Abstract
Synthetic lethality is a powerful approach to study selective cell killing based on genotype. We show that loss of Rad52 function is synthetically lethal with breast cancer 2, early onset (BRCA2) deficiency, whereas there was no impact on cell growth and viability in BRCA2-complemented cells. The frequency of both spontaneous and double-strand break-induced homologous recombination and ionizing radiation-induced Rad51 foci decreased by 2–10 times when Rad52 was depleted in BRCA2-deficient cells, with little to no effect in BRCA2-complemented cells. The absence of both Rad52 and BRCA2 resulted in extensive chromosome aberrations, especially chromatid-type aberrations. Ionizing radiation-induced and S phase-associated Rad52-Rad51 foci form equally well in the presence or absence of BRCA2, indicating that Rad52 can respond to DNA double-strand breaks and replication stalling independently of BRCA2. Rad52 thus is an independent and alternative repair pathway of homologous recombination and a target for therapy in BRCA2-deficient cells.
Keywords: DNA repair, genetic instability, chromosomal aberrations
DNA double-strand breaks (DSBs) are potentially lethal DNA lesions which may arise spontaneously during DNA replication or result from exposure to ionizing radiation or other DNA-damaging agents (1). To repair DSBs, eukaryotes have developed two DSB repair pathways: nonhomologous end joining and homologous recombination (HR) (2). HR is required for the repair of complex double-strand lesions such as crosslinks or one-ended DSBs that occur with a cleaved replication fork; nonhomologous end joining appears to have little role in the repair of these lesions. The absence of Rad51 in proliferating cells (and therefore any measurable HR) results in cell lethality. The loss of function of proteins involved in HR, such as breast cancer 2, early onset (BRCA2), will be viable only if there is a BRCA2-independent pathway for Rad51 function.
In Saccharomyces cerevisiae, the Rad52 protein plays a key role in HR (3). However, in vertebrates, knockouts of the Rad52 gene show little phenotype, with no obvious defect in HR. Rad52 knockout mice exhibit a nearly normal phenotype, and Rad52-deficient embryonic stem cells are not hypersensitive to agents that induce DSBs, either simple or complex (4, 5). In contrast, Rad51 knockout is embryonically lethal (6, 7), and depletion of Rad51 from vertebrate cells results in an accumulation of chromosome aberrations and subsequent cell death (8). These findings indicate the essential role of Rad51 in the maintenance of chromosomal DNA during the mitotic cell cycle, but the role for Rad52 in vertebrate cells is unclear.
Accumulating evidence implicates BRCA2 as an integral component of the HR machinery via the direct regulation of the assembly of Rad51 filaments and its subsequent activity in strand exchange (9–11). Biochemical studies showed that the Ustilago maydis BRCA2 ortholog, Brh2, is involved in the recruitment of Rad51 to the sites of HR; Rad51 then mediates the displacement of replication protein A (RPA) to allow the formation of the Rad51 nucleoprotein filament, the key substrate in initiating DNA strand exchange during HR (9). Now following recent papers describing its purification and biochemical analysis (12, 13), this role also has been shown for human BRCA2. In S. cerevisiae, which do not appear to have a BRCA2 homolog, Rad52 performs a role in assembling the Rad51 nucleoprotein filament similar to that of BRCA2 in mammalian cells (14–18). Furthermore, mammalian BRCA2 and yeast Rad52 share many similar activities, including interactions with Rad51 and RPA (17, 19, 20) and ssDNA-binding activity (21, 22). These observations suggest that BRCA2 and human Rad52 may provide alternative pathways for Rad51-mediated HR in mammalian cells.
Results
Rad52 Expression and Rad51 Nuclear Foci in BRCA2-Defective Cells.
Capan-1 cells, derived from a human pancreatic epithelial tumor, contain a BRCA2 6174delT mutation in one allele that encodes for a truncated form of the BRCA2 protein; the other BRCA2 allele is lost (23). EUFA423 cells are derived from a Fanconi anemia patient with complementation group D1 and have biallelic mutations (7691 insAT and 9900 insA) in BRCA2 that result in two different truncated forms of BRCA2 (24). A marked reduction in the level of Rad52 in the Capan-1 cell line was observed relative to HeLa, MCF7, and HCC1937 cells, all of which contain wild-type BRCA2 (Fig. S1A and Table S1). Full-length BRCA2 was undetectable in either Capan-1 or EUFA423 cells using an anti-BRCA2 antibody that recognizes the C terminus of the protein (Fig. S1A). Relatively little of the truncated BRCA2 proteins of EUFA423 reach the nucleus, because the nuclear localizing signal is lost (24), but the cytoplasmic levels are low also, implying that the truncated forms of BRCA2 are unstable (Fig. S1D).
