Abstract
Endoplasmic reticulum (ER)-associated degradation (ERAD) is an integral part of the ER quality-control system that removes toxic misfolded proteins via ubiquitin/proteasome-mediated degradation. Most of our knowledge on ERAD comes from biochemical and genetic studies in yeast and mammalian cells. Although ERAD is known to operate in plant cells, little is known about its molecular components and its biochemical mechanism. A genetic screen for suppressors of the Arabidopsis bri1-9, a weak dwarf mutant caused by ER retention of a structurally defective yet biochemically competent brassinosteroid (BR) receptor BRI1, resulted in identification of the EMS-mutagenized bri1 suppressor 5 (EBS5) gene that encodes an Arabidopsis homolog of the yeast Hrd3/mammlian Sel1L protein known to be involved in ERAD. Loss-of-function ebs5 mutations block the ERAD of bri1-9 and bri1-5, another ER-retained BR receptor. We showed that EBS5 complemented the ERAD defect of the yeast Δhrd3 mutant and interacted with the two mutated BR receptors in plant cells. Using a reverse genetic approach, we discovered that two Arabidopsis homologs of the yeast/mammalian Hrd1, an ER membrane-localized ubiquitin ligase, function redundantly in the ERAD of bri1-9. Together, our results revealed functional roles of two conserved ERAD components in degrading mutated/misfolded receptor-like kinases in Arabidopsis.
Keywords: plant steroid receptor, endoplasmic reticulum-associated degradation substrate-recruiting factor, E3 ligase, unfolded protein response
The proper functions of a protein strictly depend on its correct 3D structure, yet protein folding is a fundamentally error-prone process because of stochastic events, genetic errors, and cellular stresses. It was estimated that ∼30% of newly synthesized proteins in mammalian cells are inappropriately folded (1). Because misfolded proteins tend to form toxic aggregates or bind to correctly folded proteins to interfere with normal cellular processes, eukaryotic cells are equipped with a variety of mechanisms to recognize and remove folding-defective proteins. One such control system is localized in the endoplasmic reticulum (ER), the site of entry for all membrane and secretory proteins into the secretory system. The ER-mediated quality-control system delivers correctly folded proteins to their sites of action, retains incompletely/incorrectly folded proteins for additional chaperone-assisted folding, and removes terminally misfolded proteins via retrotranslocation into cytosol for ubiquitin/proteasome-mediated degradation (2), a multistep process widely known as ER-associated degradation (ERAD) (3).
Most of our knowledge on ERAD came from genetic/biochemical studies in yeast and mammalian systems (4). Yeast has at least three different ERAD pathways, known as ERAD-L, ERAD-M, and ERAD-C, to remove misfolded proteins with folding defects exposed in the ER lumen (L), ER membrane (M), and cytosol (C), respectively (5). The central component of the three ERAD pathways is an ER membrane-localized ubiquitin ligase (E3)—Hrd1 for the ERAD-L/-M pathways and Doa10 for the ERAD-C pathway—that ubiquitinates misfolded proteins (6). Both E3 ligases contain multiple transmembrane segments and a cytosolic-facing E3-catalytic RING domain, and they form multisubunit complexes with other proteins that are highly conserved between yeast and mammals (6). The two distinct E3 ligase complexes share some common components, including several cytosolic or ER membrane-anchored ubiquitin-conjugating (E2) enzymes and an ER-membrane protein, Cue1, that recruits cytosolic E2 enzymes. The Hrd1 complex contains several additional proteins, including Hrd3 [also known as Sel1L in mammalian systems (7)], an integral ER membrane protein with a large ER luminal domain and a C-terminal membrane anchor (8), and Yos9, a newly discovered ER-luminal lectin (9). It is believed that Hrd3 and Yos9 work together to select terminally misfolded proteins for ERAD with Hrd3 detecting exposed hydrophobic amino acids and Yos9 recognizing a unique Asn-linked glycan (N-glycan) structure on an ERAD substrate (10). The selected ERAD substrates are retrotranslocated through the ER membrane in a yet-to-be-characterized process, which is thought to be mediated by the Sec61 translocon, a four-transmembrane–spanning ER membrane protein Derlin, or a transmembrane E3 ligase (11), and is powered by a cytosolic Cdc48 complex composed of a homohexamer of the AAA-type ATPase Cdc48 plus its two cofactors Npl4 and Ufd1 (12). After translocation, the Cdc48 complex delivers a misfolded protein to the cytosolic proteasome for degradation.
