Abstract
The tumor suppressor p53 slides along DNA while searching for its cognate site. Central to this process is the basic C-terminal domain, whose regulatory role and its coordination with the core DNA-binding domain is highly debated. Here we use single-molecule techniques to characterize the search process and disentangle the roles played by these two DNA-binding domains in the search process. We demonstrate that the C-terminal domain is capable of rapid translocation, while the core domain is unable to slide and instead hops along DNA. These findings are integrated into a model, in which the C-terminal domain mediates fast sliding of p53, while the core domain samples DNA by frequent dissociation and reassociation, allowing for rapid scanning of long DNA regions. The model further proposes how modifications of the C-terminal domain can activate “latent” p53 and reconciles seemingly contradictory data on the action of different domains and their coordination.
Keywords: recognition, response element, transcription factor
An essential transcription factor in multicellular organisms, the tumor suppressor p53 regulates cell-cycle arrest and apoptosis. Genetic alteration and mutations of p53 have been found in more than 50% of all human cancers (1). Most of these mutations affect the core domain responsible for recognition and binding to cognate sites on DNA. In contrast to many other well characterized transcription factors, p53 contains two distinct DNA-binding domains (Fig. 1). Whereas the core domain binds DNA in a sequence-specific fashion, the C-terminal domain of p53 interacts with DNA in a manner independent of the sequence (2–4) and is subject to multiple acetylations and phosphorylations that activate p53. How the two DNA-binding domains coordinate their actions and influence dynamics of p53 has been a subject of great interest and controversy.
Fig. 1.
Multidomain structure of p53 and constructs used in the study. Tumor suppressor p53, a 393-residue long protein, is composed of four different domains: The activation domain at the N-terminal end of the protein, the sequence-specific DNA-binding domain at the core of the protein, the tetramerization domain, and the nonspecific DNA-binding domain at the C-terminal end of the protein. p53 can bind DNA through both sequence-specific (core domain) and nonspecific (C-terminal domain) protein-DNA interactions.
Initial studies of the interactions between p53 and DNA suggested that the C-terminal domain of p53 negatively regulates the binding of core domain to its specific site on DNA. Hupp et al. reported that the C-terminal domain of p53 has a remarkable effect on the ability of the core domain to bind to its target site on DNA (5). The authors reported that the deletion of the C-terminal domain, phosphorylation of a serine residue in the C-terminal domain, or the use of an antibody (PAb 421) directed to the C-terminal domain increased the binding of the core domain to short DNA oligonucleotides. Further, phosphorylation and acetylation of various residues in the C-terminal domain leads to an increase in the binding of core domain in vitro (5). In addition, modifications of the C-terminal domain are widely observed in cells in which DNA damage has activated a p53 response (6). These observations led to the hypothesis that the nonspecific interaction of the C-terminal domain with the DNA interferes with the ability of core domain to bind to the cognate site until relevant signals cause modifications in the C-terminal domain, and alleviate its negative regulation of core DNA binding (4, 5, 7).
More recent studies showed that p53 requires its C-terminal domain for efficient recognition of the target site in long or circular DNA (8). Further, the C-terminal domain is important in one-dimensional translocation of the protein along DNA (8). McKinney et al. also demonstrated that the C-terminally truncated protein is considerably less efficient at binding and transactivating targets in vivo. Taken together, these results suggest a positive regulatory role of p53′s C-terminal domain for DNA binding (8).
Here we use single-molecule imaging tools to examine the complex role of the C-terminal domain in p53 recognition. We demonstrate that seemingly contradictory earlier studies can be reconciled into a comprehensive model of p53-DNA recognition if both kinetics of the search process and competition between specific and nonspecific binding are considered at high DNA concentrations present in the cell nucleus.
Central to this mechanism is the process by which p53 searches for its sites on long genomic DNA. Such a search process was suggested to constitute multiple rounds of three-dimensional diffusion and effectively one-dimensional sliding along DNA (9). Experimental studies of other, mostly bacterial, transcription factors have confirmed this mechanism in vitro and in vivo (10, 11). Key to this process is sliding along the duplex while bound to DNA nonspecifically. Because the C terminus binds DNA nonspecifically, it is implied to be responsible for mediating sliding of p53 along DNA. Importantly, an efficient search process requires fast translocation along DNA and optimal affinity for nonspecific DNA. Excessive affinity leads to sequestration of the protein by nonspecific DNA, while insufficient affinity makes sliding rounds short and search slow. Intriguingly, while the C terminus can provide sliding, it is the core domain that recognizes the cognate sequence (12–14). While structural studies have ruled out allosteric models of direct interactions between C terminus and core domains (15, 16), interplay between the two domains remains a subject of great interest.