The Rad51 protein often is detected in multiple discrete subnuclear structures, referred to as “nuclear foci,” which participate in the DNA repair process and represent sites of ongoing HR. The ability to form Rad51 nuclear foci correlates well with the ability to carry out HR and therefore provides a useful surrogate measure of HR (25). The number of cells containing spontaneous Rad51 foci was lower in the Capan-1 cell line (8%) than in the EUFA423 cell line (15%) (Fig. S1 B and C). In response to ionizing radiation (10 Gy, 8 h) no significant increase in Rad51 foci formation (10%) was observed in Capan-1 cells. In contrast, the EUFA423 line showed a significant increase (to 30%) in the number of cells containing Rad51 foci. The difference in Rad51 foci formation in these two cell lines previously was thought to result from residual BRCA2 function in the EUFA423 cells. However, because little BRCA2 reaches the nucleus, we surmised that the measurable difference in Rad52, rather than residual BRCA2 activity, might be the explanation.
Rad52 Stimulates Rad51 Nuclear Foci Formation in BRCA2-Defective Cells.
Because of the inability to detect endogenous Rad52 foci with currently available anti-Rad52 antibodies, MCF7 cells were transfected to express stably GFP-tagged Rad52 (Rad52-GFP), previously shown to be a functional protein (26). Stably expressing Rad52-GFP clones were exposed to 10 Gy of ionizing radiation and examined by fluorescence microscopy at various time intervals over a 24-h period. In untreated cells, 6% contained spontaneous GFP-Rad52 foci, and 25% contained Rad51 foci, reflected in the zero time-point (Fig. S2A). Without exposure to ionizing radiation, no significant changes were seen in the percentage of cells containing foci. In response to ionizing radiation, the number of cells containing Rad51 foci increased to 54% at 1 h and reached 73% at 2 h. The formation of Rad52 foci increased to 39% at 2 h and peaked at 4 h (56%). After 8 h, the number of both Rad51 and Rad52 foci decreased, suggesting that the kinetics of focus formation was the same for Rad51 and Rad52. There was little colocalization of spontaneously arising Rad51 and Rad52 foci. However, in response to ionizing radiation, the majority of Rad51 and Rad52 foci appear to colocalize (Fig. S2B).
We reasoned that Rad52 may regulate the function of Rad51 in the absence of BRCA2. The EUFA423 cells, which express normal levels of Rad52, were treated with Rad52 siRNA to reduce the levels of Rad52 (Fig. S2C), resulting in a BRCA2- and Rad52-deficient cell line (similar to Capan-1). The number of EUFA423 cells containing spontaneous Rad51 foci was 21% after transfection with control siRNA (Fig. S2D and E) and was reduced to 11% in cells transfected with Rad52 siRNA. A similar trend was observed when EUFA423 cells treated with Rad52 siRNA were exposed to ionizing radiation: The number of cells containing ionizing radiation-induced Rad51 foci was reduced from 33% to 14%. Reducing Rad52 levels in EUFA423 cells, which are BRCA2 deficient, results in a defect in Rad51 foci formation similar to that seen in Capan-1 cells. These observations further suggest that there is little significant residual BRCA2 function in EUFA423 cells, despite the truncated forms of the protein.
To validate the relationship between Rad52 and Rad51 in the absence of BRCA2, we stably expressed an HA-tagged form of Rad52 in Capan-1 cells (Fig. S2F). The percentage of Capan-1 cells containing spontaneous Rad51 foci was 16% in control cells and increased to 23% in cells expressing HA-Rad52 (Fig. S2G and H). Similarly, when Capan-1 cells transfected with HA-Rad52 were irradiated, the percentage of ionizing radiation-induced Rad51 foci increased to 35%, compared with 18% in vector control cells. Establishing stable transfectants of Capan1 cells with pDR-GFP is difficult, but episomally expressed pDR-GFP cotransfected with HA-Rad52 (or control vector) shows a 2.4-fold increase in HR with Rad52. These results support our hypothesis that Rad52 plays an important role in Rad51 function in BRCA2-deficient cell lines.
Depletion of Rad52 by shRNA Is Synthetically Lethal with BRCA2.
Because the Rad51-deficient phenotype shows severe growth impairment and chromosomal damage within a few cell cycles after inactivation (6, 7), we reasoned that loss of Rad52 in BRCA2-deficient cells may show a similar phenotype. BRCA2 alone can play an important role in cellular growth, and BRCA2-knockout cells show proliferation arrest (27). However, BRCA2-deficient tumor cells have developed ways to proliferate despite loss of function of BRCA2. We examined the proliferative properties of the BRCA2-defective cell lines that express varying levels of Rad52 and found that Capan-1 cells have a reduced proliferative rate compared with EUFA423 cells (Fig. S3A).
The reduction in Rad52 expression in EUFA423 cells resulted in a decrease in the growth rate compared with the control siRNA-treated cells (Fig. S3B). In contrast, EUFA423 cells stably expressing wild-type BRCA2, treated with either control siRNA or Rad52 siRNA, showed little difference in their cellular proliferation. Cellular proliferation studies also were carried out in Capan-1 cells complemented with Rad52, which showed improved growth rates (Fig. S3C). In the Capan-1 cell line complemented with BRCA2, cell proliferation was not significantly affected by the correction of Rad52 expression. Conversely, depleting residual Rad52 in Capan-1 cells resulted in a further reduction in cell growth.