By contrast, little is known about the molecular components and biochemical mechanism of a plant ERAD process, despite the fact that similar ERAD processes do operate in plant cells to remove misfolded proteins (13, 14). An earlier study suggested the involvement of Cdc48 in the degradation of mutated barley mildew-resistance locus O (MLO) protein when expressed in Arabidopsis (15), and a recent study reported the complementation of an ERAD defect of a yeast Δder1 mutant by two maize homologs of the yeast/mammalian Derlins (16). Two genome-wide gene-expression analyses discovered up-regulation of Arabidopsis genes encoding potential homologs of the known yeast/mammalian ERAD components in response to ER stresses (17, 18). One of the major reasons for the slow progress in studying plant ERAD is the lack of convenient model proteins for forward genetic screens and gene discovery. Recent studies identified several excellent model proteins to study ER quality control/ERAD in Arabidopsis (19), including two mutant forms of a cell-surface receptor, BRI1, for the plant steroid hormone brassinosteroid (BR) (20, 21) and a transmembrane receptor, EFR, for the bacterial translational elongation factor EF-Tu (a conserved pathogen-associated molecular pattern) (22). We recently discovered that a Cys69–Tyr mutation in bri1-5 and a Ser662–Phe mutation in bri1-9 result in ER retention and subsequent ERAD of the two mutated BR receptors, causing a severe BR-insensitive dwarf phenotype in Arabidopsis (23–25). A genetic screen looking for suppressers of the bri1-9 mutant led to identification of EMS-mutagenized bri1 suppressor 1 (EBS1), the only Arabidopsis homolog of the mammalian UDP-glucose:glycoprotein glucosyltransferase (UGGT), an ER-localized protein-folding sensor capable of discriminating completely and incompletely folded glycoproteins, and EBS2, an ER-luminal chaperone-like lectin calreticulin 3 (CRT3) (23, 24).
To identify proteins involved in the ERAD of bri1-9, we performed a secondary screen looking for bri1-9–accumulating ebs mutants and identified several ERAD mutants, including two allelic ebs4 mutants defective in the biosynthesis of the N-glycan precursor. Here, we report the characterization of another ERAD mutant, ebs5, which led to the identification of two crucial components of the Arabidopsis ERAD system that degrades ER-retained cell-surface receptors.
Results
The ebs5-1 Mutation Confers BR Sensitivity to a BR-Insensitive Mutant by Blocking the ERAD of bri1-9.
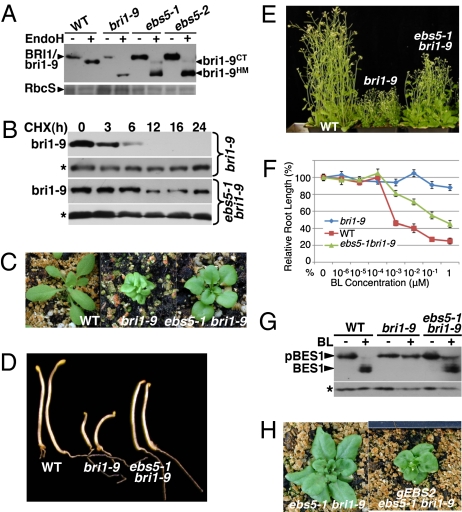
We previously reported a large-scale genetic screen for suppressors of a weak dwarf mutant bri1-9 and made a surprising discovery that its dwarf phenotype is caused by ER retention and subsequent ERAD of a structurally defective but functionally competent BR receptor (23, 24). To search for mutations that specifically affect the ERAD of bri1-9, we performed a secondary biochemical screen by immunoblotting and identified several bri1-9–accumulating ebs mutants. Two of them, ebs4-1 and ebs4-2, were found to be defective in an ER-localized mannosyltransferase involved in assembling the N-glycan precursor, resulting in transfer of smaller glycans to bri1-9, which moves faster on protein gels than the BR receptor in bri1-9 (26). This screen also identified several ebs mutations that have little effect on the molecular mass of bri1-9. Among them are several allelic ebs5 mutants, including ebs5-1 (Fig. 1A). To confirm that the increased bri1-9 abundance was caused by decreased ERAD rather than increased biosynthesis of bri1-9, we treated the ebs5-1 bri1-9 seedlings with cycloheximide (CHX), a protein biosynthesis inhibitor. As shown in Fig. 1B, although bri1-9 became nondetectable after 12 h of CHX treatment in bri1-9, the mutant BR receptor in ebs5-1 bri1-9 was quite stable even after 24 h of CHX treatment.
Fig. 1.
An ebs5 mutation suppresses the dwarf phenotype of bri1-9 by blocking the ERAD of the mutated BR receptor. (A) Immunoblot analysis of bri1-9. Total proteins extracted from 2-wk-old seedlings were treated with Endo H, separated by SDS/PAGE, and analyzed by immunoblotting with anti-BRI1 antibody. bri1-9HM and bri1-9CT denote the HM- and C-type N-glycan–carrying form of bri1-9, respectively. Coomassie blue staining of the small subunit of the Arabidopsis Rubisco (RbcS) serves as a loading control. (B) Immunoblot analysis of the bri1-9 degradation. Two-week-old seedlings were transferred into liquid 1/2 MS medium containing 180 μM CHX. Equal amounts of seedlings were removed at indicated incubation times to extract total proteins in 2× SDS sample buffer, which were analyzed by immunoblotting with the anti-BRI1 antibody. Asterisk indicates a nonspecific band for loading control. (C–E) Shown here, from left to right, are 2-wk-old (C), 5-d-old (D), and 2-mo-old (E) plants of wild type, bri1-9, and ebs5-1 bri1-9. (F) The root-growth inhibition assay. Root lengths of 7-d-old seedlings grown on BL-containing medium were measured and presented as the relative value of the average root length of BL-treated seedlings to that of untreated seedlings of the same genotype. Each data point represents the average of ∼40 seedlings of duplicated experiments. Error bars represent SE. (G) Immunoblot analysis of BES1 phosphorylation in 2-wk-old seedlings treated with or without 1 μM BL. Total protein extracts were separated by SDS/PAGE and analyzed by immunoblotting with an anti-BES1 antibody. Asterisk indicates a nonspecific band for loading control. (H) Phenotypic comparison between ebs5-1 bri1-9 and a representative ebs5-1 bri1-9 transgenic line expressing a genomic EBS2 transgene.