Results
C-Terminal Domain of p53 Translocates on DNA Much Faster than the Full-Length p53, While the Core Domain Is Unable to Slide on DNA.
Aiming to understand the role of individual domains and to investigate the molecular mechanism underlying one-dimensional diffusion of p53 protein on DNA, we visualized and quantitatively characterized the motion of individual p53 proteins in vitro along flow-stretched DNA. In previously reported single-molecule studies, we visualized the interaction between fluorescently labeled p53 and DNA, and showed that the full-length p53 is capable of a diffusive translocation along DNA (17). In order to determine the role of the individual DNA-binding domains of p53 we perform single-molecule experiments on the following three constructs: the TC domain (tetramerization domain + C-terminal domain), the NCT domain (N-terminal domain + core domain + tetramerization domain) and the full-length p53 molecule. We fluorescently labeled NCT, TC, and full-length p53 constructs and used total internal reflection fluorescence (TIRF) microscopy to visualize their movement on flow-stretched lambda phage DNA molecules.
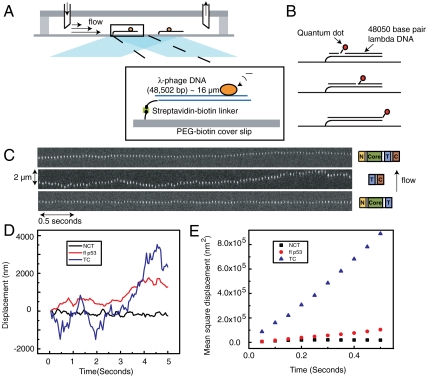
The 48.5-kb long double-stranded DNA was coupled at one end to the top surface of a microscope cover slip and hydrodynamically stretched by applying a laminar flow of aqueous buffer (Fig. 2A) (11, 17). The fluorescence emitted by the labeled protein was collected by a CCD camera and its position determined by fitting the fluorescence intensity profile to a two-dimensional Gaussian distribution (17). The protein positions for each captured frame were then tracked and linked to determine the trajectory of each protein.
Fig. 2.
Experimental set up and sliding trajectories of different p53 constructs. (A) Set up of the experiment. Several lambda-phage DNA molecules are attached to the glass bottom surface of a microfluidic flow cell through a streptavidin–biotin linker. The molecules are stretched by the drag force of a laminar flow of aqueous buffer through the flow cell. A laser is reflected off of the interface between the aqueous buffer and the glass surface generating an evanescent field decaying exponentially into the buffer. By mechanically stretching the DNA molecules within the evanescent field, the fluorophore labels of the p53 proteins on DNA can be selectively illuminated and excited while keeping the illumination from the background to a minimum. (B) To visualize the fluctuations due to DNA Brownian motion, quantum dots are attached to three different locations on lambda DNA, 14.7 kbp from the tethered point, 33,8 kbp from the tethered point, and at the end of the 48,5 kbp-long lambda DNA. (C) Kymograph of individual full-length p53, C-terminal domain (Tetramerization + C-terminal domain) and core domain (N-terminal + Core + Tetramerization domain) moving along flow-stretched DNA. All kymographs are superimposed on the same time and length axis. (D) Diffusion trajectory for the same three constructs. (E) MSD vs. time of the same trajectories.
Fig. 2C shows a time series of fluorescence images of representative full-length p53, the C-terminal, and the core domain of p53 moving along stretched DNA. Fig. 2D shows the trajectory of the same three protein constructs as determined from the images in Fig. 2C. Fig. 2E shows the Mean Square Displacement (MSD) vs. time for the three trajectories in two-dimensions. As can be seen from the trajectories and the MSD plots, the C-terminal domain is capable of translocating much faster on DNA than the full-length protein, while the core domain does not show a significant translocation on the same time scale.
To characterize dynamics of individual molecules, we measure diffusion coefficients of their one-dimensional sliding. A key challenge is to factor out drift due to the flow and fluctuations of the DNA itself. The drift was determined from individual trajectories (Fig. S1) and subtracted (see SI Text). To take into account DNA fluctuations, we developed a method that uses DNA-attached quantum dots as reference points. We attached quantum dots to three different locations on DNA (Fig. 2B; positions corresponding to one-third, two-thirds, and full length of the lambda phage DNA) and measured the trajectories of the quantum dots in the same flow condition as used in our sliding experiments. The corresponding MSD vs. t plots, both for the movement along (longitudinal) and perpendicular (transversal) to the direction of the flow, are shown in Fig. S2. The MSD of the DNA-bound quantum dots increases at short time scales, but remains constant in longer time scales as is expected for bounded diffusion.