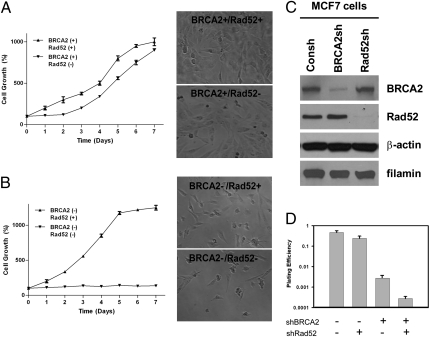
To understand the longer-term role of Rad52 inactivation in BRCA2-deficient cells, we transfected EUFA423 cells, with or without wild-type BRCA2 complementation, with a plasmid expressing a shRNA that targets Rad52. In cells with wild-type BRCA2, Rad52 depletion has no significant effect (Fig. 1A). In marked contrast, Rad52 depletion in BRCA2-deficient cells resulted in complete inactivation of cellular proliferation (Fig. 1B) with abortive attempts at cell division. We surmise that in cells that have adapted to life without BRCA2, Rad52 maintains the residual capacity for cell proliferation by maintaining a sufficient residual amount of Rad51 function.
Fig. 1.
Rad52 shows synthetic lethality with BRCA2. (A) Cellular proliferation in EUFA423-BRCA2 complemented cells transfected with sh-Rad52 or control shRNA. (Left) Cell number measuring started 5 d after transfection and 3 d after selection in hygromycin. Cells were counted on a daily basis for 7 d. (Right) Photomicrograph shows the cells in culture at day 4. (B) (Left) EUFA423 cell lines, transfected with either sh-Rad52 or control shRNA as in A. (Right) Photomicrograph at day 4. For Rad52-depleted cells, little cell growth and abortive cell divisions are seen. In A and B the data are the mean ± SE from three independent experiments. (C) MCF7 cells were transfected with shBRCA2 or shRad52 or control construct. Cell lysates of each sample were made from puromycin- or hygromycin-resistant colonies and were immunoblotted as shown. Greater than 90% depletion of the target protein was observed, without a significant effect on the nontarget protein. β-actin and Filamin were used as loading controls. (D) Plating efficiency (clonogenic survival) of MCF7 cells following relatively acute depletion of BRCA2, Rad52, or both proteins. Cells were seeded using in the appropriate selection medium to assure stable suppression of the target protein. The results shown are from three independent experiments; error bars indicate SEM.
To support these conclusions in a cell that was not already BRCA2 deficient, we depleted MCF7 cells of Rad52, BRCA2, neither, or both using shRNA (Fig. 1C). The effect of depleting BRCA2 has no effect on the measured level of Rad52, and vice versa. Colony survival after double depletion of BRCA2 and Rad52 was 0.00028 (Fig. 1D), reflecting the synthetically lethal relationship between the two proteins. The relatively acute depletion of BRCA2 also results in reduced plating efficiency (0.0032), a relative plating efficiency of 0.58%, and a slow growth rate in cell culture. However, acute BRCA2 depletion is not thought to be representative of what occurs in tumor development. MCF7 cells with control transfections have a plating efficiency of 0.55.
Rad52 Function Is BRCA2-Independent.
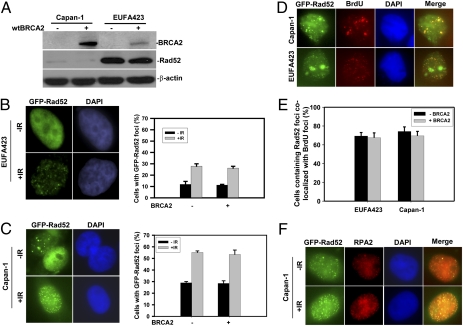
Mammalian Rad51 and Rad52 interact and together produce the most efficient activity for in vitro DNA recombination reactions (14, 22). In addition, we have shown that Rad51 and GFP-Rad52 colocalize in response to DNA damage (Fig. S2B), providing in vivo evidence that these proteins function together in the repair of DNA damage. To delineate the role of Rad52 and its relationship to BRCA2 in supporting Rad51-dependent HR, we used the two pairs of isogenic cell lines differing in their BRCA2 status. We initially determined that transfection with either pcDNA3 (empty vector) or pcDNA3-BRCA2 had no effect on the expression of Rad52 in either Capan-1 cells (which have low levels of BRCA2) and EUFA423 cells (which have normal levels of BRCA2) (Fig. 2A). To determine if the formation of Rad52 foci was dependent on BRCA2, EUFA423 cells with or without BRCA2 were transiently transfected with GFP-tagged Rad52, and either spontaneous or ionizing radiation-induced (10 Gy) Rad52 foci were scored. In untreated EUFA423/pcDNA3 cells without wild-type BRCA2, GFP-Rad52 was distributed throughout the nucleus, and only 12% of the cells contained Rad52 foci (Fig. 2B). In response to ionizing radiation, the number of EUFA423/pcDNA3 cells with Rad52 foci increased to 28%. In untreated and irradiated EUFA423/BRCA2 cells, which now express wild-type BRCA2 (Fig. 2A), the number of GFP-Rad52 foci detected was 11% and 26% respectively (Fig. 2B), indicating that both spontaneous and ionizing radiation-induced Rad52 foci formation are entirely independent of BRCA2 status. To verify this assertion in the Capan-1 cell pair, we found that in untreated and irradiated Capan-1/BRCA2 cells the number of GFP-Rad52 foci detected was 28% and 53%, respectively (Fig. 2C), not significantly different from the results in the Capan-1/pcDNA3 line. Thus, Rad52 foci can form in vivo independently of BRCA2.
