Abstract
c-Myc is frequently deregulated in human cancers. Although deregulated c-Myc leads to tumor growth, it also triggers apoptosis in partnership with tumor suppressors such as ARF and p53. Apoptosis induced by c-Myc is a critical fail-safe mechanism for the cell to protect against unrestrained proliferation. Despite the plethora of information on c-Myc, the molecular mechanism of how c-Myc induces both transformation and apoptosis is unclear. Oncogenic c-Myc can indirectly induce the expression of the tumor suppressor ARF, which leads to apoptosis through the stabilization of p53, but both c-Myc and ARF have apoptotic activities that are independent of p53. In cells without p53, ARF directly binds to c-Myc protein and inhibits c-Myc–induced hyperproliferation and transformation with a concomitant inhibition of canonical c-Myc target gene induction. However, ARF is an essential cofactor for p53-independent c-Myc–induced apoptosis. Here we show that ARF is necessary for c-Myc to drive transcription of a unique noncanonical target gene, Egr1. In contrast, c-Myc induces another family member, Egr2, through a canonical mechanism that is inhibited by ARF. We further demonstrate that Egr1 is essential for p53-independent c-Myc–induced apoptosis, but not ARF-independent c-Myc–induced apoptosis. Therefore, ARF binding switches the inherent activity of c-Myc from a proliferative to apoptotic protein without p53 through a unique noncanonical transcriptional mechanism. These findings also provide evidence that cofactors can differentially regulate specific transcriptional programs of c-Myc leading to different biological outcomes.
Keywords: oncogene, cell death
Deregulation or overexpression of the transcription factor c-Myc causes hyperproliferation and tumorigenesis and is a driving factor in the majority of human cancers (1). Although c-Myc regulates hundreds of downstream target genes involved in many different cellular processes, it is unclear which target genes mediate specific c-Myc functions (2). Apoptosis in response to deregulated c-Myc is a major fail-safe mechanism that is essential to prevent the proliferation of tumorigenic cells (3). Apoptosis induced by oncogenic c-Myc occurs through both p53-dependent and independent mechanisms that are not well understood (4, 5). A prevailing model for a p53-dependent mechanism is that the tumor suppressor ARF, which is induced by oncogenic c-Myc, causes the stabilization of the p53 protein by inhibiting its E3 ubiquitin ligase Mdm2 (6, 7). Additionally, ARF, independently of p53, binds to c-Myc directly and blocks the ability of c-Myc to activate transcription of examined canonical target genes containing a CACGTG E-box Myc binding site (EMS) and also inhibits c-Myc–induced hyperproliferation and transformation (8, 9). Despite this inhibition of canonical c-Myc activity, ARF has been shown to be essential for c-Myc to induce p53-independent apoptosis in mouse embryo fibroblasts (MEFs) (9, 10). However, the mechanism of how ARF regulates c-Myc induced p53-independent apoptosis is unknown. In this report, we examined the transcriptional consequences of the c-Myc/ARF interaction and discovered that in contrast to the ability of ARF to block c-Myc canonical target gene up-regulation, ARF is necessary for the direct transcriptional induction of Egr1 through a unique noncanonical mechanism.
The early growth response family of proteins, which are zinc finger transcription factors, have been shown to play roles in multiple pathways and processes, including differentiation, proliferation, apoptosis, and tumorigenesis, in a variety of tissues (11–14). Numerous studies suggest that Egr1, the best-characterized family member, is a tumor suppressor. For example, studies showed that the expression of Egr1 was undetectable in a majority of human breast and nonsmall cell lung carcinomas and deleted in 50% of acute myeloid leukemias (15, 16). Additionally, Egr1-null mice are more prone to skin cancer than their littermates when challenged with the two-step carcinogenesis model (17). Furthermore, several studies suggest Egr1 is an important mediator of apoptosis. The MEFs from the Egr1-null mice, as well as several other cell types with experimentally lowered levels of Egr1, are resistant to radiation-induced apoptosis (18–20). Also, Egr1 overexpression is sufficient to induce or enhance p53-dependent and p53-independent apoptosis in several different cell types (14, 21, 22). Here we show that Egr1 is essential for p53-independent c-Myc–induced apoptosis, but not ARF-independent c-Myc-induced apoptosis. Therefore, the differential transcriptional induction of Egr1 by c-Myc depending on the presence of ARF provides a mechanism for the ability of ARF to switch the inherent activity of c-Myc from a proliferative to apoptotic protein without p53.
Results
Activated c-Myc Directly Induces Egr1 in the Presence of ARF.