The ebs5-1 mutant is a very good suppressor of bri1-9. As shown in Fig. 1C, ebs5-1 bri1-9 has a larger rosetta with easily recognizable petioles, although its leaves are still round compared with bri1-9. The suppressor has a longer hypocotyl in the dark (Fig. 1D) and longer inflorescence stems (Fig. 1E). Consistent with these morphological alterations, a BR-induced root-growth inhibition assay (27) showed that increased concentrations of brassinolide (BL), the most active BR, inhibited the root elongation of both wild type and ebs5-1 bri1-9 while having a marginal effect on that of bri1-9 (Fig. 1F). We also examined the phosphorylation status of BES1, a robust biochemical marker of active BR signaling (28). Because BES1 has many predicted phosphorylation sites (29), BL treatment of the wild-type seedlings resulted in a dramatic change of the BES1 mobility on a protein gel (Fig. 1G). Fig. 1G reveals that a similar BL treatment had little effect on the mobility of the BES1 band in bri1-9 but caused a significant accumulation of nonphosphorylated BES1 in ebs5-1 bri1-9, which still contained detectable amount of phosphorylated BES1. These results thus demonstrated that ebs5-1 confers BR sensitivity to the BR-insensitive bri1-9 mutant.
This regained BR sensitivity is likely caused by escape of some bri1-9 proteins from the ER to the cell surface caused by overaccumulation of bri1-9 that saturates its ER retention system (25, 26). Consistent with this interpretation, a simple biochemical assay with the endoglycosidase H (Endo H) capable of cleaving N-linked high-mannose–type (HM-type) glycans but not Golgi-processed complex-type (C-type) N-glycans, revealed the presence of a small pool of bri1-9 carrying the C-type N-glycans indicative of ER escape (Fig. 1A). Further support for our explanation came from an experiment showing that overexpression of EBS2, a known rate-limiting ER retention factor of bri1-9 (26), inhibited the suppressing activity of ebs5-1 on bri1-9 (Fig. 1H).
ebs5 also Inhibits the ERAD of bri1-5.
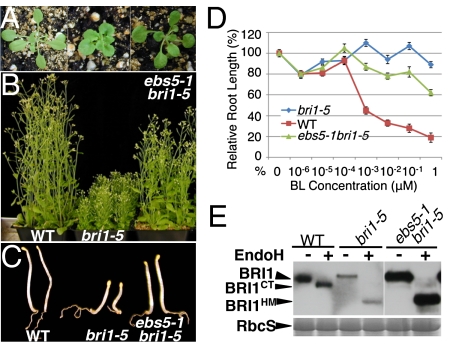
In addition to bri1-9, another mutated BR receptor, bri1-5, was recently shown to be retained in the ER and degradated by a proteasome-independent ERAD process (25). Because chemical inhibition of the bri1-5 ERAD partially rescued the bri1-5 mutation (25), we suspected that ebs5 could also suppress bri1-5. Indeed, when crossed into bri1-5, ebs5-1 was able to suppress many of the bri1-5 mutant phenotypes, including a compact rosette, a short hypocotyl in the dark, and short inflorescence stems at maturity (Fig. 2 A–C), and conferred BR sensitivity to the bri1-5 mutant (Fig. 2D). Consistently, ebs5-1 bri1-5 accumulated a much higher level of bri1-5 than the parental bri1-5 mutant and contained a small pool of C-type N-glycan–carrying bri1-5 proteins (Fig. 2E).
Fig. 2.
The ebs5-1 mutation also suppresses the ERAD of bri1-5. (A–C) Shown here are plants of wild type, bri1-5, and ebs5-1 bri1-5 grown in soil for 2 wk (A), 2 mo (B), or on 1/2 MS medium in the dark for 5 d (C). (D) The root-growth inhibition assay of the BR sensitivity of the 2-wk-old seedlings of wild type, bri1-5, and ebs5-1 bri1-5. Each data point represents the relative average root length of ∼40 seedlings grown on 1/2 MS medium containing varying BL concentrations to that of nontreated seedlings of the same genotype. Error bars represent SE. (E) Immunoblot analysis of BRI1/bri1-5 abundance in 2-wk-old seedlings of wild type, bri1-5, and ebs5-1 bri1-5. Coomassie blue staining of a duplicate gel showing the relative amount of RbcS served as loading control.
