Abstract
Although expression of non–protein-coding RNA (ncRNA) can be altered in human cancers, their functional relevance is unknown. Ultraconserved regions are noncoding genomic segments that are 100% conserved across humans, mice, and rats. Conservation of gene sequences across species may indicate an essential functional role, and therefore we evaluated the expression of ultraconserved RNAs (ucRNA) in hepatocellular cancer (HCC). The global expression of ucRNAs was analyzed with a custom microarray. Expression was verified in cell lines by real-time PCR or in tissues by in situ hybridization using tissue microarrays. Cellular ucRNA expression was modulated with siRNAs, and the effects on global gene expression and growth of human and murine HCC cells were evaluated. Fifty-six ucRNAs were aberrantly expressed in HepG2 cells compared with nonmalignant hepatocytes. Among these ucRNAs, the greatest change was noted for ultraconserved element 338 (uc.338), which was dramatically increased in human HCC compared with noncancerous adjacent tissues. Although uc.338 is partially located within the poly(rC) binding protein 2 (PCBP2) gene, the transcribed ncRNA encoding uc.338 is expressed independently of PCBP2 and was cloned as a 590-bp RNA gene, termed TUC338. Functional gene annotation analysis indicated predominant effects on genes involved in cell growth. These effects were experimentally demonstrated in both human and murine cells. siRNA to TUC338 decreased both anchorage-dependent and anchorage-independent growth of HCC cells. These studies identify a critical role for TUC338 in regulation of transformed cell growth and of transcribed ultraconserved ncRNA as a unique class of genes involved in the pathobiology of HCC.
Keywords: liver cancer, hepatocarcinoma, exaptation, transposon
Hepatocellular carcinogenesis involves a complex interaction of genes resulting in variable modulation of key pathways involved in tumor cell growth. By using molecular techniques for global genomic profiling, the transcriptome in hepatocellular cancers (HCCs) has been described, and several genes that are differentially activated have been identified. The major focus of attention in these efforts has been on the characterization of expression of protein-coding genes and their use for determining clinical outcomes (1–9). However, the majority of the human genome consists of non–protein-coding RNA (ncRNA). Increasing evidence points to an important functional or regulatory role of ncRNA in cellular processes as well as a contribution of aberrant ncRNA expression to disease phenotypes.
Along with the highly abundant transfer and ribosomal RNAs, ncRNAs include microRNAs that modulate mRNA expression, small nucleolar RNAs that guide chemical modification of RNA molecules, siRNAs that account for the interference pathway, Piwi-RNAs that are linked to transcriptional gene silencing of retrotransposons, and long ncRNAs, whose role is still unknown (10, 11). The role played by the ncRNA genome in malignant transformation and tumor growth in HCC is being increasingly recognized. We and others have recently provided data from profiling studies in which several microRNAs were identified and shown to be involved in the modulation of cell proliferation and apoptosis (12–15). Recent studies revealing the presence of several hundred long transcribed ncRNAs raise the possibility that many ncRNAs contributing to cancer remain to be discovered (16). Other than microRNAs, however, only a handful of ncRNAs have been implicated in hepatocarcinogenesis (17–20). For the most part, the function of these ncRNAs is unknown. Sequence conservation across species has been postulated to indicate that a given ncRNA may have a cellular function (21, 22). A genome-wide survey identified several hundred ncRNAs with a size >200 bp that showed a remarkable conservation with 100% identity across the human, mouse, and rat genomes (23). These highly conserved ncRNAs have been named ultraconserved elements and are conserved across many other species as well, with 99% of these RNAs showing high levels of conservation within the dog, 97% within the chicken, and 67% within the fugu genomes. Their wide distribution in the genome and lack of natural variation in the human population suggested that these ultraconserved regions have a biological function that is essential for normal cells (24, 25).
Recent genome-wide expression profiling studies showed that transcribed ultraconserved RNAs (ucRNAs) exhibit distinct profiles in various human cancers (26, 27). Aberrant expression of specific ucRNA has been linked with leukemia and several solid tumors. The ultraconserved element 73 (uc.73), for example, modulates apoptosis and cell proliferation in colorectal cancer cells (26). A correlation of some ucRNA with clinical prognostic factors, such as Myc amplification, has been reported in neuroblastoma (28). Although rare, single-nucleotide polymorphisms in ultraconserved regions have been reported in familial breast cancer, chronic lymphocytic leukemia, and colorectal cancer (29, 30). The functional role of ucRNAs in human cancer behavior and development is unknown, and our aims were to evaluate their expression and potential involvement in growth regulation in human HCC.