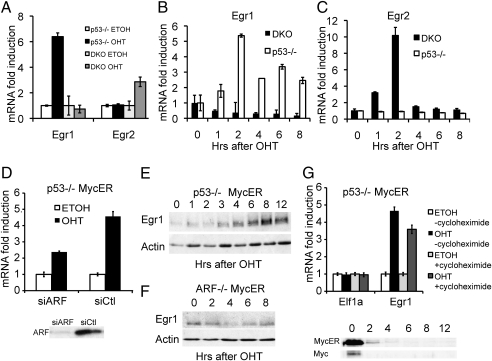
To examine whether there are c-Myc target genes that are differentially regulated as a result of the presence of ARF, we performed microarray analyses comparing c-Myc estrogen receptor (c-MycER)-inducible gene expression between p53/ARF double KO (DKO) MEFs having no ARF with p53−/− MEFs having high levels of endogenous ARF. We used genetically defined MEFs without p53 to avoid effects caused by ARF activation of p53. Gene expression was analyzed 2 h following activation of the chimeric c-MycER protein by hydroxytamoxifen (OHT) to enrich for direct targets. Interestingly, through these microarray analyses we identified all the Egr transcription factor family members as c-Myc–responsive genes (Table S1). Two of the family members (Egr2 and Egr3) were up-regulated by c-Myc in DKO MycER MEFs, but not in p53−/− MycER MEFs, which was previously observed for the canonical c-Myc target genes nucleolin, eIF4E, and htert (9). Surprisingly, the other two family members (Egr1 and Egr4) were up-regulated by c-Myc in p53−/− but not DKO-MycER MEFs, suggesting that their inductions are dependent on ARF expression. The differential regulation of Egr1 and Egr2 in p53−/− and DKO-MycER MEFs following c-Myc activation was verified by real-time RT-PCR (Fig. 1A). Further, time course analyses of Egr1 induction by c-MycER demonstrated that Egr1 mRNA levels were induced by c-Myc within 1 to 2 h in the p53−/− MycER MEFs, but not in the DKO-MycER MEFs (Fig. 1B). Conversely, Egr2 mRNA levels were induced in the DKO, but not the p53−/− MycER MEFs (Fig. 1C).
Fig. 1.
c-Myc differentially induces Egr1 and Egr2 depending on the presence of ARF. (A) Microarray verification by real-time RT-PCR of Egr1 and Egr2 mRNA levels following 2 h of OHT treatment in p53−/− and DKO MycER MEFs. (B and C) Time course analyses following MycER activation of mRNA levels of Egr1 (B) and Egr2 (C) in p53−/− and DKO MycER MEFs (± OHT) as measured by real-time RT-PCR. Results are reported as the mean of the relative mRNA levels of Egr1 or Egr2 to β-actin at each time point normalized to time 0 to give the relative fold induction ± SD. (D) Upper: Real-time RT-PCR analysis of Egr1 mRNA levels following c-Myc activation with OHT in p53−/− MycER MEFs treated first with siARF or control SMART pool. (D) Lower: Immunoblot shows ARF protein levels following siRNA treatment. (E and F) Immunoblot analyses of Egr1 protein levels following OHT activation in p53−/− MycER MEFs (E) and ARF−/− MycER MEFs (F). (G) c-Myc induces Egr1 mRNA in the absence of protein synthesis. Real-time RT-PCR analysis of Egr1 or Elf1a in p53−/− MycER MEFs (± OHT, 2 h) treated first with cycloheximide or mock treated with DMSO for 30 min to inhibit protein synthesis. Results are graphed as the mean of the relative mRNA fold induction over mock-treated (ETOH) controls ± SD. Immunoblot shows c-Myc and c-MycER protein following cycloheximide addition.
Differential regulation of Egr1 and Egr2 by c-Myc in the presence of ARF suggests different mechanisms of induction and biological outcomes. As ARF is essential for p53-independent c-Myc–induced apoptosis, the ARF-dependent Egr1 putative target gene was further characterized. To determine the necessity of ARF for induction of Egr1 by c-Myc, ARF protein expression was silenced by siRNA, as confirmed by immunoblot analysis, in p53−/− MycER MEFs (Fig. 1D Lower). Egr1 induction by c-Myc activation was reduced in the ARF siRNA treated cells compared with cells treated with control siRNA (Fig. 1D Upper), suggesting that ARF is necessary for c-Myc–driven Egr1 induction. Additionally, we confirmed the induction of Egr1 mRNA levels by c-Myc in another cell line, c-myc−/− Rat1 fibroblasts (H016) expressing c-MycER, which has low levels of ARF expression (9). Again, real-time RT-PCR confirmed that Egr1 levels are increased by OHT activation of c-MycER, but not by mock treatment (Fig. S1). Further, we also examined the effects of c-Myc activation on Egr1 protein expression. Immunoblot analyses revealed that activated c-MycER induced the expression of Egr1 protein in p53−/− MycER MEFs within 3 to 4 h, with maximal induction by 8 h (Fig. 1E), but failed to induce Egr1 levels in ARF−/− MycER MEFs (Fig. 1F). Egr1 levels were also not induced by OHT treatment alone in p53−/− MEFs with vector (Fig. S2A), nor by activated c-MycER in DKO MEFs (Fig. S2B), indicating that Egr1 protein levels are increased by activated c-Myc only in the presence of ARF.