Fig. 2.
Rad52 expression and function is independent of BRCA2. (A) Capan-1 and EUFA423 cells were transfected with a plasmid containing wild-type BRCA2 or with an empty vector and then were analyzed by Western blotting. Nuclear extracts were probed for BRCA2 (Ab-1) and whole-cell extracts for Rad52 (5H9). (B) Ionizing radiation-induced GFP-Rad52 foci were detected in the EUFA423 cell line with or without WT-BRCA2. EUFA423 cells were transiently transfected with GFP-Rad52, treated 48 h later with 10 Gy of ionizing radiation, and incubated for 8 h. The percentage of cells with Rad52 foci is shown (mean and SE from three independent experiments).There was no significant difference in Rad52 foci with or without BRCA2 (P = 0.69 without ionizing radiation; P = 0.55 with ionizing radiation). (C) Ionizing radiation-induced GFP-Rad52 foci in Capan-1 cells with or without WT-BRCA2, as in B. There was no significant difference in Rad52 foci with or without BRCA2 (P = 0.52 without ionizing radiation; P = 0.37 with ionizing radiation). (D) Capan-1 and EUFA423 cells were transiently transfected with GFP-Rad52 and then were incubated for 2 h in the presence of BrdU, followed by immunofluorescence using an antibody to BrdU. (E) The percentage of Capan-1 and EUFA423 cells (treated as in D) containing Rad52 foci that also colocalized with BrdU foci. Bars represent SEM, based on three independent experiments, with nonsignificant differences. (F) Capan-1 cells were transiently transfected with GFP-Rad52 and 48 h later were treated with 10 Gy of ionizing radiation or sham treated and were incubated for 8 h before fixation for immunofluorescence studies using RPA2 antibody and DAPI.
More than 70% of GFP-Rad52 foci colocalized with regions of BrdU incorporation in Capan-1 and EUFA423 cells (Fig. 2 D and E). The BRCA2 status of these cells did not alter the percentage of cells in which Rad52 and BrdU colocalized. Colocalization of GFP-Rad52 and RPA2 was observed in Capan-1/GFP-RAD52 cells, either with or without ionizing radiation exposure (Fig. 2F). Taken together, the results indicate that Rad52 forms nuclear foci in S phase associated with DNA damage, suggesting that they are located with stalled replication forks, and that this localization does not depend on functional BRCA2. The independence of Rad52 from BRCA2 supports the overall concept that the two proteins function in independent pathways of Rad51-dependent HR.
Rad52 Depletion Impairs HR in BRCA2-Defective Cell Lines.
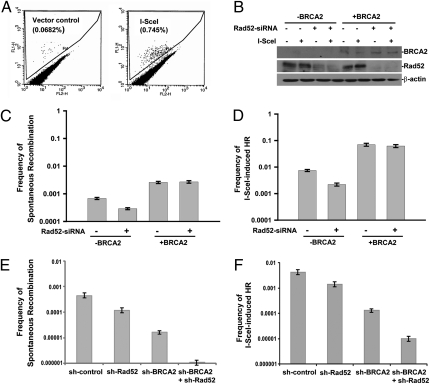
The central role of Rad51 in HR is well documented, but the relative roles of Rad52 and BRCA2 in Rad51-dependent HR are not well delineated in mammalian cells. To address this issue, we depleted Rad52 in a BRCA2-proficient or -deficient background and measured Rad51-dependent HR using the pDR-GFP recombination reporter (28). Two independently isolated clones of EUFA423/pDR-GFP were used to measure HR. Prescreening showed that HR could be detected in these clones by flow cytometry (Fig. 3A). The EUFA423/pDR-GFP clones were transiently transfected with Rad52 siRNA alone, HA-BRCA2 alone, or Rad52 siRNA and HA-BRCA2. Twenty-four hours later, the cells were transfected with either pCMV-I-SceI-3xNLS expression vector (to induce DSBs) or a control plasmid. The period of protein depletion thus matched the period of measuring the cumulative frequency of HR. The frequencies of I-SceI–induced homology-mediated repair and spontaneous recombination were reduced by more than twofold in BRCA2-defective cells in which Rad52 expression levels were reduced (Fig. 3 B–D). In BRCA2-corrected EUFA423/pDR-GFP cells, however, knockdown of Rad52 had no detectable effect on HR (Fig. 3 C and D).