Results and Discussion
Aberrant Expression of Selected ucRNAs in Malignant Hepatocytes.
Genome-wide expression profiling identified 56 ucRNAs, representing 11% of all ucRNAs analyzed, that were aberrantly and significantly (P < 0.05) expressed in malignant HepG2 cells compared with nonmalignant human hepatocytes (Fig. 1A). Of these, 33 were increased by 1.3- to 6.9-fold, whereas 23 were decreased by 0.8- to 0.3-fold in malignant hepatocytes. Exonic ucRNAs were not selectively enriched in malignant cells, and the proportion of exonic regions in aberrantly expressed ucRNA (29% exonic, 42% nonexonic) was similar to those of all ultraconserved regions (Fig. 1B). The greatest change was noted for ultraconserved element 338 (uc.338), which was increased in expression by 6.9 ± 0.9-fold (P = 0.001), and we thus focused our attention on this ucRNA.
Fig. 1.
ucRNAs are aberrantly expressed in malignant hepatocytes. (A) Genome-wide expression profiling was performed in HepG2 malignant cells and normal human hepatocytes (HH). Fifty-six ucRNAs were aberrantly expressed in malignant hepatocytes with P < 0.05, with 12 ucRNAs increased and 7 decreased by greater than twofold. The ratio of expression of these ucRNAs in malignant cells relative to nonmalignant cells is plotted against the P value. Selected ucRNAs with a greater than threefold change in expression are annotated. (B) The genomic locations of the ucRNA as exonic, nonexonic, or possibly exonic relative to protein-coding genes is depicted for all ucRNAs and for the group of ucRNAs that are aberrantly expressed in malignant hepatocytes. Selective enrichment of a specific group of ucRNA based on their genomic relationship to known protein-coding genes was not observed.
uc.338 Is Increased in Expression in HCC Cell Lines.
By real-time PCR, a striking increase in uc.338 expression by 2.2- to 5.1-fold was observed in several HCC cell lines compared with nonmalignant human hepatocytes (Fig. 2). We next determined uc.338 expression in a panel of human cancer cell lines, including biliary, pancreatic, colorectal, prostatic, and breast cancers. In most cells, the expression of uc.338 was reduced or comparable to the expression in normal hepatocytes. Interestingly, all cholangiocarcinoma cells showed very low levels of uc.338 expression, suggesting that uc.338 may differentiate between primary liver cancers arising from different liver epithelia. Thus, uc.338 is increased in HCC cells and might be a promising marker for HCC.
Fig. 2.
uc.338 is overexpressed in HCC cells lines. RNA was extracted from different cell lines and uc.338 expression evaluated by quantitative real-time-PCR. The expression of uc.338 was normalized to that of RNU6. Bars represent the mean and SEM of four samples. uc.338 expression was increased in all HCC cell lines compared with normal human hepatocytes (HH). *P < 0.05 relative to human hepatocytes. Nonmalignant epithelial cells (hatched bars): HE, hepatocytes; BE, biliary epithelia; PE, prostatic epithelia. Malignant cells (solid bars): CCA, cholangiocarcinoma; PaC, pancreatic cancer; CRC, colorectal cancer; PC, prostate cancer; BC, breast cancer.
uc.338 Expression Is Increased in Human HCC Tissues.
We next studied uc.338 expression in HCC tissues with in situ hybridization. We analyzed 221 HCC samples in two tissue microarrays. The arrays also included 169 cases of adjacent noncancerous liver tissue, with cirrhosis present in 97 cases. uc.338 expression was classified based on the percentage of cells with detectable expression as follows: negative (<5%), weak (5–19%), moderate (20–49%), or strong (≥50%) (Fig. S1). uc.338 expression was detected in 170 cases (77%), with a moderate to strong expression in 62% of these cases (Fig. 3A). The mean uc.338 expression was 4.0 ± 1.5% in noncirrhotic liver, 15.0 ± 4.5% in cirrhotic liver, and 24.0 ± 5.7% in HCC tissues (Fig. 3B). Adequate paired tumoral and adjacent nontumoral tissue for analysis was available from 156 cases. Of these, uc.338 expression was increased in 62%, was unchanged in 24%, or was decreased in 14% of HCC compared with adjacent tissues (Fig. 3C). Consistent increases in uc.338 expression (score > 2.0) were noted with HCC although a reduction in uc.338 expression (score < −2.0) occurred sporadically (Fig. 3D). Within tumor cells, uc.338 expression was predominantly nuclear (Fig. 4A). Similarly, the nuclear/cytoplasmic ratio of uc.338 expression in HepG2 and Huh-7 cells was 17 and 27, respectively (Fig. 4B).