The relatively rapid induction of Egr1 mRNA in p53−/− MycER MEFs by activated c-MycER suggests that it is a direct target. To test if c-Myc can induce Egr1 mRNA levels in the absence of protein synthesis, p53−/− MycER MEFs were treated with cycloheximide to inhibit translation before c-MycER activation. Cycloheximide treatment alone caused Elf1a, a gene not regulated by c-Myc (23), and Egr1 mRNA levels to increase (Fig. S3), as previously observed with many transcripts (24). However, activation of c-MycER in the presence of cycloheximide increased Egr1 levels by approximately fourfold in 2 h versus cycloheximide treatment alone, suggesting that c-Myc can induce Egr1 without de novo protein synthesis (Fig. 1G), even with relatively modest levels of c-MycER remaining (Fig. 1G Lower). In contrast, activated c-MycER did not enhance levels of Elf1a in the presence or absence of cycloheximide (Fig. 1G). This suggests that Egr1 is a direct transcriptional target of c-Myc. However, c-Myc may also control the levels of Egr1 by other mechanisms, such as inhibition of mRNA degradation. To determine whether c-Myc influenced Egr1 mRNA degradation, real-time RT-PCR analysis was performed following transcriptional inhibition with actinomycin D treatment in H016 cells expressing c-MycER. The decay of Egr1 mRNA levels was the same with c-MycER activation or mock treatment (Fig. S4). Therefore, activation of c-MycER primarily increases transcription of Egr1 in cells with ARF expression, rather than enhancing Egr1 mRNA stability.
Endogenous c-Myc Regulates Egr1 Levels.
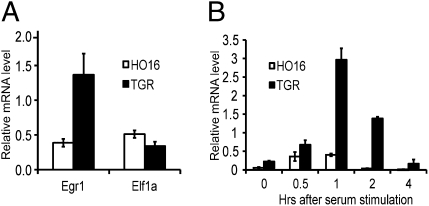
To determine whether endogenous c-Myc regulates Egr1 expression, c-myc−/− (HO16) and parental WT (TGR) Rat1 cells were harvested and the relative expression of Egr1 and a negative control Elf1a was determined with real-time RT-PCR. Egr1 was expressed substantially higher in the cells with c-Myc, unlike Elf1a, which was expressed equally in the two cell lines (Fig. 2A), suggesting that Egr1 expression is controlled by endogenous c-Myc. As the Egr genes are also known to be immediate early genes like c-Myc that are induced by serum (25), we examined the influence of endogenous c-Myc expression on the serum induction of Egr1. The TGR and HO16 cells were made quiescent by serum deprivation and then the expression of Egr1 was followed after serum stimulation. Egr1 levels were substantially increased and sustained to a greater extent in the cells with c-Myc compared with cells without c-Myc (Fig. 2B). These results suggest that Egr1 gene expression is controlled by endogenous c-Myc and that c-Myc is necessary for the full serum induction of Egr1.
Fig. 2.
Endogenous c-Myc is necessary for the full expression and induction of Egr1. (A) Real-time RT-PCR analysis of Egr1 mRNA levels in logarithmically growing c-myc−/− (H016) and parental (TGR) rat1 cells. (B) Real-time RT-PCR analysis of Egr1 mRNA levels following serum stimulation of serum-starved H016 and TGR cells. Results are graphed as the mean of the relative mRNA levels of Egr1 to Tbp ± SD.
c-Myc and ARF Are Recruited to and Regulate the Egr1 Promoter.
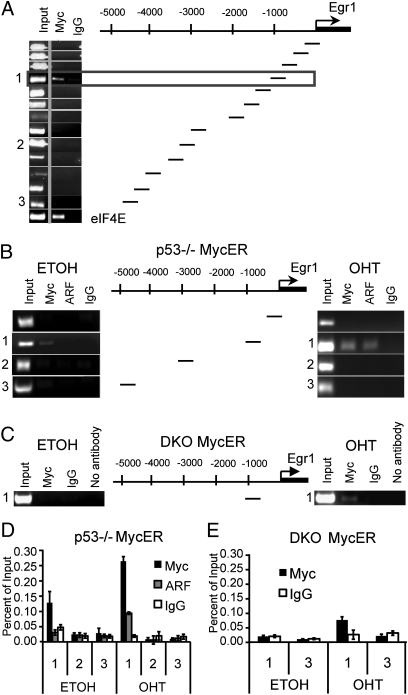
If Egr1 is a direct target as suggested by the preceding data, then c-Myc must be recruited to the Egr1 promoter. However, as the proximal promoter of Egr1 (≥15 kb in both directions) does not contain any canonical E-box Myc sites (CACGTG), we used a scanning ChIP approach to identify the c-Myc binding region. ChIP was performed using p53−/− MycER MEFs with partially overlapping primers spanning 5 kb upstream of the start site. We found that activated c-Myc was recruited to one region of the Egr1 promoter (−904 to −1,319; Fig. 3A). As a positive control, primers for the c-Myc canonical target gene eIF4E promoter were used (Fig. 3A). As Egr1 induction by c-MycER is dependent on the presence of ARF, we determined whether ARF is also recruited to the Egr1 promoter. ChIP analysis revealed that ARF was recruited to the same region as c-Myc on OHT activation, but not to other regions of the promoter (Fig. 3B Right). However, ARF was not detected at the Egr1 promoter without OHT activation (Fig. 3B Left), suggesting that ARF is recruited to the Egr1 promoter only upon c-Myc activation, as previously observed with canonical target genes (9). This observation agrees with the finding that ARF is mostly nucleolar in p53−/− MEFs until c-Myc activation causes ARF to be translocated to the nucleoplasm (9, 10). Conversely, to determine whether c-Myc can be recruited to the Egr1 promoter without ARF, we used DKO-MycER MEFs. ChIP analysis revealed that activated c-MycER was recruited to the Egr1 promoter without ARF (Fig. 3C Right). In contrast to Egr1, Egr2 is induced by c-MycER in DKO-MycER MEFs and the Egr2 promoter contains three putative canonical CACGTG sites. ChIP analysis using DKO-MycER MEFs revealed that activated c-Myc was recruited to the canonical CACGTG in the Egr2 promoter located at −2,400 (Fig. S5A), but not to the other two CACGTG sequences. As a positive control, primers for the eIF4E promoter were used (Fig. S5B).