Fig. 3.
Requirement for Rad52 in Rad51-mediated HR in BRCA2-deficient cells. (A) EUFA423 cells containing the recombination substrate, pDR-GFP, were transiently transfected with the I-SceI restriction endonuclease plasmid or with vector alone and 48 h later were analyzed for the percentage of green fluorescent cells. (B) EUFA423/pDR-GFP cells were transiently cotransfected with a combination of BRCA2 plasmid and Rad52 siRNA as indicated and were incubated for 24 h. The cells then were transiently transfected with the I-SceI restriction endonuclease plasmid or with vector alone. After an additional 48 h, the cells were harvested for Western blotting using BRCA2 (Ab-2) and Rad52 (5H9). (C and D) The frequency of (C) spontaneous recombinations and (D) I-SceI break-induced recombination (HR) were observed by flow cytometry. The bars represent the results of three or more independent experiments; error bars show SEM. Differences between control siRNA and Rad52 siRNA in BRCA2-deficient cells are significant: P = 0.0019 for spontaneous recombination, and P = 0.0025 HR, using an unpaired t test on logarithmic transformed data. (E and F) Frequency of (E) spontaneous recombinations and (F) I-SceI break-induced HR of MCF7 cells depleted of Rad52 or BRCA2, neither, or both using shRNA. The mean and SEM of three independent experiments is shown. The reduction in spontaneous recombination and HR for Rad52 depletion in BRCA2-deficient cells is highly significant: P < 0.001.
To support this observation, we used MCF7/pDR-GFP cells, which have been used previously to measure HR, and additionally transfected them with shRNA to Rad52, BRCA2, neither, or both to establish pooled colonies of each functional genotype. The effect of depleting Rad52 in a wild-type background was small (Fig. 3 E and F), whereas the effect in a BRCA2-deficient background was an ∼20-fold reduction, which was highly significant (P < 0.001). The effect was observed for both spontaneous recombination and I-SceI–induced recombination. In addition, ionizing radiation-induced Rad51 foci showed a significant effect of depleting Rad52 only in a BRCA2-deficient background (Fig. S4). Taken together, these data suggest that Rad52 can play an important role in Rad51-dependent HR in mammalian cells when BRCA2 is inactive.
Chromosomal Instability in BRCA2-Rad52–Deficient Cells.
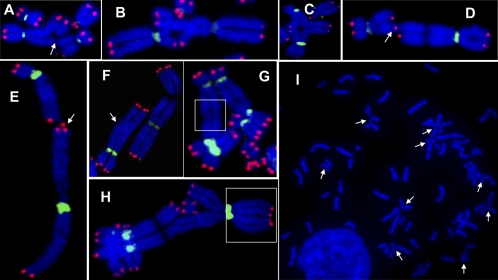
Because Rad52 has a function in both spontaneous recombination and homology-mediated recombinational repair that is independent of BRCA2, we determined whether the roles of BRCA2 and Rad52 were the same in relation to chromosomal damage. The frequency of spontaneous chromosome and chromatid aberrations was determined using FISH to view chromosomal structure in BRCA2-defective cells with Rad52 depleted by siRNA. A comparison between EUFA423 cells (BRCA2 defective) and EUFA423 cells treated with Rad52 siRNA (Table 1 and Fig. 4 A–E) revealed that the BRCA2-Rad52–deficient cells exhibited the highest frequency of spontaneous and X-ray–induced chromatid and chromosomal breaks, particularly affecting triradial and quadriradial structural aberrations. In addition, specifically in the BRCA2-Rad52–defective cells, “meiotic prophase-like” paired chiasmata and “meiotic synapsis structures” were found (Fig. 4 F–H). Paired chiasmata represent a strand exchange between nonsister chromatids (Fig. 4F); this type of chromosomal aberration is exceedingly rare in somatic cells. Cells also were irradiated (2 Gy) 72 h after siRNA treatment, and chromosomal aberrations induced by ionizing radiation were measured 24 h later. As with spontaneous aberrations, the double-defective BRCA2-Rad52 cells exhibited greater levels of ionizing radiation-induced chromatid-type aberrations than did the BRCA2 single mutant (Table 1 and Fig. 4I). These results further indicate the involvement of Rad52 in repairing ionizing radiation-induced DSBs by HR in the absence of BRCA2.
Table 1.