Molecular Cloning of the EBS5 Gene.
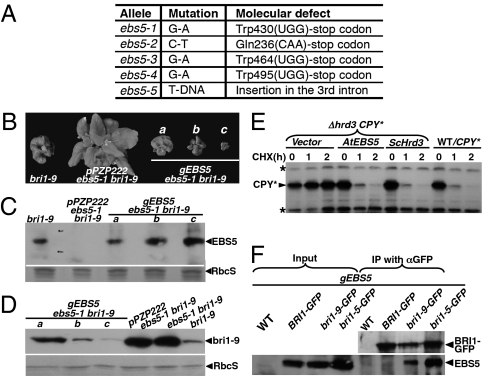
To understand how ebs5 mutations inhibit ERAD of these mutated/misfolded receptors, we cloned the EBS5 gene. PCR-based genetic mapping located the EBS5 locus within an 820-kb region on chromosome I (Fig. S1). Analysis of the annotated genes within this region identified an Arabidopsis protein that is highly similar to the yeast Hrd3/mammalin Sel1L known to be a critical ERAD component (Figs. S1 and S2). Sequence analysis of the At1g18260 revealed a G-A single-nucleotide change between ebs5-1 bri1-9 and wild type, causing a nonsense mutation of Trp430 (TGG-TGA) (Fig. 3A).
Fig. 3.
EBS5 encodes the Arabidopsis homolog of the yeast Hrd3/mammalian Sel1L protein. (A) Identified nucleotide change and predicted molecular defects of five ebs5 alleles. (B) Shown here are pictures of bri1-9 and four transgenic ebs5-1 bri1-9 lines carrying the pPZP222 vector or a genomic EBS5 transgene. (C) Immunoblot analysis of EBS5 abundance in 2-wk-old seedlings of bri1-9 and 4 transgenic ebs5-1 bri1-9 lines shown in B. (D) Immunoblot analysis of the bri1-9 abundance in 2-wk-old seedlings of bri1-9, ebs5-1 bri1-9, and four ebs5-1 bri1-9 transgenic lines shown in B. For C and D, equal amounts of total proteins extracted from 2-wk-old seedlings were separated by SDS/PAGE and analyzed by immunoblotting with anti-EBS5 (C) or anti-BRI1 (D). Coomassie blue staining of a duplicate gel showing the relative amount of RbcS served as loading control. (E) Immunoblot analysis of the CPY* abundance in the wild-type yeast cells transformed with a CPY* plasmid or the yeast Δhrd3 CPY* strain transformed with the indicated plasmid. Equal amounts of total proteins extracted from yeast cells of midlog growth phase treated with or without CHX were separated by SDS/PAGE and analyzed by immunoblotting with anti-HA antibody. Asterisk indicates two cross-reacting bands used for loading control. (F) Coimmunoprecipitation of bri1-5/bri1-9 with EBS5. Equal amounts of total proteins and anti-GFP immunoprecipitates from control/coinfiltrated tobacco leaves were separated by SDS/PAGE and analyzed by immunoblotting with anti-GFP or anti-EBS5 antibodies.
The identity of At1g18260 as the EBS5 gene was further confirmed by three additional experiments. First, sequence analyses revealed a nonsense mutation within At1g18260 in each of three other allelic ebs5 mutants (Fig. 3A). Second, we identified a transfer DNA (T-DNA) insertional mutant of At1g18260 (ebs5-5) and crossed it into bri1-9 and bri1-5, and we found that the ebs5-5 mutation suppressed both bri1 mutants (Fig. S3). Consistent with the detected mutations, immunoblot analysis revealed no detectable EBS5 protein in the ebs5-1, ebs5-2, and ebs5-5 mutants (Fig. S4). Third, the suppressive effect of ebs5-1 on both the bri1-9 dwarfism and bri1-9 ERAD could be inhibited by expression of an At1g18260 genomic transgene in ebs5-1 bri1-9 (Fig. 3 B–D). Interestingly, increased At1g18260 expression caused a more severe dwarf phenotype than bri1-9 and decreased the bri1-9 abundance below that of the parental bri1-9 mutant (Fig. 3 B–D), suggesting that EBS5 is a rate-limiting component of the bri1-9 ERAD pathway.
EBS5 Is an ER-Localized Glycoprotein That Is Up-Regulated by the Arabidopsis Unfolded Protein Response Pathway.