Fig. 3.
uc.338 is overexpressed in human HCC tissues. uc.338 expression was evaluated in a total of 221 HCCs, 72 cases of noncirrhotic liver tissues, and 97 cases of cirrhotic adjacent liver tissues. Paraffin-embedded, formalin-fixed liver tissues were incubated with LNA–anti-uc.338. (A) uc.338 expression was classified as negative, weak, moderate, or strong based on the percentage of cells with detectable staining for uc.338. The proportion of cases of HCC, cirrhotic liver, or noncirrhotic liver within each class is depicted in the columns. (B) The mean and 95% confidence intervals of uc.338 expression in noncirrhotic liver, cirrhotic liver, and HCC tissues is shown. *P < 0.05. (C) uc.338 expression was compared between 156 HCCs and their corresponding adjacent liver tissues. Picture of representative cases are shown. (D) An expression score was derived as the ratio of the difference in expression between HCC and adjacent liver tissues to the SD of uc338 in all tissues and plotted with the size of the bubble representing the number of cases.
Fig. 4.
Nuclear expression of uc.338 in HCC cells. (A) Paraffin-embedded, formalin-fixed liver tissues were incubated with LNA–anti-uc.338. uc.338 was frequently detected in nuclei of HCC in situ. (B) RNA was extracted from the nuclear and the cytoplasmic fraction of liver cells and uc.338 expression evaluated by real-time-PCR. Bars represent the mean and SEM of relative expression of uc.338 from two experiments performed in four replicates. *P < 0.05.
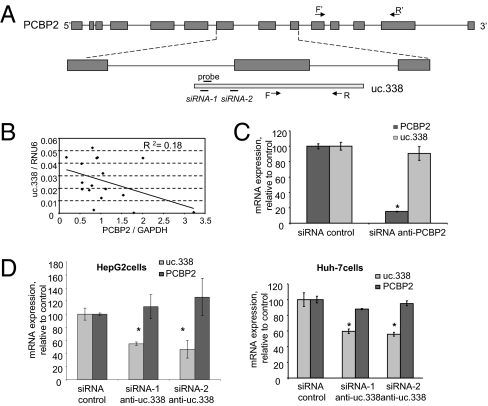
uc.338 Expression Is Regulated Independently of the Poly(rC) Binding Protein 2 (PCBP2) Gene.
uc.338 consists of 223 nt that are highly conserved throughout the species. In humans, the uc.338 ultraconserved region is located partly within the exon of the PCBP2 gene on chromosome 12 (Fig. 5A). To evaluate the potential interrelationships between PCBP2 and uc.338 transcription, we first examined the expression of PCBP2 by real-time PCR in normal and HCC cell lines. The primers used spanned a genomic region in exons 10–13 of PCBP2 that was distant from that of uc.338 (Fig. 5A). PCBP2 expression was not increased in any of the HCC cell lines, with the exception of Huh-7 cells, and PCBP2 expression did not correlate with that of uc.338 in all samples tested (Fig. 5B). Furthermore, uc.338 expression was unchanged in HepG2 cells transfected with siRNA against PCBP2 despite an 85% reduction in PCBP2 mRNA expression (Fig. 5C). We next evaluated the effect of uc.338 on PCBP2 expression in HepG2 and Huh-7 cells but did not observe any effect of reduction in uc.338 expression on PCBP2 mRNA expression (Fig. 5D). These observations confirm that uc.338 is not expressed as part of the PCBP2 gene.
Fig. 5.
uc.338 and PCBP2 are independently regulated. (A) Schematic representation of the partial exonic location of uc.338 within the PCBP2 gene. The exons of PCBP2 are indicated by dark gray boxes, and uc.338 is depicted as the light gray box. The location of the uc.338 forward (F) and reverse (R) primers used for real-time-PCR and probe used for in situ hybridization are shown. With the exception of the forward primer, these primers are located within the PCBP2 intronic region. Of the siRNAs targeting uc.338, siRNA-1 is entirely intronic, whereas siRNA-2 overlaps a few nucleotides of the coding sequence of PCBP2. F′ and R′ indicate primers used for detection of PCBP2. (B) Expression of uc.338 is plotted against PCBP2 mRNA expression in samples of normal and HCC cell lines. There is no linear correlation between uc.338 and PCBP2 gene expression (P = 0.08). (C) HepG2 cells were transfected with siRNA against PCBP2 or siRNA control. Bars represent the mean of two independent experiments performed in four replicates. *P < 0.05 compared with control siRNA. (D) The expression of uc.338 was decreased in HepG2 and Huh-7 cells by using two different siRNAs against uc.338. After 48 h, RNA was collected and uc.338 and PCBP2 expression were evaluated by real-time PCR. Bars represent the mean and SEM of two experiments performed in four replicates. *P < 0.05 relative to siRNA control. These data indicate that uc.338 does not regulate the expression of PCBP2 and that the expression of uc.338 is independent of PCBP2.