Spontaneous and radiation-induced chromosome aberrations in BRCA2- and Rad52-deficient cells
| Functional state of BRCA2/Rad52 in EUFA423 cells | Chromatid breaks | Chromosome break | Telomere end associations | Dicentrics | Radials | Diplochromosomes |
| +/+ | 0.07/0.42 | 0.01/0.09 | 0.07/0.18 | 0.01/0.23 | 0/0.04 | 0.005/0 |
| +/− | 0.19/0.51 | 0.03/0.13 | 0.12/0.42 | 0.02/0.30 | 0/0.11 | 0.01/0 |
| −/+ | 0.24/0.51 | 0.05/0.16 | 0.15/0.46 | 0.06/0.40 | 0.17/0.10 | 0.01/0 |
| −/− | 0.31/2.02 | 0.12/0.36 | 0.20/0.40 | 0.10/0.50 | 0.18/0.66 | 0.15/0 |
| Statistics: BRCA2–/+ vs. −Rad52 | P = 0.38/<0.0001 | P = 0.126/0.002 | NS | NS | NS P = <0.0001 | NS |
Numeric values are the average number of aberrations per cell. Spontaneous aberrations are derived from the analysis of 200 metaphase spreads and are shown as the first result for each cell type. One hundred metaphase spreads were analyzed in cells exposed to 2 Gy of ionizing radiation, shown as the second result for each cell type. Statistical comparisons of Poisson distribution metrics are shown for BRCA2-deficient cells, comparing cells with Rad52 to cells in which Rad52 is depleted (row 3 versus row 4). NS, nonsignificant.
Fig. 4.
Severe chromosomal fragility in BRCA2- and Rad52-deficient cells. EUFA423 cells were transiently transfected with Rad52 siRNA and incubated for 72 h. Then metaphase chromosome spreads were prepared for FISH analysis. Centromere-specific PNA probes were labeled with fluorescein isothiocyanate (green), and telomere-specific PNA probes were labeled with the fluorochrome Cy3 (red). Representative images are as follows: (A) Chromatid break (arrow); (B) triradial and (C) quadriradial structures; (D) telomere fusions (arrow); (E) Chromosome end association (arrow); (F) mitotic crossover between nonsister chromatids (arrows); (G) mitotic chiasma (in box); and (H) meiotic synapsis between nonsister chromatids (in box). (I) For ionizing radiation-induced chromosome aberrations, EUFA423 cells were treated initially with Rad52 siRNA, then were irradiated with 2 Gy, and were incubated for a further 24 h. Metaphase chromosome spreads were prepared and stained with DAPI. Chromosome aberrations (chromatid breaks, chromosome breaks, triradials, and quadriradials) are shown by the arrows.
Discussion
In this paper we have shown that human Rad52 supports Rad51-dependent HR, both in replicating cells and in response to double-strand breaks, specifically in the absence of BRCA2. Multiple lines of evidence, including Rad51 foci, cellular proliferation, spontaneous and DSB-induced direct-repeat gene conversion, and chromosomal aberrations all support the notion that Rad52 plays a role in mediating HR in BRCA2-defective cells. More importantly, inactivation of Rad52 in BRCA2-deficient cells results in synthetic lethality, which makes Rad52 a tumor-specific target for therapy in BRCA2-deficient tumors. Both cell types die as a result of chromosomal aberrations that prevent a viable completion of cell division (i.e., postmitotic death).
Rad51-Dependent HR by Rad52 in BRCA2-Deficient Cells.
Yeast Rad52 is required for single-strand annealing (SSA) and Rad51-dependent strand-exchange events (29, 30). In contrast, mammalian Rad52 has been shown to affect homologous recombinational repair by SSA but not Rad51-dependent HR (31). Mouse cells deficient in RAD52 have been shown to exhibit only a mild gene-targeting defect (6, 7) in contrast to other proteins known to play a role in HR. Our results suggest that in the presence of wild-type BRCA2, Rad52 has a small effect on the frequency of HR, whereas in the absence of BRCA2, silencing Rad52 results in a significant impairment in both replication-associated HR and homology-mediated repair triggered by I-SceI breaks. Depletion of Rad52 in BRCA2-deficient cells also resulted in a decrease of ionizing radiation-induced formation of Rad51 foci, further supporting the role of Rad52 in Rad51-dependent HR in the absence of BRCA2.
Synthetic Lethality of Rad52 and BRCA2.
Because Rad52 is required for cellular proliferation in BRCA2-defective cells, these observations suggest that silencing of Rad52 could cause BRCA2-defective tumor cells, but not normal cells, to stop proliferating and to be sensitized to a variety of genotoxic agents. Therefore, inactivation of Rad52 could be an attractive strategy for improving cancer therapy in the BRCA-defective subgroup of cancers. The Rad52 gene is not documented to be mutated or inactivated in human tumors, making it a viable potential target. Other synthetically lethal relationships have been reported for Rad52 with X-ray repair complementing defective repair in Chinese hamster cells 3 (XRCC3) (32) and for BRCA2 with poly (ADP-ribose) polymerase family, member 1 (PARP1) (33).
Rad52 Function Is BRCA2-Independent.