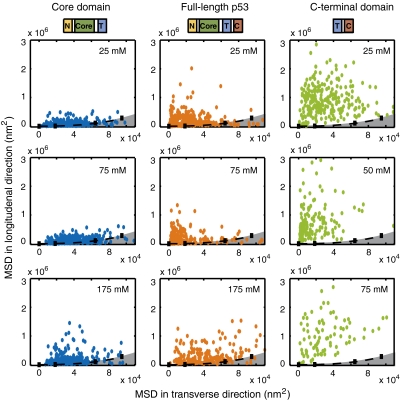
Fig. 3 compares the MSD at time t = 0.5 s of individual proteins (core domain, full-length protein, and C-terminal domain) in the longitudinal direction to its MSD in the transverse direction and in different salt concentrations. The black dashed line indicates the MSD of bound quantum dots on different locations on DNA and the gray area under the dashed line represents the intrinsic DNA fluctuations (Fig. S2). The MSDs obtained for the single-molecule trajectories using the core domain lie close to the MSD of the DNA fluctuations, suggesting that core domain is incapable of moving along DNA. But, for full-length p53 as well as the C-terminal domain, the MSD in the longitudinal direction is distinctly larger than that of the DNA fluctuations, suggesting diffusive movement of those constructs on DNA.
Fig. 3.
Mobility of different p53 constructs. MSD for drift-corrected trajectories of different constructs compared to MSD of DNA fluctuations in time 0.5 s and different salt concentration for core domain (NCT) (Blue), full-length p53 (Red), and C-terminal (TC) domain (Green) of p53. The gray area represents the MSD as a result of the DNA fluctuations measured by the quantum dot attached to DNA in both longitudinal and transverse direction. Full-length protein and C-terminal domain have higher MSD in the longitudinal direction, suggesting a one-dimensional diffusional translocation along DNA. For the core domain this is only the case at high salt concentration.
The diffusion coefficients for the three different protein constructs are determined at 75 mM total salt concentration after subtracting the effect of DNA fluctuation (see SI Text) and shown in Table 1. The C-terminal domain is capable of translocating much faster on DNA than the full-length protein, while the core domain displays a diffusion coefficient that is an order of magnitude smaller than either the full-length protein or the C-terminal domain.
Table 1.
Diffusion coefficient of different p53 constructs
| Mean diffusion coefficient ± Standard error of the mean in 75 mM total salt concentration | ||
| Full-length p53 | C-terminal domain (TC) | Core domain (NCT) |
| (1.62 ± 0.17) × 105 nm2/ sec | (7.76 ± 0.98) × 105 nm2/ sec | (2.39 ± 0.48) × 104 nm2/ sec |
| (1.40 ± 0.15) × 106 bp2/ sec | (6.71 ± 0.58) × 106 bp2/ sec | (2.07 ± 0.42) × 105 bp2/ sec |
Mean diffusion coefficient and standard error of the mean for full-length p53, TC domain, and NCT domain of p53. All measurements are in 75 mM total salt concentration. The C-terminal domain moves much faster on DNA than the full-length protein, while the core-domain is almost immobile on DNA.
The Core Domain of p53 Translocates on DNA via a Hopping Mechanism.
Two distinct mechanisms are suggested for a protein that diffuses along DNA—sliding and hopping. A sliding protein remains in contact with DNA while translocating along it. On the other hand, a hopping protein is suggested to make microscopic associations and dissociations on and off the DNA. To differentiate between these mechanisms, we use the dependence of the diffusion coefficient on the salt concentration in the solvent. It is expected that the diffusion coefficient of sliding is independent of the salt concentration. However, in the case of hopping, the fraction of time spent off DNA depends on protein affinity for nonspecific DNA. A higher salt concentration reduces proteins affinity for nonspecific DNA making it dissociate more frequently, thus spending more time in solution subject to three-dimensional diffusion and thus yielding a higher diffusion coefficient (18).
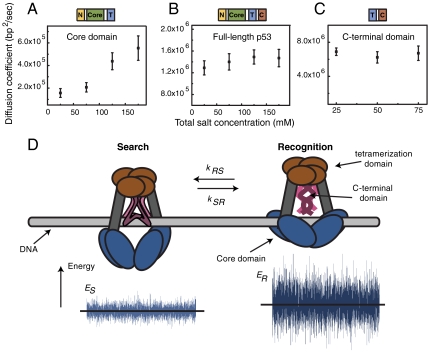
In earlier work, we observed that the diffusion coefficient of full-length p53 protein is independent of salt concentration. Those data suggested a sliding mechanism in which the protein moves along the DNA while maintaining constant contact with the duplex. To understand better the mechanism of sliding of the p53 protein on DNA, we measured the diffusion coefficient for the core domain, C-terminal domains, and full-length p53 in (total) salt concentrations ranging from 25 mM to 175 mM. Fig. S3 A–C shows the distributions of diffusion coefficients for the three constructs and Fig. 4 A–C shows the means and standard errors of the means of the distributions. The widths of the distributions reflect both uncertainties due to the short length of the photo-bleaching-limited trajectories and the intrinsic heterogeneity within the population of the studied single molecules. Also, the short length of some trajectories results in apparent negative diffusion coefficients. However, the large number of molecules present in the distributions allows us to determine their means with high precision. In particular, we are able to detect small shifts as a function of salt concentration. At low salt concentration, the diffusion coefficients of core domain are negligibly small, suggesting that under these conditions the core domain is effectively immobile on the DNA. At higher salt concentrations, the mean diffusion coefficient increases, suggesting a hopping mechanism for translocation of core domain along DNA. The residence time of the core domain on DNA can be calculated in different salt concentrations by comparing our observed experimental diffusion coefficient with that obtained for a protein freely diffusing in solution (see SI Text). These values suggest that the core domain spends only 10-4–10-3 of the total time in solution in 75 mM total salt concentration and thus is bound to the nonspecific DNA the majority of the time (> 99.9%).