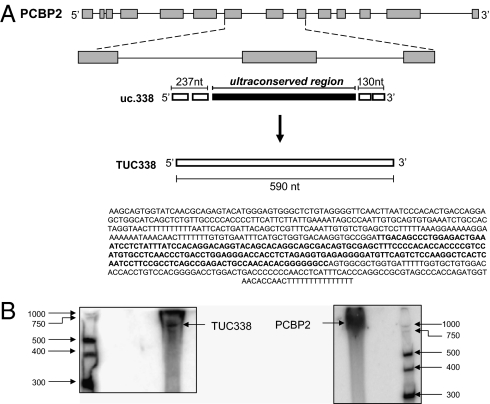
Identification of the Transcript Encoding uc.338.
Having shown that uc.338 is transcribed independently of PCBP2, we proceeded to clone the transcript encoding this ultraconserved element. Rapid amplification of cDNA ends (RACE) was performed to characterize the 5′ end and the 3′ end of this transcript, which we termed TUC338. HepG2 RNA was retrotranscribed with the SMARTerScribe RT that exhibits terminal transferase activity upon reaching the end of an RNA template and adds residues to the first-strand cDNA. The SMARTer oligo contains a terminal stretch of modified bases that anneal to the extended cDNA tail, allowing the oligo to serve as a template for the RT. Thus, a complete cDNA copy of the original RNA with an additional SMARTer sequence at the end is generated. To study TUC338, we used intronic primers that would not recognize the PCBP2 coding sequence (Fig. S2). No products were produced with the antisense intronic (ASI) primer, suggesting that TUC338 is not encoded in antisense. Conversely, a few bands were produced after amplification with sense intronic (SI) primer, with the larger bands being ∼500 nt. A nested PCR with the nested sense intronic (nSI) primer produced a single defined band of ∼500 nt that was further sequenced, leading to the characterization of the 5′ end of TUC338. As control, we used the sense exonic (SE) primer that produced a >800-nt band by recognizing coding sequence PCBP2 (Fig. S3). These findings further showed that the TUC338 transcript is different from the PCBP2 transcript and characterized the 5′ end of TUC338. The 3′ RACE studies identified 130 nt at the 3′ end downstream of the ultraconserved sequence identified by Bejerano et al. (23) (Fig. S4). In conclusion, the uc.338 ultraconserved element is part of a 590-nt-long transcript, TUC388, that is transcribed independently on PCBP2 (Fig. 6).
Fig. 6.
Cloning of the transcript including uc.338. (A) Schematic representation of the transcript including uc.338 (TUC338) in relation to PCBP2 gene. By performing 5′ and 3′ RACE, we identified 237 nt upstream and 130 nt downstream of the ultraconserved region that was reported by Bejerano et al. (23). The complete sequence of the TUC338 transcript is reported. (B) Northern blotting analysis for TUC338 and PCBP2 was performed as described in Materials and Methods. TUC338 was expected to be 590 nt long, and PCBP2 was ∼1,200 nt long.
Functional Expression Analysis of TUC338-Regulated Genes.
To gain insight into the functional role of TUC338, we performed gene annotation enrichment analysis of genome-wide mRNAs that were changed in expression after TUC338 inhibition using siRNA. Functional annotation analysis identified the top four significantly overrepresented cellular process gene classifications (and number of genes) as transcription (569), cell cycle (248), ubiquitin cycle (225), and cell division (115), whereas the top four overrepresented molecular function classifications were ligase activity (159), protein binding (1,810), nucleotide binding (774), and ATP binding (638). The top four significantly overrepresented GenMAPP pathway gene classifications were cell cycle/KEGG (56), mRNA processing reactome (63), RNA transcription reactome (28), and G1 to S cell-cycle reactome (41). These data suggested that TUC338 could modulate cellular processes involved in cell growth.
