, London, England) and obtained through Bruce Walcheck (University of Minnesota, Minneapolis, MN). All animal experimentation was approved by the University of Minnesota Institutional Animal Care and Use Committee.
Flow Cytometry.
All fluorochrome- and biotin-conjugated antibodies were purchased from eBioscience, BD BioScience, R&D Systems, or BioLegend. Flow cytometry samples were prepared by staining single cell suspensions with antibodies in FACS buffer for 30 min at 4 °C and washing cells two times with FACS buffer. Flow cytometry data were collected using an LSR-II cytometer (BD Biosystems) and analyzed using FlowJo software (Tree Star).
Immunofluorescence.
Frozen sections were presoaked in FACS buffer for 10 min and stained with anti-IgM–FITC and biotinylated anti-IgD, the latter revealed with Cy3 Streptavidin; 20× images were obtained with a Leica DM5500B automated upright microscope.
Mixed Bone Marrow Chimeras.
Recipient mice were lethally irradiated (1,000 rads, using a Cs source) and reconstituted with donor cells that were depleted for T cells. Specifically, (C57BL/6 × B6.SJL)F1 (CD45.1/CD45.2) was used as control bone marrow (BM) mixed at a 1:1 ratio with either Cre+Klf2+/+ or Cre+Klf2Fl/Fl (CD45.2) experimental BM, and 107 total BM T cell-depleted cells were injected into B6.SJL (CD45.1) hosts. Cre+ mice were either CD19-Cre or Vav-Cre as indicated. Reconstitution progressed for at least 2 mo time before mice were used for experiments.
Sorting.
For microarray analysis, B cells were purified through a negative selection B-cell isolation kit on MACS columns (Miltenyi Biotec) from (C57BL/6 × B6.SJL)F1 (CD45.1/CD45.2) control and CD19CreKlf2Fl/Fl (CD45.2) splenocytes. Isolated B cells were mixed in equal amounts, and FO B cells of each genotype were sorted based on their congenic markers on a FACSVantage or FACSAria (BD Biosciences) followed by RNA preparation. Alternatively, FO B cells were sorted from individual CD19CreKlf2+/+ and CD19CreKlf2Fl/Fl animals for RNA preparation for RT-PCR and in vitro proliferation experiments. Purity was >95% for the target follicular cell population.
RNA Purification.
RNA was isolated from B cells using the RNeasy kit (Qiagen) according to the manufacturer's recommendations (on-column DNase digestion was included). RNA was quantified using a NanoDrop 2000 Spectrophotometer (Thermo Scientific) and Agilent 2100 Bioanalyzer.
Real-Time PCR.
cDNA and RT control was generated using the SuperScriptIII Platinum Two-Step gRT-PCR kit (Invitrogen). PCR products were amplified using SYBR green master mix (Roche) on an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Primer sequences can be obtained by contacting the corresponding author (S.C.J.).
Proliferation Assays.
To enrich for FO B cells, biotinylated CD9 and CD93 antibodies (BD Bioscience and eBioscience, respectively) were added to B cell-negative enrichment antibodies (Miltenyi Biotech). Cells were washed and then incubated with antibiotin magnetic particles, and cells were depleted by MACS. For the experiments shown, B-cell purity was greater than 95%, greater than 86% follicular phenotype, and less than 7% MZ phenotype. Enriched follicular cells were labeled with CFSE (Molecular Probes) and plated (in triplicate) at 2.5 × 105 cells/200 μL supplemented Roswell Park Memorial Institute (RPMI) 1640 media in 96-well plates. F(ab′)2 anti-IgM (Jackson ImmunoResearch) at 10 μg or 1 μg/mL with or without 10 μg/mL anti-CD40 (BioXCell) or LPS at 5 μg/mL (Sigma) was used for stimulation. Cells were analyzed by flow cytometry after staining for CD19, and DAPI (Invitrogen) was used to exclude dead cells. Up-regulation of AnnexinV (eBioscience) was monitored at day 2 of stimulation, whereas analysis of CFSE dye dilution and cell accumulation was determined at day 3. AccuCheck Counting Beads (Invitrogen) were used to determine cell counts.
Statistical Methods.
SDs and P values were calculated using Prism software (Graphpad Software Inc.) through Student unpaired t tests or two-way ANOVA analyses where appropriate. Symbols indicate significance levels (*P < 0.05; **P < 0.01; ***P ≤ 0.001; ****P < 0.0005).
Supplementary Material
Acknowledgments
We thank the Jamequist laboratory members for critical input, especially Sam Dunmire for performing the gene expression cluster analysis, Michael Cancro and Jean Sholz for discussions and practical advice, and Pam Baker for timely provision of RT-PCR equipment for these studies. This work was supported by National Institutes of Health Predoctoral Training Grant T32 AI07313 (to G.T.H.) and Grant R37 AI38903 (to S.C.J.).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013168108/-/DCSupplemental.