Fig. 3.
c-Myc and ARF are recruited to the Egr1 promoter at a noncanonical binding site. (A) Chromatin prepared from p53−/− MycER MEFs (+ OHT) was subjected to immunoprecipitation using anti-Myc or IgG followed by PCR by using the indicated panel of partially overlapping primers spanning 5 kb of the Egr1 promoter. The box indicates a region (−904 to −1,319) to which c-Myc is recruited. eIF4E primers spanning an established EMS binding sequence were used as a positive control. (B) ChIPs performed as in A except MycER was either activated with OHT treatment (Right) or mock treated with ethanol (Left) for 6 h and chromatin was subjected to immunoprecipitation with anti-Myc, anti-ARF, or IgG. (C) Chromatin prepared from DKO MycER MEFs (± OHT) was subjected to immunoprecipitation using anti-Myc, IgG, or no antibody followed by PCR using the indicated primers that span the binding site in the Egr1 promoter. (D) ChIPs as in B in p53−/− MycER MEFs ± OHT except purified DNA was subjected to real-time PCR. Results are reported as the mean of the percent of input ± SD. The numbers below the x axis indicate the region of the Egr1 promoter being amplified as demonstrated in A and B. (E) Quantitative ChIPs as in D but in DKO MycER MEFs ± SD.
To quantitatively determine the relative amounts of c-Myc and ARF recruited to the Egr1 promoter under different conditions, we used real-time PCR to analyze the ChIP assays. We verified that c-Myc and ARF were both recruited to the Egr1 promoter after MycER activation, but ARF was not detected without activation of c-Myc in p53−/− MycER MEFs (Fig. 3D). Without ARF, activated c-MycER was still recruited to the Egr1 promoter in DKO-MycER MEFs (Fig. 3E), but at lower levels compared with p53−/− MycER MEFs (Fig. 3D), suggesting that ARF enhances the recruitment of c-Myc to the Egr1 promoter. Without OHT activation we detected low levels of c-Myc at the Egr1 promoter in p53−/− MycER MEFs (Fig. 3D), but not in DKO-MycER MEFs (Fig. 3E), suggesting that c-MycER is partially active without OHT in the p53−/− MycER MEFs. Taken together, the results suggest that the interaction of activated c-Myc with ARF enhances the recruitment of c-Myc to the Egr1 promoter, but that ARF alone cannot be detected.
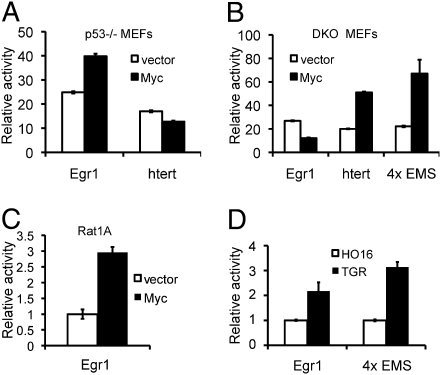
To further examine the regulation of the Egr1 promoter by c-Myc and ARF we performed luciferase assays with a 2.5-kb fragment of the Egr1 promoter containing the putative c-Myc binding site identified earlier. The Egr1 promoter was induced in p53−/− MEFs overexpressing c-Myc (Fig. 4A), demonstrating that c-Myc activates the Egr1 promoter. In contrast, the Egr1 promoter was slightly inhibited in DKO MEFs expressing c-Myc (Fig. 4B), confirming the dependence on ARF for the c-Myc activation of the Egr1 promoter. As previously reported (9), the htert promoter and the 4XEMS promoter were both induced by c-Myc in DKO MEFs (Fig. 4B), but the htert promoter induction was blocked in p53−/− MEFs with high ARF expression (Fig. 4A). In addition, the Egr1 promoter was induced by c-Myc in Rat1a cells (Fig. 4C), confirming that this regulation is not cell type-specific. To determine whether the Egr1 promoter is regulated by endogenous c-Myc, the activity of the Egr1 luciferase promoter was compared between TGR (c-myc WT) and HO16 (c-myc−/−) Rat1 cells. The Egr1 promoter was more active in TGR cells than in the HO16 cells, similar to the levels observed with the artificial canonical 4XEMS promoter (Fig. 4D). In comparison with the endogenous Egr1 expression induced by c-MycER (Fig. 1B), the activity of the Egr1 promoter induced by c-Myc in transient luciferase assays is relatively modest, suggesting that the chromatin environment and/or the different assay conditions influence the regulation of the Egr1 promoter by c-Myc. Taken together these results suggest that exogenous and endogenous c-Myc induce the Egr1 promoter by a unique ARF-dependent noncanonical transcriptional mechanism, unlike other c-Myc target gene promoters containing the canonical CACGTG binding site that are inhibited by ARF.