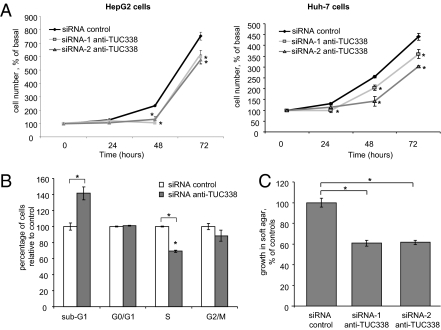
TUC338 Modulates Cell Growth in Human Hepatocytes.
We assessed anchorage-dependent cell growth after transfection with either siRNA to TUC338 or scrambled nucleotide control siRNA. Compared with control siRNA, siRNA-1 and siRNA-2 reduced cell proliferation in HepG2 as well as in Huh-7 cells (Fig. 7A). Compared with controls, there was a reduction of cells in S phase (P < 0.0001) in cells transfected with siRNA to TUC338 (Fig. 7B). Cancer is characterized by the acquisition of cellular traits that enhance cell growth under adverse microenvironmental conditions. Therefore, we examined anchorage-independent growth by examining growth in soft agar assays. Compared with controls, siRNA to TUC338 reduced soft agar growth of HepG2 cells by 40.0 ± 2.0% (Fig. 7C).
Fig. 7.
TUC338 modulates cell growth in HCC cells. (A) HepG2 and Huh-7 cells were transfected with siRNAs against TUC338 or control siRNA for 48 h and then plated. After 24, 48, and 72 h, cells were counted by trypan blue staining. Mean values of three independent experiments with SEM are represented. *P < 0.05 compared with control. Cell viability after transfection with siRNA-1 was not significantly different from that with siRNA-2 (P > 0.05 at each time point in each of the cell lines). (B) HepG2 cells were transfected with siRNA-2 anti-TUC338 or control siRNA for 48 h, and analysis of cell-cycle distribution was performed by flow cytometry. Compared with controls, there was a reduction of 30% in S phase and of 12% in G2/M phase for cells transfected with siRNA against TUC338. Bars represent the mean and SEM of three experiments. *P < 0.05 compared with controls. (C) HepG2 cells were transfected with siRNA anti-TUC338 or control siRNA by nuclear transfection for 48 h and then plated in agar in 96-well plates. Anchorage-independent growth was assessed fluorometrically after 7 d. Bars represent the mean and SEM of two experiments performed in seven replicates. *P < 0.05.
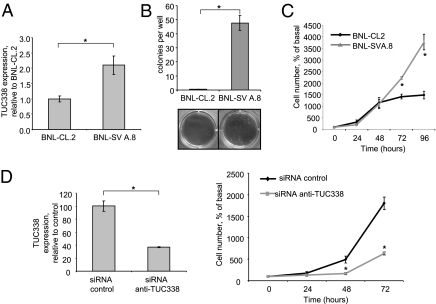
TUC338 Modulates Cell Growth in Mouse Hepatocytes.
The sequence conservation of ucRNAs across diverse species suggests that these RNAs may participate in essential roles that may be similar across species. To examine cross-species similarities in the effects of TUC338, we studied the effect of this gene in modulating transformed cell growth in murine cells. First, we examined the effect of cell transformation on TUC338 expression in BNL-CL.2 embryonic mouse hepatocytes. Compared with parental BNL-CL.2 cells, the expression of TUC338 was increased by 2.1-fold in BNL-SVA.8 cells, which are derived from BNL-CL.2 by SV40 transformation (Fig. 8A). BNL-SVA.8 cells have increased anchorage-dependent and anchorage-independent growth in soft agar compared with BNL-CL.2 cells (Fig. 8 B and C). Knockdown of TUC338 in BNL-SVA.8 cells with siRNA caused a dramatic reduction in cell proliferation. At 72 h, cell proliferation was reduced by 65.0 ± 2.0% (P = 0.0001) in BNL-SVA.8 cells transfected with siRNA to TUC338 compared with control siRNA (Fig. 8D). These data, indicating a relationship among TUC338 expression, cell transformation, and cell growth in mouse hepatocytes, are similar to those observed in humans.
Fig. 8.
