Homologous recombination (HR) is a key pathway to repair complex DNA damage, such as DNA gaps, double-strand breaks, and interstrand cross-links (1) (Fig. 1). Moreover, HR supports the recovery of stalled or broken replication forks and ensures faithful chromosome segregation during meiosis. The RAD52 gene in the budding yeast Saccharomyces cerevisiae encodes the lynchpin of HR in this organism. Hence, the genes of the HR pathway are called the RAD52 epistasis group. However, much to everybody's surprise, the RAD52 gene knockout in mice exhibits minimal to no phenotypes in recombination, repair, and meiosis (2). Instead, the breast and ovarian tumor suppressor protein BRCA2 maintains a central HR function, a protein that is missing in budding yeast. Analyzing HR in BRCA2-deficient human cells, a study reported in PNAS (3) redefines human RAD52 by showing that it plays a critical role in HR in BRCA2-deficient cells. This important result mandates a reexamination of vertebrate RAD52 and its functions, which opens perplexing mechanistic conundrums.
Fig. 1.
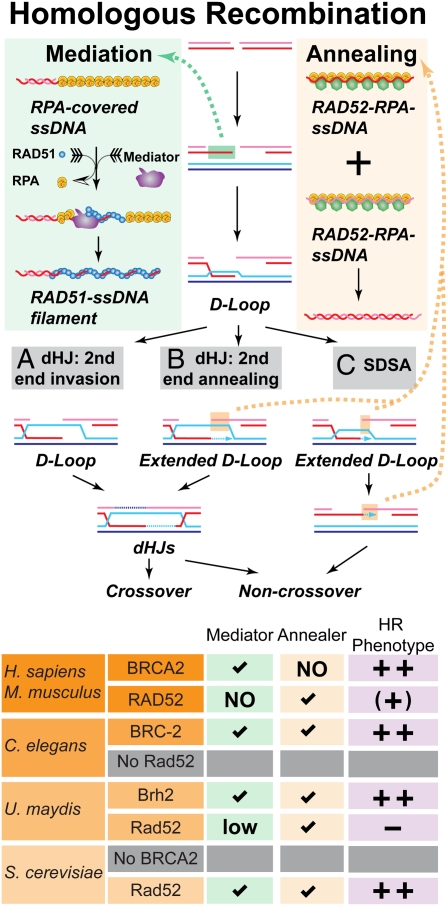
Roles of BRCA2 and RAD52 in homologous recombination. Upper: Schematic representation of the mediator and annealing functions during recombination involving (A) invasion of both ends, (B) invasion of the first end and second end capture by annealing, or (C) invasion of the first end, D-loop disruption, and annealing of the extended first end to the second end (SDSA, synthesis-dependent strand annealing). Lower: BRCA2 and RAD52 functions and phenotypes in selected organisms. Checkmark denotes presence of the biochemical activity, − denotes absence of phenotype, ++ denotes a strong and (+) a weak phenotype.
In the study (3), Feng et al. manipulate the BRCA2 and RAD52 status by mutation, complementation, or RNA-mediated knockdown in three different human cell lines expressing either normal or truncated BRCA2 proteins. They show that lower RAD52 protein levels cause decreased levels of RAD51 foci and HR in BRCA2-deficient cells. These data greatly expand on a recent observation of a negative genetic interaction between mutants in the Ustilago maydis BRCA2 and RAD52 homologs (4). As noted by Feng et al. (3), these results have interesting implications for tumor therapy and suggest a more important role of RAD52 protein in human HR than previously thought.
Analyzing the proliferation rates in their cell lines, Feng et al. (3) show that normal levels of RAD52 expression are required for proliferation in cells with no or low BRCA2 function. This identifies a vulnerability of BRCA2-deficient cells besides inhibition of poly(ADP-ribose) polymerase (5), because depletion of RAD52 in such cells causes a strong proliferation defect (synthetic lethality) associated with severe chromosomal fragility. This identifies RAD52 as a potential therapeutic target not only in BRCA2-deficient cells (3) but possibly also in BRCA2 revertants that become treatment resistant (6, 7).
Why are the results by Feng et al. (3) so interesting from a mechanistic point of view? Biochemical analysis shows that the S. cerevisiae Rad52 protein performs two critical reactions in HR: (i) it mediates the assembly of Rad51 filament on replication protein A (RPA)-coated ssDNA (8–10), and (ii) it performs the annealing step in second end capture and synthesis-dependent strand annealing (11) (Fig. 1). This dual function readily explains why rad52 mutants display even more extreme phenotypes than defects in the Rad51 protein, which performs the signature reactions of HR: homology search and DNA strand invasion (1). Surprisingly, in organisms containing a BRCA2 homolog, such as U. maydis, chicken, and mice, RAD52 inactivation causes minimal or no HR and DNA repair defects (2, 4, 12). Caenorhabditis elegans and Drosophila melanogaster seem to lack a RAD52 homolog entirely (Fig. 1).
So is RAD52 irrelevant for HR in BRCA2-containing organisms? The expectation horizon from budding yeast rad52 mutants was so high that the absence of strong phenotypes in the mouse RAD52 mutants left this gene without much interest. However, the findings by Feng et al. (3) in BRCA2-deficient cells force another look at RAD52 and its role in HR in vertebrates. Already the initial reports in RAD52-deficient mouse or chicken DT40 cells demonstrate a small but significant reduction in gene targeting (2, 12). Now Feng et al. (3) show that RAD52 depletion in BRCA2-complemented EUFA423 cells causes an increase in damage-induced chromosomal abnormalities, such as telomere end associations and radials. The reduction in the development of T-cell lymphomas in ATM-deficient mice by the RAD52 single knockout also indicates a significant in vivo function of mouse RAD52 (13). In conclusion, RAD52 mutants in vertebrates do have HR-related phenotypes, although they are much more subtle than expected from the yeast paradigm.