At1g18260, consisting of six exons and five introns (Fig. S1), is predicted to encode a polypeptide of 678 aa containing a signal peptide, nine Sel1-like repeats known to be related to the tetratricopeptide repeat (30), and a near C-terminal transmembrane anchor (Fig. S2). The Arabidopsis genome encodes another Hrd3/Sel1L homolog (At1g73570, referred to as AtSel1B in this study) that plays no role in ERAD of bri1-5 or bri1-9 (Figs. S5 and S6). Consistent with a recent organelle proteomic study (31), EBS5 is mainly localized in the ER (Fig. S7). Based on its homology with the Hrd3/Sel1L proteins, we predicted that the majority of the EBS5 protein is in the ER lumen with a very small C-terminal tail of 36 aa in the cytosol. Our sequence analysis also predicted three potential N-glycosylation sites: Asn298, Asn335, and Asn626 (Fig. S2). As expected, the abundance of EBS5 is increased, but its molecular mass is decreased by tunicamycin (Fig. S8), a glycosylation inhibitor widely used to activate the so-called unfolded protein response that increases the production of ER resident proteins to promote protein folding and stimulate ERAD (32).
EBS5 Complemented the Yeast Δhrd3 Mutation.
To test whether EBS5 is a functional ortholog of the yeast Hrd3 protein, we performed a yeast complementation experiment. We replaced the coding region of the yeast ALG9 in the previously reported pYEp352-yALG9 expression plasmid (26) with that of Hrd3 or EBS5 to generate pYEp352-Hrd3 and pYEp352-EBS5, respectively, and deleted the yALG9 coding region to create a vector control. The resulting plasmids were individually transformed into the yeast Δhrd3 cells expressing carboxypeptidase Y (CPY*), an ER-localized form of the vacuolar carboxypeptidase C that undergoes ERAD in yeast cells (33). As shown in Fig. 3E, the expression of both yeast Hrd3 and the Arabidopsis EBS5 resulted in detectable degradation of the CPY* with a similar kinetics of CPY* degradation in the corresponding wild-type yeast cells. By contrast, the stability of CPY* remained more or less the same during the 2-h CHX incubation period in the Δhrd3 cells transformed with the vector control. These results indicate that the Arabidopsis EBS5 is a likely ortholog of the yeast Hrd3 protein.
EBS5 Interacts with the Mutated BR Receptors.
It was generally believed that the yeast Hrd3/mammalian Sel1L functions as an ERAD substrate-recruiting factor that recognizes misfolded proteins for the Hrd1 E3 ligase complex (11). We thus predicted that bri1-9 or bri1-5, but not the wild-type BRI1, should interact with EBS5. To directly test this hypothesis, we coexpressed the GFP-tagged bri1-5, bri1-9, or BRI1 with EBS5 in tobacco leaves by agroinfiltration-mediated transient expression (34) and performed a coimmunoprecipitation experiment using the infiltrated leaves with an anti-GFP antibody that successfully brought down all three GFP-tagged proteins. The resulting immunoprecipitates were analyzed by immunoblotting with the anti-EBS5 antibody to test the bri1-EBS5 interaction. Fig. 3F revealed the presence of EBS5 in the anti-GFP immunoprecipitates of the infiltrated leaves coexpressing EBS5 and bri1-5/bri1-9–GFP but not from the EBS5-BRI1-GFP-coexpressing leaves. Our results thus demonstrated an interaction between EBS5 and the two mutated BRI1 proteins in plant cells.
Simultaneous Elimination of Two Arabidopsis Homologs of the Yeast Hrd1 also Suppresses the bri1-9 Mutation.
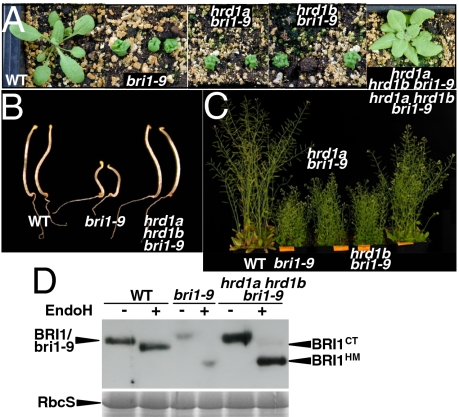
In yeast, Hrd3 binds stoichiometrically to the ER membrane-localized E3 ligase Hrd1 (35). Using the yeast/human Hrd1 as query, we performed a BLAST search against the annotated Arabidopsis proteins and identified two Hrd1 homologs (At3g16090 and At1g65040, renamed AtHrd1A and AtHrd1B, respectively) that contain six putative transmembrane segments and a predicted RING domain (Fig. S9). Phylogenetic analysis revealed that the two Hrd1 homologs are clustered together with Hrd1 homologs from other eukaryotic organisms and suggested that the two Arabidopsis Hrd1 homologs arose from a recent gene-duplication event (Fig. S10). To test whether the two AtHrd1 proteins are involved in the bri1-9 ERAD, we identified a T-DNA insertional mutant for either AtHrd1 gene by searching the Ecker's SIGnAL T-DNA database (36) (Fig. S11) and crossed the resulting hrd1a (SALK_032914) and hrd1b (SALK_061776) mutations into bri1-9. As shown in Fig. 4 A–C, neither single mutant suppressed the bri1-9 phenotype; however, simultaneous elimination of the two AtHrd1 genes led to a phenotypic suppression of the bri1-9 mutant. Consistently, immunoblot analysis showed that the hrd1a hrd1b double mutation inhibits the bri1-9 ERAD (Fig. 4D). As expected from the increased bri1-9 accumulation in the ER, our Endo H assay detected the presence of a small pool of bri1-9 carrying the C-type N-glycan indicative of ER escape (the last lane in Fig. 4D). We thus concluded that the two Arabidopsis Hrd1 homologs function redundantly in the ERAD of a mutated BR receptor.