TUC338 modulates cell growth in mouse hepatocytes. BNL-CL.2 are nonmalignant embryonic mouse hepatocytes. BNL-SVA.8 cells are derived from BNL-CL.2 after SV40 transformation, and they exhibit transformed cell growth. (A) TUC338 expression was assessed by real-time-PCR and normalized to that of RNU6. Bars represent the mean and SEM of three experiments. (B) BNL-CL.2 and BNL-SVA.8 were plated in soft agar, and colonies were counted after 4 wk. Representative pictures are shown along with the mean and SEM of three independent experiments. *P < 0.05. (C) BNL-CL.2 and BNL-SVA.8 cells were plated in 6-well plates, and cell number was counted by trypan blue staining at different time points. Mean and SEM derived from three independent experiments are shown. *P < 0.05 compared with BNL-CL.2. (D) BNL-SVA.8 cells were transfected with siRNA-1 anti-TUC338 or siRNA control for 48 h. Bars represent mean and SEM of four replicates. At the indicated times, cells were counted after trypan blue staining. Mean and SEM from three independent experiments are represented. *P < 0.05 compared with siRNA control.
TUC338 Modulates Progression Through the Cell Cycle.
The functional genomic expression analysis showed enrichment in genes involved in cell-cycle progression from phase G1 to phase S in response to inhibition of TUC338. Moreover, inhibition of TUC338 in HCC reduced the number of cells in S phase (Fig. 7). Thus, we assessed the effect of TUC338 inhibition on expression of several proteins involved in the G1/S checkpoint in HepG2 and Huh7 cells. S-phase progression can be modulated by CDK4/6-cyclin D1 mechanisms, and alterations in cyclin D are prominent in HCC. After inhibition of TUC338, we observed an increase in expression of the tumor suppressor p16INK4a and an associated reduction of CDK4, CDK6, and cyclin D1 (Fig. 9). Indeed, altered expression of these cell-cycle regulatory proteins is associated with HCC growth (31–33). Although S-phase progression can also be modulated by CDK2/cyclin E, effects on cell growth after inhibition of TUC338 were noted in Huh7 cells, which do not express p21 and cyclin E (34–36), making it unlikely that the effects of TUC338 on cell-cycle progression involved these proteins.
Fig. 9.
TUC338 expression modulates expression of cell-cycle regulatory proteins. HepG2 and Huh7 cells were serum-starved for 48 h before transfection with either control siRNA or anti-TUC338 siRNA. Cells were collected 48 h after transfection, and protein lysates were obtained. Lysates were also obtained from untransfected cells and normal human hepatocytes (HH). Western blotting was performed for the indicated cell-cycle–associated proteins, and their expression was quantitated by densitometry and normalized to that of vinculin. The expression relative to cells transfected with a control siRNA is reported along with representative immunoblots. PCNA, proliferating cell nuclear antigen.
Conclusions
We hope that this demonstration of a functional role of ucRNAs in epithelial cell growth modulation will stimulate exploration of similar roles and functions of other ncRNA transcripts that are being reported in increasing numbers from the FANTOM studies and other sequencing and bioinformatic genomic analyses (37). Although transcribed ucRNAs were described several years ago, and sequence conservation across species is highly suggestive of functional roles, elucidation of the potential physiological roles of these ncRNAs has remained elusive. Much of the attention in the field has focused on the evolutionary significance (38, 39). Recent studies have identified the involvement of these ucRNAs and other ncRNAs in human cancers (26). We have extended these studies to identify aberrantly expressed ucRNAs in HCC as an initial step to examine and understand their cellular role in hepatic neoplasia. Our data indicate an important contribution of ucRNAs to tumor cell growth in hepatic epithelia and implicate these RNAs as a previously uncharacterized group of genes that are involved in liver cancers.
Our studies identified a ncRNA transcript, TUC338, that incorporates the highly ultraconserved region designated as uc.338. The sequence of TUC338 overlaps with that of PCBP2, an RNA binding protein involved in mRNA processing in humans (40–42). Interestingly, exonic ultraconserved regions are frequently associated with RNA processing, such as RNA binding or RNA splicing, suggesting a potential role as regulators of RNA. Despite the exonic location of TUC338 within PCBP2, these two genes are independently expressed. We speculate that TUC338 functions as a component of a self-regulating gene network that participates in development or in the fine-tuning of regulation of expression of several genes or proteins. The nuclear localization of TUC338 is consistent with such a role as a modulator of gene expression. Based on our findings showing a role for TUC338 in human cancer, further investigation into the potential roles of TUC338 and effects on hepatic gene expression are clearly warranted.