Fig. 4.
c-Myc activates the Egr1 promoter in the presence of ARF. (A and B) Reporter constructs for Egr1 (2.5 kb upstream), htert, and 4XEMS promoters were transiently transfected into p53−/− MEFs (A) and DKO MEFs (B) with exogenous c-Myc or vector control and a thymidine kinase renilla luciferase (pRL-TK) transfection control. The mean of the relative luciferase reporter activity to pRL-TK is reported ± SD. (C) The Egr1 reporter construct and pRL-TK were transfected into Rat1A cells that constitutively express c-Myc or vector. Relative luciferase activity was determined as described earlier and then normalized to the vector control. (D) The Egr1 reporter construct or 4XEMS were transfected into c-Myc–null (H016) and WT (TGR) cells and relative luciferase activity was determined and results were normalized to the activity in c-myc−/− cells.
Egr1 Is Necessary for c-Myc–Induced p53-Independent Apoptosis.
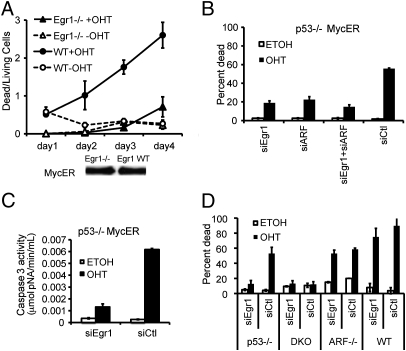
As c-Myc induces both apoptosis and Egr1 expression in an ARF-dependent manner and because Egr1 is necessary for apoptosis in several cell types (13, 14), we examined whether Egr1 might be involved in p53-independent c-Myc–mediated apoptosis. We obtained Egr1−/− MEFs and parental WT MEFs and generated lines that express comparable levels of c-MycER (Fig. 5A Lower). Activated c-MycER induced apoptosis in the WT MEFs, but failed to induce apoptosis in the Egr1−/− MEFs, suggesting that Egr1 expression is necessary for c-Myc–induced apoptosis (Fig. 5A Top). To confirm this result using another approach, we treated p53−/− MycER MEFs with Egr1 siRNA or control siRNA. Egr1 protein expression was effectively inhibited by Egr1 siRNA for up to 5 d with or without OHT treatment (Fig. S6A). Inhibiting Egr1 expression significantly reduced c-Myc–induced apoptosis in p53−/− MycER MEFs using a pool of Egr1 siRNA oligomers (Fig. 5B) or individual Egr1 siRNA oligomers (Fig. S6B). In addition, to confirm the necessity of ARF for p53-independent c-Myc–induced apoptosis, we inhibited ARF protein expression by using ARF siRNA (Fig. 1D). The reduction of c-Myc–induced apoptosis by inhibition of ARF expression was comparable to the reduction observed with inhibition of Egr1 expression (Fig. 5B). The combined inhibition of both ARF and Egr1 expression did not further reduce c-Myc–induced apoptosis (Fig. 5B), suggesting that ARF and Egr1 function in the same pathway. Reduced apoptosis resulting from the loss of Egr1 expression was confirmed by a decrease in caspase-3 activation (Fig. 5C).
Fig. 5.
Egr1 and ARF are necessary for c-Myc–induced p53-independent apoptosis. (A) Egr1−/− and parental WT MEFs expressing equal levels of c-MycER (Lower) were assayed for apoptosis (± OHT) in low serum by counting living and dead cells. (B) p53-/− MycER MEFs (± OHT) treated with siEgr1, siARF, siEgr1, and siARF, or control SMART pool no. 1 (siCtl) for 24 h were assayed for apoptosis in low serum 3 d after OHT activation. (C) Caspase-3 activity was determined in siEgr1 or siCtl-treated p53-/− MycER MEFs ± OHT for 3 d as described in Materials and Methods. (D) p53−/− MycER, DKO-MycER, ARF−/− MycER, and WT-MycER MEFs treated with siEgr1 or siCtl were assayed for apoptosis as in B.
To compare the effects of Egr1 inhibition on c-Myc–induced apoptosis in MEFs with different genetic backgrounds, we treated p53−/−, DKO, ARF−/−, and WT-MycER MEFs with Egr1 siRNA. Confirming the results shown in Fig. 5B, the inhibition of Egr1 expression by siRNA significantly reduced c-Myc–induced apoptosis in p53−/− MycER MEFs (Fig. 5D). Also, as previously shown, activation of c-MycER did not induce apoptosis in MEFs lacking p53 and ARF, and inhibition of Egr1 expression had no effect on the cells (Fig. 5D). However, activation of c-MycER in both ARF−/− and in WT MycER MEFs did cause apoptosis (Fig. 5D). This confirms that c-Myc can induce apoptosis independently of ARF in cells with p53, which has been previously shown (26). Importantly, inhibition of Egr1 by siRNA had no effect on p53-dependent, ARF-independent c-Myc–induced apoptosis observed in ARF−/− MEFs (Fig. 5D) or apoptosis caused by staurosporine treatment (Fig. S7), suggesting that reducing Egr1 levels does not cause a general defect in apoptosis, but rather Egr1 is specifically necessary for c-Myc–induced, p53-independent apoptosis. Finally, to determine whether Egr1 is capable of inducing apoptosis without activated c-Myc or p53, we generated p53−/− MEFs expressing an Egr1-ER fusion protein (Fig. S8 Lower). Upon 4 d of activation with OHT, Egr1-ER efficiently induced apoptosis in low serum (Fig. S8 Upper). Overall, these results suggest that Egr1 is necessary and sufficient for mediating p53-independent and ARF-dependent c-Myc–induced apoptosis.