Fig. 4.
The two Arabidopsis homologs of the yeast/mammalian Hrd1 E3 ligase function redundantly in promoting ERAD of bri1-9. (A) Pictures of 3-wk-old soil-grown seedlings of wild type, bri1-9, hrd1a bri1-9, hrd1b bri1-9, and hrd1a hrd1b bri1-9. (B) Pictures of 5-d-old dark-grown seedlings of wild type, bri1-9, and hrd1a hrd1b bri1-9. (C) Pictures of 2-mo-old soil-grown mature plants of the wild type and bri1-9, hrd1a bri1-9, hrd1b bri1-9, and hrd1a hrd1b bri1-9 mutants. (D) Immunoblot analysis of the bri1-9 abundance in wild type, bri1-9, and hrd1a hrd1b bri1-9 triple mutant. Equal amounts of total proteins extracted from 2-wk-old seedlings were treated with Endo H for 1 h at 37 °C, separated by SDS/PAGE, and analyzed by immunoblotting with anti-BRI1. Coomassie blue staining of a duplicate gel showing the relative amount of RbcS served as loading control.
Discussion
Despite many reports on the existence of ERAD-like processes in plant cells to remove misfolded/mutated proteins, little is known about the molecular components of a plant ERAD pathway. Using both forward and reverse genetic approaches, we identified two components of an Arabidopsis ERAD mechanism that degrades two ER-localized mutated BR receptors. Our sequence and phylogenetic analyses showed that EBS5/At1g18260 is closely related to the yeast Hrd3 and mammalian Sel1L, and At3g16090 and At1g65040 are two Arabidopsis homologs of the yeast/mammalian Hrd1 E3 ligase. Our functional study revealed that the Arabidopsis EBS5 gene was able to complement the ERAD defect of the yeast Δhrd3 mutant, and loss-of-function ebs5 mutations blocked ERAD of both bri1-5 and bri1-9. Consistent with our sequence analysis, we discovered that simultaneous elimination of the two Arabidopsis Hrd1 homologs also prevented the ERAD of bri1-9. These results thus demonstrated the functional roles of two highly conserved components of the eukaryotic ERAD systems to eliminate ER-retained cell-surface proteins in Arabidopsis. It is worth noting that our discoveries not only revealed evolutionary conservation of the ERAD systems in eukaryotes but also uncovered the functional difference between the plant Hrd3/Hrd1 ligase complex and its mammalian homolog. A recent study claimed that the mammalian Sel1L/Hrd1 ligase was required to remove misfolded luminal-soluble proteins but plays no role in degrading their membrane-attached variants carrying the same luminal defects (37). It will be interesting to see whether the Arabidopsis Hrd1/Hrd3 ligase complex is also required to remove misfolded luminal-soluble proteins such as a mutated CRT-GFP fusion protein (38).
It is well established that the yeast Hrd3 has two physiological functions: regulating the stability of Hrd1 and selecting ERAD substrates. Deletion of the yeast Hrd3 results in rapidly degradation of Hrd1, which could be rescued by expression of an N-terminal truncated form of Hrd3 that was unable to support ERAD (35). The Hrd3 directly binds misfolded proteins in the ER lumen and interacts with Yos9 (10), an ER-luminal lectin that recognizes a unique N-glycan ERAD signal carrying an exposed α1,6 mannose residue (39), thus providing a bipartite recognition mechanism to commit a terminally misfolded glycoprotein for ERAD (10, 40). Our coimmunoprecipitation experiment showing that EBS5 interacted with mutated bri1 proteins but not with the wild-type BRI1 strongly suggested that EBS5 was capable of recognizing the folding state of a glycoprotein; however, it remains to be tested whether EBS5 also regulates the stability of the two Arabidopsis Hrd1 homologs and interacts with an Arabidopsis homolog of the yeast Yos9 protein in degrading the mutated/misfolded receptor kinases.