The reason for the extremely high conservation of uc.338 across species as divergent as rat, mouse, human, dog, and fugu remains enigmatic. uc.338 shows 80% similarity to a short repeat within the Sarcopterygii, which is also present in the “living fossil” coelacanth, showing a remarkably ancient conservation of this genetic element (23). Mobile elements, or transposons, drive genome evolution, and at least 50% of our genome originates from characterized transposon-derived DNA (23, 24). Thus, uc.338 may have originated as a result of exaptation, in which an evolutionarily ancient transposable element became fixed within the genome, in this case, within the TUC338 ncRNA. The extent to which ultraconserved regions or exapted repeats contribute to cancer pathogenesis remains unknown. Our findings suggest that, at least in the case of uc.338, exaptation can mediate a cellular role important in cancer cell behavior. It is tempting to speculate that exaptation of the uc.338 ultraconserved element resulted in acquiring a function that served the host that may be unrelated to tumorigenesis but that the enhanced expression of uc.338 after cell transformation promotes cell growth. Further investigation into the potential role of exapted genes and transposons in the pathogenesis of human HCC and in other diseases is clearly warranted. The discrepancy in expression of TUC338 between normal and malignant hepatocytes and cholangiocytes, the two major hepatic epithelial cell types, is noteworthy. These differences indicate tissue specificity in TUC338 expression within the same organ. These differences suggest that functional effects of TUC338 may be specific to hepatocytes, thereby increasing the attractiveness of exploring TUC338-mediated cellular growth regulatory pathways as therapeutic targets to modulate HCC growth. In conclusion, our studies showing the aberrant expression of TUC338 in transformed hepatocytes and its functional role in modulating growth may form the basis for further investigation of this previously uncharacterized ncRNA as a therapeutic target for selected HCC.
Materials and Methods
Additional materials and methods are available in SI Materials and Methods.
ucRNA Expression Profiling.
RNA was extracted from three separate biological samples for each analysis by using TRIzol reagent (Invitrogen). Total RNA (5 μg) was reverse-transcribed with biotin end-labeled random oligonucleotide primers, and cDNA was hybridized to a custom microarray (OSU-CCC 4.0), which includes sense and antisense probes to all 481 human ultraconserved sequences reported by Bejerano et al. (23), each spotted in duplicate. Biotin-containing transcripts were detected with streptavidin–Alexa647 conjugate and scanned and analyzed by using an Axon 4000B scanner and GenePix 6.0 software (Axon Instruments). The mean fluorescence intensity of replicate spots were subtracted from background and normalized by using the global median method. We selected ucRNAs measured as present in all of the three samples. Differentially expressed ucRNAs were identified by using the Class Comparison Analysis of BRB tools version 3.6.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html). The criterion for inclusion of a gene in the gene list was P < 0.05.
In Situ RNA Hybridization.
A locked nucleic acid (LNA) probe with complementarity to a 22-bp section of uc.338 identical to that used for Northern blot analysis was labeled with 5′-digoxigenin and synthesized by Exiqon. Tissue sections on the tissue microarray were digested with 2 mg/mL pepsin and in situ hybridization performed as described (43). Negative controls included omission of the probe and the use of a scrambled LNA probe. Each sample was classified by two independent reviewers based on the percentage of cells with detectable uc.338 expression as follows: negative (<5%), weak (5–19%), moderate (20–49%), or strong (≥50%). An expression score was derived as the difference in percentage of cells that expressed uc.338 in HCC and in the corresponding adjacent liver divided by the SD of the percentage of uc.338 expression across all samples analyzed.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Schmittgen for assistance with primer design. Grant support was provided by an American Italian Cancer Foundation fellowship (to N.V.), the National Institutes of Health (to T.P. and C.M.C.), and the Ohio State University Comprehensive Cancer Center (to T.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011098108/-/DCSupplemental.