Discussion
Our results suggest that there is a unique mechanism of c-Myc transcriptional regulation, whereby ARF binds with c-Myc at promoters and selectively and differentially induces c-Myc target genes. We propose that the differential regulation of c-Myc transcriptional activity by ARF allows for different biological outcomes. Previously, we demonstrated that ARF inhibits well established target genes with canonical Myc binding sites (9), as exemplified by Egr2 shown here. In contrast, we have now established that ARF is necessary for c-Myc to directly induce transcription of a noncanonical target gene, Egr1. In support of this finding, Egr1 was recently found to be induced by c-Myc in a microarray using B cells (27). Several previously identified canonical c-Myc target genes can induce apoptosis, including ODC and MT-MC1 (28, 29); however, it has not been shown conclusively that loss of a canonical direct target gene disrupts c-Myc–induced apoptosis independently of p53. In this report we identify Egr1 as a c-Myc target gene that mediates p53-independent, ARF-dependent c-Myc–induced apoptosis.
As ARF-dependent noncanonical induction of Egr1 is essential for c-Myc to induce apoptosis independently of p53, ARF binding essentially switches the inherent activity of c-Myc to an apoptotic protein through transcriptional regulation. As c-Myc is recruited to the Egr1 promoter without ARF, albeit at lesser amounts than with ARF, we propose that ARF may not only enhance recruitment of c-Myc to the Egr1 promoter, but also influences the transcriptional activity of c-Myc after DNA binding. Substantial control of c-Myc target genes after DNA binding is supported by the previous observation that c-Myc was directly recruited to approximately 3,000 genes in human B cells, but only 406 were induced by activated c-Myc (30).
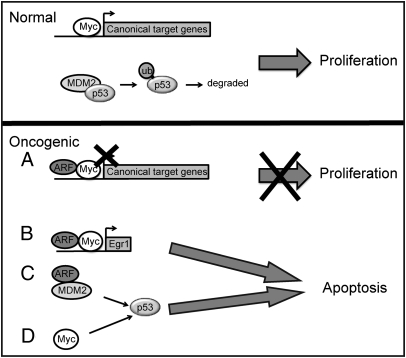
As summarized in Fig. 6, our model is that under normal physiological low ARF conditions, c-Myc induces canonical target genes, such as cyclin D2, cdk4, nucleolin, eIF4E, and Egr2, which stimulate cell cycle progression and cell growth. Under normal conditions, p53 levels are also low as a result of Mdm2-mediated degradation of p53 protein. Upon oncogenic activation, c-Myc causes both an increase in ARF expression and a relocalization of ARF from the nucleolus to the nucleoplasm, independently of p53 (9). In a direct feedback mechanism, ARF binds with c-Myc to inhibit canonical c-Myc target gene induction and proliferation while inducing noncanonical expression of Egr1 and Egr1-mediated apoptosis. In cells with WT p53, elevated ARF also inhibits Mdm2 activity, leading to p53 protein stabilization and p53-induced apoptosis. In cells that lack functioning ARF, but have a WT p53, c-Myc induces p53-dependent apoptosis through less defined mechanisms (7, 31). Considering that p53 expression is lost in half of all human tumors, this unique c-Myc-ARF-Egr1 apoptosis pathway has the potential to be exploited for future therapeutic agents as an alternative to p53 reactivation.
Fig. 6.
Mechanisms mediating c-Myc–induced apoptosis. Upper: During normal physiological low-ARF conditions c-Myc drives expression of canonical target genes, leading to proliferation. Further, without ARF expression, Mdm2 mediates degradation of p53, thereby preventing a block in cell cycle progression. Lower: Expression of ARF is induced by oncogenic stress such as deregulated c-Myc. (A) ARF then directly interacts with c-Myc and inhibits canonical target gene expression necessary for proliferation. (B) Conversely, ARF is necessary for c-Myc to induce the noncanonical target Egr1, which then mediates c-Myc–induced apoptosis independently of p53. (C) ARF induced by oncogenic c-Myc also binds to and inhibits Mdm2 activity, leading to p53 stabilization and p53-dependent apoptosis. (D) c-Myc–induced apoptosis can also occur independently of ARF through alternate p53-dependent mechanisms. Failure to trigger ARF or p53 pathways by oncogenic c-Myc leads to unregulated proliferation and tumorigenesis.
Materials and Methods
Cell Culture, Transfection, and Retroviral Infection.