Our study revealed that elimination of EBS5 had little effect on plant growth and development in the BRI1+ background under our standard growth conditions (Fig. S12). This effect is unlikely caused by the presence of a second Sel1L/Hrd3 homolog, AtSel1B, in Arabidopsis because AtSel1B failed to complement the ebs5 mutation when driven by the EBS5 promoter (Figs. S5 and S6). Our result is thus quite different from what was observed in mammalian systems. A recent study reported that mice homozygous mutant for Sel1L were embryonic lethal because of impaired ERAD and ER homeostasis (41). We suspected that the failure of a null ebs5 mutation on plant growth could be attributable to high folding efficiency of plant secretory and membrane proteins compared with the mammalian systems. This interpretation is consistent with earlier findings that Arabidopsis mutants defective in the UGGT/CRT3-mediated quality control exhibit no noticeable growth defect under normal growth conditions but fail to mount a plant defense response against bacterial translation elongation factor EF-Tu (23, 24, 42, 43), a well-studied pathogen-associated molecular pattern (22), because of incomplete folding/ERAD of the EF-Tu receptor EFR. Alternatively, elimination of EBS5 or the two Hrd1 homologs could slow down the EBS5/Hrd1-mediated ERAD process, but the resulting accumulation of misfolded proteins in the ER often leads to activation of the unfolded protein response, an ER stress-signaling process that up-regulates the production of ER chaperones/folding enzymes and components of several ERAD processes to assist protein folding and removal of terminally misfolded proteins (32). Indeed, our immunoblot analyses showed that a null ebs5 mutation in the BRI1+ background did activate the Arabidopsis unfolded protein response (Fig. S12), suggesting involvement of EBS5 in ERAD of many misfolded proteins. We suspect that the ebs5 mutation could affect plant growth and development under certain stress conditions that reduce the folding efficiency of plant proteins and increase ER accumulation of misfolded proteins. Identification of these stressed growth conditions will not only reveal the physiological function of the Hrd3/Hrd1-mediated ERAD pathway but will also provide opportunities to study possible crosstalk mechanisms between the ER stress-signaling process and other signaling pathways that protect plants from various biotic/abiotic stresses.
Experimental Procedures
Plant Materials and Growth Conditions.
All Arabidopsis mutants/transgenic lines used in this study are in the Col-0 ecotype, except bri1-9 (Ws-2) for cloning EBS5 and bri1-5 (Ws-2) for genetic analysis. T-DNA insertional mutants (SALK_109430, SALK_061776, and SALK_032914) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Methods for seed sterilization and conditions for plant growth were described previously (44). The root-growth inhibition assay on BR-containing medium was performed as previously described (27).
Map-Based Cloning of EBS5.
The ebs5-1 bri1-9 (Col-0) was crossed with bri1-9 (Ws-2), and the resulting F1 plants were self-fertilized to generate F2 mapping populations. Genomic DNA isolated from 100 F2 ebs5-1 bri1-9 plants and the molecular markers listed in Table S1 were used for PCR-based genetic mapping to locate the EBS5 locus into a 820-kb region on the top of chromosome I. The genomic DNA fragment of At1g18260 was independently amplified from four individual ebs5-1 bri1-9 seedlings, sequenced, and compared with the Col-0 reference sequence to identify mutations in ebs5 mutants.
Generation of Transgenic Constructs/Plants and Transient Expression in Tobacco Leaves.
A 4.5-kb genomic fragments of At1g18260 was amplified from the bacterial artificial clone T10O22 and cloned into pPZP222 (45) to make the pPZP222-gEBS5 plasmid that was fully sequenced to ensure no PCR error. The gEBS2, pBRI1-BRI1:GFP, pBRI1-bri1-9:GFP, and pBRI1-bri1-5:GFP constructs were previously described (23, 24). These transgenes were mobilized into the Agrobacterium GV3101 strain, and the resulting Agrobacterial cells were used to transform the ebs5-1 bri1-9 mutants by the vacuum-infiltration method (46) or infiltrated into leaves of 3-wk-old tobacco (Nicotiana benthamiana) plants via a previously described protocol (34).
Yeast Complementation Assay.
The yeast Δhrd3 strain WQY6 carrying the pDN436 plasmid that encodes a HA-tagged CPY* was provided by A. Chang (University of Michigan). The coding regions of EBS5 and Hrd3 were individually amplified by RT-PCR from Arabidopsis and yeast cells and replaced that of the yeast ALG9 from the pYEp352-ScALG9 expression plasmid to create pYEp352-EBS5 and pYEp352-ScHRD3, respectively, by using the strategy previously described (26, 44). The plasmids were sequenced to ensure no PCR error and were individually transformed into the WQY6 cells via a previously described protocol (47). Yeast cells of midlog phase were treated with 100 μg/mL CHX; removed after 0, 1, and 2 h from culture tubes; collected by centrifugation; resuspended in 1× yeast extraction buffer (0.3 M sorbitol, 0.1 M NaCl, 5 mM MgCl2, and 10 mM Tris, pH 7.4); lysed by vortexing with glass beads; mixed with 2× SDS buffer; and boiled for 10 min. The resulting supernatants were separated on 10% SDS/PAGE gels and analyzed by immunoblotting with an anti-HA antibody (10A5; Invitrogen).
Immunoblot and Coimmunoprecipitation Assays.