References
- 1.Teufel A, et al. Genetics of hepatocellular carcinoma. World J Gastroenterol. 2007;13:2271–2282. doi: 10.3748/wjg.v13.i16.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27:55–76. doi: 10.1055/s-2006-960171. [DOI] [PubMed] [Google Scholar]
- 3.Midorikawa Y, Makuuchi M, Tang W, Aburatani H. Microarray-based analysis for hepatocellular carcinoma: From gene expression profiling to new challenges. World J Gastroenterol. 2007;13:1487–1492. doi: 10.3748/wjg.v13.i10.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olgun E, Roberts LR. A new, effective and high-yield approach for identifying liver tumor suppressors. Genome Med. 2009;1:26. doi: 10.1186/gm26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoshida Y, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-β gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang SM, Ooi LL, Hui KM. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res. 2007;13:6275–6283. doi: 10.1158/1078-0432.CCR-06-2236. [DOI] [PubMed] [Google Scholar]
- 8.Zucman-Rossi J, Laurent-Puig P. Genetic diversity of hepatocellular carcinomas and its potential impact on targeted therapies. Pharmacogenomics. 2007;8:997–1003. doi: 10.2217/14622416.8.8.997. [DOI] [PubMed] [Google Scholar]
- 9.Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: Classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127(5 Suppl 1):S51–S55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 10.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasuda J, Hayashizaki Y. The RNA continent. Adv Cancer Res. 2008;99:77–112. doi: 10.1016/S0065-230X(07)99003-X. [DOI] [PubMed] [Google Scholar]
- 12.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gramantieri L, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 14.Fornari F, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 15.Ladeiro Y, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 16.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matouk IJ, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matouk IJ, et al. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur J Gastroenterol Hepatol. 2009;21:688–692. doi: 10.1097/meg.0b013e328306a3a2. [DOI] [PubMed] [Google Scholar]
- 19.Oliva J, et al. The regulation of non-coding RNA expression in the liver of mice fed DDC. Exp Mol Pathol. 2009;87:12–19. doi: 10.1016/j.yexmp.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panzitt K, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 21.Brosius J. Waste not, want not—transcript excess in multicellular eukaryotes. Trends Genet. 2005;21:287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 23.Bejerano G, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 24.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 25.Katzman S, et al. Human genome ultraconserved elements are ultraselected. Science. 2007;317:915. doi: 10.1126/science.1142430. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 27.Lujambio A, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mestdagh P, et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene. 2010;29:3583–3592. doi: 10.1038/onc.2010.106. [DOI] [PubMed] [Google Scholar]
- 29.Catucci I, et al. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 2009;30:544–545. doi: 10.1093/carcin/bgn289. author reply 546. [DOI] [PubMed] [Google Scholar]
- 30.Wojcik SE, et al. Non-coding RNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 2010;31:208–215. doi: 10.1093/carcin/bgp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marozin S, et al. Cell cycle progression or translation control is not essential for vesicular stomatitis virus oncolysis of hepatocellular carcinoma. PLoS ONE. 2010;5:e10988. doi: 10.1371/journal.pone.0010988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azechi H, et al. Disruption of the p16/cyclin D1/retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology. 2001;60:346–354. doi: 10.1159/000058531. [DOI] [PubMed] [Google Scholar]
- 33.Das SK, Hashimoto T, Kanazawa K. Growth inhibition of human hepatic carcinoma HepG2 cells by fucoxanthin is associated with down-regulation of cyclin D. Biochim Biophys Acta. 2008;1780:743–749. doi: 10.1016/j.bbagen.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Tsuji T, et al. Cyclin E overexpression responsible for growth of human hepatic tumors with p21WAF1/CIP1/SDI1. Biochem Biophys Res Commun. 1998;242:317–321. doi: 10.1006/bbrc.1997.7958. [DOI] [PubMed] [Google Scholar]
- 35.Li K, Lin SY, Brunicardi FC, Seu P. Use of RNA interference to target cyclin E-overexpressing hepatocellular carcinoma. Cancer Res. 2003;63:3593–3597. [PubMed] [Google Scholar]
- 36.Koga H, et al. Involvement of p21(WAF1/Cip1), p27(Kip1), and p18(INK4c) in troglitazone-induced cell-cycle arrest in human hepatoma cell lines. Hepatology. 2001;33:1087–1097. doi: 10.1053/jhep.2001.24024. [DOI] [PubMed] [Google Scholar]
- 37.Kawaji H, et al. The FANTOM web resource: From mammalian transcriptional landscape to its dynamic regulation. Genome Biol. 2009;10:R40. doi: 10.1186/gb-2009-10-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derti A, Roth FP, Church GM, Wu CT. Mammalian ultraconserved elements are strongly depleted among segmental duplications and copy number variants. Nat Genet. 2006;38:1216–1220. doi: 10.1038/ng1888. [DOI] [PubMed] [Google Scholar]
- 39.Gardiner EJ, Hirons L, Hunter CA, Willett P. Genomic data analysis using DNA structure: An analysis of conserved nongenic sequences and ultraconserved elements. J Chem Inf Model. 2006;46:753–761. doi: 10.1021/ci050384i. [DOI] [PubMed] [Google Scholar]
- 40.Czyzyk-Krzeska MF, Bendixen AC. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood. 1999;93:2111–2120. [PubMed] [Google Scholar]
- 41.Du Z, Fenn S, Tjhen R, James TL. Structure of a construct of a human poly(C)-binding protein containing the first and second KH domains reveals insights into its regulatory mechanisms. J Biol Chem. 2008;283:28757–28766. doi: 10.1074/jbc.M803046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimura K, Kano F, Murata M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA. 2008;14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nuovo GJ, et al. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4:107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.