Cos-7, p53−/− MEFs, DKO MEFs, and ARF−/− MEFs were cultured in DMEM with 10% calf serum (CS). H016, TGR, Egr1−/−, and WT MEFs were maintained in DMEM with 10% FBS. All cells were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol with the indicated plasmids. Cells were subjected to analysis approximately 48 h after transfection. The p53−/−, DKO, ARF−/−, H016 MycER, and vector cells were generated using the retroviral expression vector pBabehygro-MycER and the Egr1 WT and Egr1−/− MEFs were generated using pBabepuro-MycER as described previously (9).
RNA Interference.
p53−/−, ARF−/−, DKO, and WT MEFs expressing MycER were seeded at 2 to 4.5 × 106 cells per 10-cm dish and treated with a final concentration of 100 nM of Egr1 SMARTpool, CDKN2A (ARF) SMARTpool, or control Non-Targeting Pool no. 1 siRNA purchased from Dharmacon using Dharmafect Reagent 4 according to the manufacturer's instructions. Approximately 24 h later, cells were trypsinized and seeded.
Immunoblot Analysis.
Cell lysates were prepared in antibody buffer (20 mM Tris, pH 7.4, 50 mM NaCl, 0.5% Triton X-100, 0.5% deoxycholic acid (DOC), 0.5% SDS, 1 mM EDTA, 0.1 M PMSF, 10 mg/mL aprotinin, 2 mg/mL leupeptin). The proteins were resolved by 10 or 15% SDS/PAGE, subjected to immunoblot analysis using anti-Mycfl (Millipore), anti-Egr1 (C-19; Santa Cruz Biotechnology), anti-ARF (Millipore), and monoclonal anti–β-actin (Sigma-Aldrich) using enhanced chemiluminescence for detection.
Quantitative Real-Time RT-PCR.
MEFs expressing c-MycER were harvested at the indicated times following treatment with 2 μM hydroxytamoxifen (OHT). Lysates, RNA, and cDNA were prepared, real-time PCR was performed, and results were calculated as detailed previously (32). For mRNA level analyses in HO16 and TGR cells, the cells were shifted to media containing 0.1% FBS for 48 h before stimulation with 20% FBS. Cells were harvested at the indicated times and the mRNA levels were determined as described earlier. All real-time RT-PCR analyses were performed in triplicate with the primers listed in SI Materials and Methods and the results are reported as mean ± SD relative to actin levels.
ChIP.
Sixteen hours after p53−/− MycER and DKO MycER MEFs were plated at 6 × 106 cells per 150-mm dish, they were treated with 2 μM OHT or ethanol for 6 h. Cross-linking and ChIP was performed as detailed by Farnham et al. (http://www.genomecenter.ucdavis.edu/farnham/protocol.html) with anti–c-Myc (N-262x; Santa Cruz Biotechnology), anti-ARF (CDKN2A; GeneTex), and normal rabbit IgG (Upstate). Purified DNA was subjected to PCR amplification by using specific primer sets listed in SI Materials and Methods. PCR products were subjected to analysis on a 1.5% agarose gel. Quantitative ChIP analyses were performed as described earlier except the purified DNA was subjected to real-time PCR using the primers listed in SI Materials and Methods. A standard curve was used to calculate the relative starting quantity of each sample. The percent of input was calculated by dividing the relative starting quantity from each immunoprecipitation (Myc, ARF, IgG) by the relative starting quantity of the input and then by multiplying by 100. The results are reported as the mean ± SD from triplicate samples.
Luciferase Assays.
p53−/− MEFs and DKO MEFs were transfected with 2 μg of Myc expression vector or empty vector, 1.9 μg of reporter plasmid, and 0.1 μg of pRL-TK internal control. Luciferase assays were carried out 48 h after transfection according to the manufacturer's instructions (dual-luciferase reporter assay system; Promega). Results were normalized for expression of pRL-TK and are reported as the mean ± SD from triplicate samples. For the assays in HO16, TGR, and Rat1a cells, 3.9 μg of reporter plasmid and 0.1 μg of pRL-TK or pRL-SV40 were transfected into the cells. Luciferase assays were performed, calculated, and reported as described earlier.
Apoptosis Assays.
Two days after plating Egr1−/− MycER and WT MycER MEFs at 1 × 105 cells per well in six-well dishes, the cells were shifted into media containing 0.5% FBS with or without 2 μM OHT (added daily). The numbers of floating (apoptotic) and attached (living) cells were determined in triplicate at the indicated times with a hemacytometer. Results are reported as a ratio of dead to living cells over time. For apoptosis assays performed on RNAi-treated cells, p53−/− MycER, ARF−/− MycER, DKO-MycER, and WT-MycER MEFs were treated with either siGENOME SMARTpool targeting Egr1 or siGENOME control SMARTpool no. 1 (Dharmacon). After 24 h the cells were seeded at 2 × 105 cells per well in six-well dishes in media containing 2% CS with or without 2 μM OHT (added daily). The numbers of living and dead cells were determined as described earlier in triplicate in at least three different experiments with two different polyclonal cell lines and reported as the number of dead cells divided by number of total cells multiplied by 100 (i.e., percent dead). Apoptosis was confirmed with an activated caspase-3 colorimetric assay (Sigma-Aldrich) performed according to manufacturer instructions.