A key finding suggesting that there are two independent pathways of Rad51-dependent repair (Fig. S5) was the observation that Rad52 foci can be formed independently of BRCA2. Complementing BRCA2-deficient cells with BRCA2 had no impact on the ability to form Rad52-Rad51 foci. Yeast cells, which have no identifiable BRCA2, are defective in recombination when Rad52 null and display increased sensitivity to DNA damage (3). In contrast, Rad52-null mice are viable with no major phenotype, and inactivation of Rad52 in chicken B cells (DT40) leads to only a slight reduction in targeted chromosomal integration, without a detectable increase in radiation sensitivity (4, 5). Biochemical studies have demonstrated that yeast and human Rad52 have both similar and different activities. Rad52 can stimulate DNA-strand exchange by the Rad51 protein (14, 22), although RPA was used in the strand-exchange reactions in only one of the reports (14). Recent papers comparing the role of Rad52 and purified BRCA2 in biochemical reactions have shown that BRCA2 can displace RPA efficiently and promote Rad51 filament formation and strand exchange, whereas both yeast and human Rad52 carry out this reaction less efficiently (12). Both proteins can interact with Rad51 and RPA, interactions that are necessary for efficient HR (20, 30). However, the ability of human Rad52 to displace RPA from ssDNA is limited, suggesting there may be differences between yeast and human Rad52 (34, 35). Rad52 may facilitate second-end capture, a function similar to SSA, which would cooperate with BRCA2-Rad51 function in stimulating strand exchange. However, the in vivo results imply that Rad52 can stimulate Rad51 function in the absence of BRCA2.
Immunofluorescence cytology experiments in vertebrate cells demonstrate a coordinated response of Rad52 and Rad51 to DNA damage (26, 36). BRCA2 and Rad52 also share a number of conserved functional domains, such as Rad5- binding domains, an RPA-interaction domain, and a DNA-binding domain. The apparent paradox between the biochemical role of Rad52 and its apparent dispensability in mammalian cells can be explained by the observations reported in this paper that Rad52-Rad51–dependent repair can occur independently of BRCA2. In U. maydis, a recent report suggested that the BRCA2 homolog was dependent on Rad52 for its toxicity to UV light (37).
We have demonstrated that spontaneous Rad52 foci form predominantly in S phase and that knockdown of Rad52 in BRCA2-defective cells results in a significant decrease in the spontaneous HR frequency, suggesting that Rad52 supports recombinational repair driven by arrested or collapsed replication forks. A recent report also suggested that Rad52 responds predominantly to replication stress, with reduced Rad52 mobility (measured by fluorescence recovery after photobleaching) after treatment with hydroxyurea (38). In contrast, a BRCA2-interacting protein, BRCA2 and cyclin-dependent kinase inhibitor 1A (CDKN1A)-interacting protein (BCCIP) was not affected significantly, suggesting that Rad52 is specific to S-phase events and is separate from the BRCA2-containing protein complex. The original report of reduced Rad51 foci observed in BRCA2-deficient cells also observed that the residual foci were formed in S phase (39). Thus, Rad52 provides an alternative recombinational repair pathway for mammalian cells, particularly during S phase, which helps keep the cells proliferating and avoids the major chromosomal abnormalities that can develop in the absence of BRCA2.
Rad52 Is a Potential Target for Tumor Therapy.
Current evidence suggests that both the BRCA1 and BRCA2 tumor suppressor genes are essential for the efficient repair of DSBs and particularly for avoiding replication- or postreplication-associated damage to chromosome structure (27, 40). The major chromosomal defect found in Rad52/BRCA2-defective cells further supports the notion that Rad52 provides an important alternate pathway for repairing replication-associated damage by HR in the absence of BRCA2. However, the comparison of BRCA2-defective cells with or without Rad52 also suggests that Rad52 permits the formation of potentially oncogenic chromosomal changes in BRCA2-defective cells, because the cells are less viable in the absence of Rad52.
In summary, our data support the view that there are two pathways of Rad51-dependent HR in mammalian cells: the Rad52 pathway as found in yeast cells, and the BRCA2 pathway found in most vertebrates but also in many plant species. Why these two pathways evolved and what type of DNA lesion they show a preference for is an interesting and open question. The potentially toxic effects of inactivating Rad52 in BRCA2-defective tumors could provide a treatment strategy in this subgroup of tumors, with Rad52 being synthetically lethal with BRCA2.
Materials and Methods
Additional details are available in SI Materials and Methods.
Cells, Plasmids, and Transfections.
MCF7, EUFA423, and HeLa cell lines were maintained in DMEM. Capan-1 and HCC1937 cell lines were cultured in Iscove's modified Dulbecco's medium (Sigma).
Rad52 and BRCA2 Knockdown.
Commercially available siRNA and shRNA were used according to the manufacturer's recommendations.
Microscopy and Immunofluorescence.
Cells were seeded into four-chamber tissue-culture slides (Fisher) and incubated overnight. Cells were stained by fixing in 10% formaldehyde solution at room temperature for 30 min, followed by 10-min permeabilization in PBS containing 0.5% Triton X-100. Commercially available antibodies were used for staining as previously described (41).