References
- 1.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: A functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 2.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 3.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 4.Weinreich MA, et al. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21 WAF1/CIP1. Oncogene. 2004;23:8088–8096. doi: 10.1038/sj.onc.1207996. [DOI] [PubMed] [Google Scholar]
- 6.Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc–dependent pathway. Nat Immunol. 2001;2:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- 7.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 8.Schuh W, Meister S, Herrmann K, Bradl H, Jack HM. Transcriptome analysis in primary B lymphoid precursors following induction of the pre-B cell receptor. Mol Immunol. 2008;45:362–375. doi: 10.1016/j.molimm.2007.06.154. [DOI] [PubMed] [Google Scholar]
- 9.Glynne R, Ghandour G, Rayner J, Mack DH, Goodnow CC. B-lymphocyte quiescence, tolerance and activation as viewed by global gene expression profiling on microarrays. Immunol Rev. 2000;176:216–246. doi: 10.1034/j.1600-065x.2000.00614.x. [DOI] [PubMed] [Google Scholar]
- 10.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 11.Arbones ML, et al. Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity. 1994;1:247–260. doi: 10.1016/1074-7613(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 12.Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niiro H, Clark EA. Regulation of B-cell fate by antigen-receptor signals. Nat Rev Immunol. 2002;2:945–956. doi: 10.1038/nri955. [DOI] [PubMed] [Google Scholar]
- 14.Pillai S, Cariappa A, Moran ST. Positive selection and lineage commitment during peripheral B-lymphocyte development. Immunol Rev. 2004;197:206–218. doi: 10.1111/j.0105-2896.2003.097.x. [DOI] [PubMed] [Google Scholar]
- 15.Hardy RR. B-1 B cell development. J Immunol. 2006;177:2749–2754. doi: 10.4049/jimmunol.177.5.2749. [DOI] [PubMed] [Google Scholar]
- 16.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: Innate B and T lymphocytes. Nat Rev Immunol. 2001;1:177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 17.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava B, Quinn WJ, 3rd, Hazard K, Erikson J, Allman D. Characterization of marginal zone B cell precursors. J Exp Med. 2005;202:1225–1234. doi: 10.1084/jem.20051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cinamon G, et al. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nat Immunol. 2004;5:713–720. doi: 10.1038/ni1083. [DOI] [PubMed] [Google Scholar]
- 21.Lo CG, Xu Y, Proia RL, Cyster JG. Cyclical modulation of sphingosine-1-phosphate receptor 1 surface expression during lymphocyte recirculation and relationship to lymphoid organ transit. J Exp Med. 2005;201:291–301. doi: 10.1084/jem.20041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato S, Ono N, Steeber DA, Pisetsky DS, Tedder TF. CD19 regulates B lymphocyte signaling thresholds critical for the development of B-1 lineage cells and autoimmunity. J Immunol. 1996;157:4371–4378. [PubMed] [Google Scholar]
- 23.Boes M. Role of natural and immune IgM antibodies in immune responses. Mol Immunol. 2000;37:1141–1149. doi: 10.1016/s0161-5890(01)00025-6. [DOI] [PubMed] [Google Scholar]
- 24.Martin F, Kearney JF. B-cell subsets and the mature preimmune repertoire. Marginal zone and B1 B cells as part of a “natural immune memory.”. Immunol Rev. 2000;175:70–79. [PubMed] [Google Scholar]
- 25.Lu TT, Cyster JG. Integrin-mediated long-term B cell retention in the splenic marginal zone. Science. 2002;297:409–412. doi: 10.1126/science.1071632. [DOI] [PubMed] [Google Scholar]
- 26.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 28.Mackay F, et al. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med. 1999;190:1697–1710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer-Bahlburg A, Andrews SF, Yu KO, Porcelli SA, Rawlings DJ. Characterization of a late transitional B cell population highly sensitive to BAFF-mediated homeostatic proliferation. J Exp Med. 2008;205:155–168. doi: 10.1084/jem.20071088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melchers F, et al. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol Rev. 2000;175:33–46. [PubMed] [Google Scholar]
- 31.Dingjan GM, et al. Bruton's tyrosine kinase regulates the activation of gene rearrangements at the lambda light chain locus in precursor B cells in the mouse. J Exp Med. 2001;193:1169–1178. doi: 10.1084/jem.193.10.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kin NW, Crawford DM, Liu J, Behrens TW, Kearney JF. DNA microarray gene expression profile of marginal zone versus follicular B cells and idiotype positive marginal zone B cells before and after immunization with Streptococcus pneumonia. J Immunol. 2008;180:6663–6674. doi: 10.4049/jimmunol.180.10.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sierro F, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci USA. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliver AM, Martin F, Gartland GL, Carter RH, Kearney JF. Marginal zone B cells exhibit unique activation, proliferative and immunoglobulin secretory responses. Eur J Immunol. 1997;27:2366–2374. doi: 10.1002/eji.1830270935. [DOI] [PubMed] [Google Scholar]
- 35.Hoek KL, et al. Follicular B cell trafficking within the spleen actively restricts humoral immune responses. Immunity. 2010;33:254–265. doi: 10.1016/j.immuni.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkelmann R, et al. B cell homeostasis and plasma cell homing controlled by Krüppel-like factor 2. Proc Natl Acad Sci USA. 2011;108:710–715. doi: 10.1073/pnas.1012858108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.