Two-week-old Arabidopsis seedlings treated with or without BL (Chemiclones) or CHX (Sigma) were ground in liquid N2, dissolved in 2× SDS buffer, and boiled for 10 min. After centrifugation, supernatants were used directly for immunoblotting or incubated with or without 1,000 U of Endo Hf in 1× G5 buffer (New England Biolabs) for 1 h at 37 °C, and the treated samples were separated by 7% or 10% SDS/PAGE and analyzed by Coomassie blue staining to serve as loading control or by immunoblotting with anti-BRI1 antibody. For tobacco transient expression, 1 g of control/infiltrated tobacco leaves were ground in liquid N2, dissolved in the extraction buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 5 mM EDTA, 0.2% Triton X-100, 10% glycerol, and the protease inhibitor mixture containing 1 mM PMSF plus 2 μg/mL each of aprotinin, leupeptin, and pepstatin A), and centrifuged for 10 min at 5,000 × g. The supernatant was directly analyzed by SDS/PAGE and immunoblotting with anti-BRI1 or anti-EBS5 antibody (see SI Experimental Procedures for generation of anti-EBS5 antibody) or was incubated with a polyclonal anti-GFP antibody (TP401; Torrey Pines Biolabs) for 1 h followed by 1-h incubation with protein A agarose beads (Invitrogen) to precipitate GFP-tagged proteins. The immunoprecipitates were washed three times with the extraction buffer, separated by SDS/PAGE, and analyzed by immunoblotting using a monoclonal anti-GFP (MMS-118P; Covance) or anti-EBS5 antibody.
Supplementary Material
Acknowledgments
We are grateful to the Arabidopsis Biological Resource Center at Ohio State University for supplying cDNA/genomic clones and T-DNA insertional mutants of EBS5, AtSel1B, and the two Arabidopsis Hrd1 homologs; F. Tax for seeds of bri1–9 (Ws-2) and bri1–5; J. Chory (Salk Institute for Biological Studies, La Jolla, CA) for anti-BRI1 antibody; Y. Yin for anti-BES1 antibody; R. Boston for anti-maize CRT serum; A. Chang (University of Michigan) for the yeast Δhrd1 strain and the CPY* plasmid; and T. Tzfira for the pSITE03-RFP-HDEL plasmid. We thank members of the J.L. laboratory for helpful discussions. This work was supported by National Institutes of Health Grant GM060519 (to J.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013251108/-/DCSupplemental.
References
- 1.Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 2.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 3.McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: Dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vembar SS, Brodsky JL. One step at a time: Endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostova Z, Tsai YC, Weissman AM. Ubiquitin ligases, critical mediators of endoplasmic reticulum-associated degradation. Semin Cell Dev Biol. 2007;18:770–779. doi: 10.1016/j.semcdb.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehara K, Kawaguchi S, Ng DT. The EDEM and Yos9p families of lectin-like ERAD factors. Semin Cell Dev Biol. 2007;18:743–750. doi: 10.1016/j.semcdb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 11.Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raasi S, Wolf DH. Ubiquitin receptors and ERAD: A network of pathways to the proteasome. Semin Cell Dev Biol. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Ceriotti A, Roberts LM. Endoplasmic reticulum-associated protein degradation in plant cells. In: Robinson DG, editor. The Plant Endoplasmic Reticulum. Berlin, Heidelberg: Springer; 2006. pp. 75–98. [Google Scholar]
- 14.Vitale A, Boston RS. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic. 2008;9:1581–1588. doi: 10.1111/j.1600-0854.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 15.Müller J, et al. Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell. 2005;17:149–163. doi: 10.1105/tpc.104.026625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirst ME, Meyer DJ, Gibbon BC, Jung R, Boston RS. Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol. 2005;138:218–231. doi: 10.1104/pp.105.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez IM, Chrispeels MJ. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15:561–576. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamauchi S, Nakatani H, Nakano C, Urade R. Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 2005;272:3461–3476. doi: 10.1111/j.1742-4658.2005.04770.x. [DOI] [PubMed] [Google Scholar]
- 19.Saijo Y. ER quality control of immune receptors and regulators in plants. Cell Microbiol. 2010;12:716–724. doi: 10.1111/j.1462-5822.2010.01472.x. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 22.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 23.Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin H, Hong Z, Su W, Li J. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106:13612–13617. doi: 10.1073/pnas.0906144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell. 2008;20:3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong Z, et al. Mutations of an α1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell. 2009;21:3792–3802. doi: 10.1105/tpc.109.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mora-García S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, et al. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittl PR, Schneider-Brachert W. Sel1-like repeat proteins in signal transduction. Cell Signal. 2007;19:20–31. doi: 10.1016/j.cellsig.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 31.Dunkley TP, et al. Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 33.Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 34.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 35.Gardner RG, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 37.Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandizzi F, et al. ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 2003;34:269–281. doi: 10.1046/j.1365-313x.2003.01728.x. [DOI] [PubMed] [Google Scholar]
- 39.Quan EM, et al. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 41.Francisco AB, et al. Deficiency of suppressor enhancer Lin12 1 like (SEL1L) in mice leads to systemic endoplasmic reticulum stress and embryonic lethality. J Biol Chem. 2010;285:13694–13703. doi: 10.1074/jbc.M109.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nekrasov V, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA. 2009;106:15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 46.Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Chang A. Heat shock response relieves ER stress. EMBO J. 2008;27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.