How does mammalian RAD52 act: as a mediator or annealer (Fig. 1)? Several genetic results favor the model proposed by Feng et al. (3) that RAD52 defines an alternative mediator pathway to BRCA2. They show that BRCA2 status does not affect RAD52–RAD51 focus formation and that restoring RAD52 expression enhances RAD51 focus formation in BRCA2-deficient cells. Although it is formally possible that RAD52 acts upstream of BRCA2, the alternative mediator model is more consistent with additional synthetic phenotypes involving RAD52. In chicken DT40 cells, RAD52 disruption is lethal with a defect in XRCC3, a RAD51 paralog that forms a complex with RAD51C required for RAD51 filament formation/stability (14). In addition in U. maydis, rad52 mutants show a synthetic enhancement of UV and IR sensitivity with a mutant in the only RAD51 paralog gene, REC2 (4). However, human RAD52 protein lacks mediator activity in reconstituted biochemical reactions (15). Possibly, RAD52 performs its mediator function in conjunction with other proteins, perhaps the RAD51 paralog complex of RAD51B/C/D-XRCC2, whose inactivation also shows a synthetic defect with an XRCC3 mutation in chicken DT40 cells (16) just like a RAD52 defect (14).
Last, the analysis of HR in humans poses another interesting conundrum: which protein is responsible for the annealing steps in mammalian HR [(b) and (c) in Fig. 1]? RAD52 is capable of performing this reaction in vitro (15) and in vivo (17), but the single mutant or RAD52 depletion lacks the expected phenotypes (IR sensitivity, strong HR phenotype) (2, 3, 17). BRCA2 mutants or depletion exhibits strong HR and DNA repair phenotypes (3, 17). However, unlike its U. maydis and C. elegans homologs (18, 19), the human BRCA2 protein is incapable of annealing RPA-coated ssDNA, although it displays strong mediator activity (15, 20). Several possible solutions can be entertained. The most radical would be that HR in vertebrates does not involve an annealing step [i.e., no synthesis-dependent strand annealing pathway; (c) in Fig. 1] but involves second end strand invasion
Lower RAD52 protein levels cause decreased levels of RAD51 foci and HR in BRCA2-deficient cells.
[(a) in Fig. 1]. The resulting double Holliday junctions (dHJs) will put a considerable burden on the dHJ dissolution pathway involving BLM-TOPO3α-RMI1-RMI2 (21). The high incidence of anaphase bridges in mammalian cells may be an indication of failure of dissolving the frequent dHJs (22). Alternatively, there might be a cofactor that endows BRCA2 with the ability to anneal RPA-coated ssDNA. Finally, an entirely novel protein might be involved. Interestingly, all five human RecQ-like proteins have been reported to anneal ssDNA, but this activity is inhibited by RPA, and the biological significance of these activities remains unclear (23–27).
In sum, human RAD52 functions to anneal RPA-coated ssDNA, and the important in vivo role of human RAD52 in BRCA2-deficient cells uncovered by Feng et al. (3) may entail RAD51 stimulation not involving classical mediation but possibly the RPA-independent effects seen in earlier biochemical studies (28, 29). The study by Feng et al. (3) forces renewed attention to RAD52 in mammalian HR, with interesting implications not only for the fundamental HR mechanism but also for anticancer therapy.
Acknowledgments
Work on homologous recombination in W.-D.H.’s laboratory is supported by National Institutes of Health Grants GM58015 and CA92276 and Department of Defense Grant BC083684/W81XWH-09-1-0116. J.L. was supported by a postdoctoral fellowship from the Tobacco-Related Disease Research Program (17FT-0046).
Footnotes
The authors declare no conflict of interest.
See companion article on page 686.
References
- 1.Heyer WD. Biochemistry of eukaryotic homologous recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Berlin: Springer-Verlag; 2007. pp. 95–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rijkers T, et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Z, et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci USA. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojic M, Mao NH, Zhou QW, Lisby M, Holloman WK. Compensatory role for Rad52 during recombinational repair in Ustilago maydis. Mol Microbiol. 2008;67:1156–1168. doi: 10.1111/j.1365-2958.2008.06116.x. [DOI] [PubMed] [Google Scholar]
- 5.Martin SA, Lord CJ, Ashworth A. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev. 2008;18:80–86. doi: 10.1016/j.gde.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Edwards SL, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 7.Sakai W, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 9.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 10.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 11.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi-Iwai Y, et al. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treuner K, Helton R, Barlow C. Loss of Rad52 partially rescues tumorigenesis and T-cell maturation in Atm-deficient mice. Oncogene. 2004;23:4655–4661. doi: 10.1038/sj.onc.1207604. [DOI] [PubMed] [Google Scholar]
- 14.Fujimori A, et al. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 2001;20:5513–5520. doi: 10.1093/emboj/20.19.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonetani Y, et al. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res. 2005;33:4544–4552. doi: 10.1093/nar/gki766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petalcorin MIR, Sandall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J Mol Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 19.Mazloum N, Zhou QW, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu LJ, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 22.Chan KL, Palmai-Pallag T, Ying SM, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 23.Sharma S, et al. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 24.Garcia PL, Liu YL, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Machwe A, Lozada EM, Xiao L, Orren DK. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 29.New JH, Kowalczykowski SC. Rad52 protein has a second stimulatory role in DNA strand exchange that complements replication protein-A function. J Biol Chem. 2002;277:26171–26176. doi: 10.1074/jbc.M203670200. [DOI] [PubMed] [Google Scholar]