Immunoblotting.
For Western blot analysis, whole-cell extracts or nuclear fractions were used as described previously (41).
Measurement of Homologous Recombination.
The plasmid pDR-GFP was stably integrated into the EUFA423 cell line to establish clones as described previously (28). EUFA423-pDR-GFP cells at 80% confluence in a 75-cm2 flask were transfected with 2 μg of HA-BRCA2 plasmid and 5 μL of 20 μM Rad52 siRNA using a Nucleofector (Amaxa) according to the manufacturer's instructions. Twenty-four hours later, the cells were transfected with the pCMV-I-SceI-3xNLS expression vector or a control vector by Lipofectamine 2000. After an additional 48 h, the cells were harvested for the measurement of HR frequency by flow cytometry (FACScan; Becton Dickinson) as previously described (25).
Cell Number and Plating Efficiency.
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to measure cell number. Plating efficiency was measured in MCF7 cells by seeding 1 × and 5 × 102, 103, and 104 cells into 6-cm dishes. Colonies (>50 cells) were counted by fixing and staining with crystal violet.
Chromosomal Aberrations.
Metaphase chromosome spreads were prepared from exponentially growing cells after treatment with colcemid (0.1 μg/mL) for 6 h using standard procedures (27). Centromere and telomere probe combinations were used to identify associations between two human chromosomes. Centromere-specific peptide nucleic acid (PNA) probes were labeled with fluorescein isothiocyanate (green), and telomere-specific PNA probes were labeled with the fluorochrome Cy3 (red) (42).
Supplementary Material
Acknowledgments
This work was supported by US Public Health Service Grants CA107640 (to S.N.P.) and CA123232 (to T.K.P.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 441.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010959107/-/DCSupplemental.
References
- 1.Iliakis G, et al. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet Genome Res. 2004;104:14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- 2.Haber JE. DNA recombination: The replication connection. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 3.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaguchi-Iwai Y, et al. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rijkers T, et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuzuki T, et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonoda E, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 10.Xia F, et al. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci USA. 2001;98:8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 15.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 16.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 17.Sung P. Mediating repair. Nat Struct Mol Biol. 2005;12:213–214. doi: 10.1038/nsmb0305-213. [DOI] [PubMed] [Google Scholar]
- 18.Hays SL, Firmenich AA, Massey P, Banerjee R, Berg P. Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol Cell Biol. 1998;18:4400–4406. doi: 10.1128/mcb.18.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong JM, Ionescu D, Ingles CJ. Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. Oncogene. 2003;22:28–33. doi: 10.1038/sj.onc.1206071. [DOI] [PubMed] [Google Scholar]
- 20.Park MS, Ludwig DL, Stigger E, Lee SH. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 21.Reddy G, Golub EI, Radding CM. Human Rad52 protein promotes single-strand DNA annealing followed by branch migration. Mutat Res. 1997;377:53–59. doi: 10.1016/s0027-5107(97)00057-2. [DOI] [PubMed] [Google Scholar]
- 22.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 23.Goggins M, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 24.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitao H, Yuan ZM. Regulation of ionizing radiation-induced Rad52 nuclear foci formation by c-Abl-mediated phosphorylation. J Biol Chem. 2002;277:48944–48948. doi: 10.1074/jbc.M208151200. [DOI] [PubMed] [Google Scholar]
- 27.Patel KJ, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 28.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 31.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimori A, et al. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 2001;20:5513–5520. doi: 10.1093/emboj/20.19.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 34.Nimonkar AV, Sica RA, Kowalczykowski SC. Rad52 promotes second-end DNA capture in double-stranded break repair to form complement-stabilized joint molecules. Proc Natl Acad Sci USA. 2009;106:3077–3082. doi: 10.1073/pnas.0813247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Maizels N. Coordinated response of mammalian Rad51 and Rad52 to DNA damage. EMBO Rep. 2000;1:85–90. doi: 10.1093/embo-reports/kvd002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojic M, Mao N, Zhou Q, Lisby M, Holloman WK. Compensatory role for Rad52 during recombinational repair in Ustilago maydis. Mol Microbiol. 2008;67:1156–1168. doi: 10.1111/j.1365-2958.2008.06116.x. [DOI] [PubMed] [Google Scholar]
- 38.Wray J, Liu J, Nickoloff JA, Shen Z. Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Res. 2008;68:2699–2707. doi: 10.1158/0008-5472.CAN-07-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 40.Boulton SJ. Cellular functions of the BRCA tumour-suppressor proteins. Biochem Soc Trans. 2006;34:633–645. doi: 10.1042/BST0340633. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Ma Z, Treszezamsky A, Powell SN. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat Struct Mol Biol. 2005;12:902–909. doi: 10.1038/nsmb991. [DOI] [PubMed] [Google Scholar]
- 42.Pandita RK, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol. 2006;26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.