Supplementary Material
Acknowledgments
We thank G. Zambetti for DKO MEFs, E. Ruley and C. Eischen for p53−/− MEFs, D. Mercola and E. Adamson for Egr1−/− and WT MEFs, C. Sherr for ARF−/− MEFs, J. Sedivy for HO16 and TGR cells, B. Christy for Egr1-CAT, R. Eisenman for 4XEMS-Luc, and J. Svaren for pCB6-HA-Egr1. We also thank M. Gregory and M. Eilers for critical review of the manuscript. This work was supported by National Institutes of Health Grants R01 CA109586, R01 CA125760, and P50 CA095103 (to S.R.H.) and by Ruth L. Kirschstein National Research Service Award Training Grant CA009385-25 (to D.N.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008848108/-/DCSupplemental.
References
- 1.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 2.Cohen DE, Prochownik EV. A functional hierarchy for c-Myc target genes? Lessons from MT-MC1. Cell Cycle. 2006;5:392–393. doi: 10.4161/cc.5.4.2484. [DOI] [PubMed] [Google Scholar]
- 3.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 4.Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol Cell Biol. 2001;21:7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomerantz J, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 7.Zindy F, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta A, et al. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J Biol Chem. 2004;279:36698–36707. doi: 10.1074/jbc.M312305200. [DOI] [PubMed] [Google Scholar]
- 9.Qi Y, et al. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- 10.Gregory MA, Qi Y, Hann SR. The ARF tumor suppressor: Keeping Myc on a leash. Cell Cycle. 2005;4:249–252. [PubMed] [Google Scholar]
- 11.Boyle KB, et al. The transcription factors Egr1 and Egr2 have opposing influences on adipocyte differentiation. Cell Death Differ. 2009;16:782–789. doi: 10.1038/cdd.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins S, et al. Opposing regulation of T cell function by Egr-1/NAB2 and Egr-2/Egr-3. Eur J Immunol. 2008;38:528–536. doi: 10.1002/eji.200737157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virolle T, et al. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat Cell Biol. 2001;3:1124–1128. doi: 10.1038/ncb1201-1124. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Baron V, Mercola D, Mustelin T, Adamson ED. A network of p73, p53 and Egr1 is required for efficient apoptosis in tumor cells. Cell Death Differ. 2007;14:436–446. doi: 10.1038/sj.cdd.4402029. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, et al. Concurrent down-regulation of Egr-1 and gelsolin in the majority of human breast cancer cells. Cancer Genomics Proteomics. 2007;4:377–385. [PubMed] [Google Scholar]
- 16.Joslin JM, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krones-Herzig A, et al. Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer Res. 2005;65:5133–5143. doi: 10.1158/0008-5472.CAN-04-3742. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed MM, et al. EGR-1 induction is required for maximal radiosensitivity in A375-C6 melanoma cells. J Biol Chem. 1996;271:29231–29237. doi: 10.1074/jbc.271.46.29231. [DOI] [PubMed] [Google Scholar]
- 19.Krones-Herzig A, Adamson E, Mercola D. Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence. Proc Natl Acad Sci USA. 2003;100:3233–3238. doi: 10.1073/pnas.2628034100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muthukkumar S, et al. Role of EGR-1 in thapsigargin-inducible apoptosis in the melanoma cell line A375-C6. Mol Cell Biol. 1995;15:6262–6272. doi: 10.1128/mcb.15.11.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zagurovskaya M, et al. EGR-1 forms a complex with YAP-1 and upregulates Bax expression in irradiated prostate carcinoma cells. Oncogene. 2009;28:1121–1131. doi: 10.1038/onc.2008.461. [DOI] [PubMed] [Google Scholar]
- 22.Das A, et al. Ionizing radiation down-regulates p53 protein in primary Egr-1-/- mouse embryonic fibroblast cells causing enhanced resistance to apoptosis. J Biol Chem. 2001;276:3279–3286. doi: 10.1074/jbc.M008454200. [DOI] [PubMed] [Google Scholar]
- 23.Knoepfler PS, et al. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemaire P, Revelant O, Bravo R, Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci USA. 1988;85:4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christy B, Nathans D. Functional serum response elements upstream of the growth factor-inducible gene zif268. Mol Cell Biol. 1989;9:4889–4895. doi: 10.1128/mcb.9.11.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan J, et al. Time-dependent c-Myc transactomes mapped by Array-based nuclear run-on reveal transcriptional modules in human B cells. PLoS ONE. 2010;5:e9691. doi: 10.1371/journal.pone.0009691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Packham G, Cleveland JL. Ornithine decarboxylase is a mediator of c-Myc-induced apoptosis. Mol Cell Biol. 1994;14:5741–5747. doi: 10.1128/mcb.14.9.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin X, Grove L, Rogulski K, Prochownik EV. Myc target in myeloid cells-1, a novel c-Myc target, recapitulates multiple c-Myc phenotypes. J Biol Chem. 2002;277:19998–20010. doi: 10.1074/jbc.M200860200. [DOI] [PubMed] [Google Scholar]
- 30.Zeller KI, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pusapati RV, et al. ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc Natl Acad Sci USA. 2006;103:1446–1451. doi: 10.1073/pnas.0507367103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Boone D, Hann SR. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad Sci USA. 2008;105:18794–18799. doi: 10.1073/pnas.0806879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.