Fig. 4.
Disentangling sliding and hopping: a two-state model of p53 search on DNA. (A) Diffusion coefficient of core domain increases with salt concentrations suggesting a hopping mechanism for the core domain along DNA. The error bars in the figure are the standard error of the mean. The diffusion coefficient for full-length protein (B) and C-terminal domain (C) stays constant with salt concentrations suggesting a sliding mechanism for these protein constructs. (D) Cartoon demonstrating the two different DNA-binding modes when the p53 protein is translocating along DNA. In the search mode, the protein is only bound to DNA with its C-terminal domain resulting in fast sliding along DNA. In the recognition mode, the protein is bound to DNA with both C-terminal and core domain resulting in a slower translocation along DNA. The energy landscape for recognition mode ER has a higher variance than the energy landscape of the search mode ES resulting in a slower rate of translocation along DNA. p53 combines the two different binding modes when searching for its target site on DNA.
The C-Terminal Domain of p53 Translocates on DNA via a Sliding Mechanism.
Fig. S3C shows distributions of the diffusion coefficient of the C-terminal domain diffusing along DNA in different salt concentration. Fig. 4C shows the dependence of the mean and standard error of the mean of the diffusion coefficient, on salt concentration. The diffusion coefficient of the C-terminal domain of p53 is independent of salt concentration and remains constant over the range of 25 mM to 75 mM total salt concentration, suggesting a sliding mechanism for translocation of the p53 C-terminal domain on DNA.
Discussion
The Sequence-Specific Core Domain of p53 Experiences a Rugged Energy Landscape, While the Nonspecific C-Terminal Domain Slides on a Smooth Energy Landscape.
Our single-molecule data suggest that both full-length p53 and its C-terminal domain diffuse rapidly along DNA. Further, we demonstrate that the core domain is essentially immobilized on nonspecific DNA. These observations suggest a model in which the p53 protein slides on DNA via its C-terminal domain. The core domain, however, does not constantly maintain contact with DNA, but rather stochastically associates and dissociates on and off the DNA.
Results of the single-molecule experiments are in very good agreement with the theory of one-dimensional/three-dimensional facilitated diffusion (9) (19) (20). First, our recent theoretical study (9) predicted that the diffusion coefficient of sliding depends strongly on the ability of the protein to bind DNA in a sequence-specific manner. A DNA-binding domain with a high sequence-specificity is predicted to experience strong sequence-dependent binding energy, even on noncognate DNA, and thus unable to slide along such rugged energy landscape. However, a domain that binds with moderate specificity (∼1 kBT sequence-specific energy) or nonspecifically is expected to have a relatively smooth sliding landscape and can slide fast (Fig. S4). In agreement with the theory, the C-terminal domain that binds DNA nonspecifically demonstrates a rapid translocation with σ = 0.6 kBT, where sigma is a measure of the sequence-specific binding energy, while the sequence-specific core domain diffuses very slowly with σ > 2 kBT (see SI Text).
Full-Length p53 Moves on DNA Through a Two-State Mechanism of Search and Recognition.
Theoretical studies (9, 20) suggest a two-state search mechanism, which provides a rationale for our single-molecule measurements and demonstrates how the DNA-binding domains of p53 are coordinated. The two-state mechanism suggests that both fast search and sequence-specific recognition can be achieved if the protein has two distinct conformational states: a search state characterized by largely nonspecific binding and fast sliding, and a recognition state in which a protein binds DNA in a sequence-specific manner while unable to slide. Simulations and analytical treatment demonstrated that the target site can be rapidly found and recognized if the protein spends most of the time in the search state while frequently interrogating DNA by going into the recognition state. These two states and fast transitions between them have been observed in a range of DNA-binding proteins (19, 21) and correspond to different conformations of the same DNA-binding domain. The multidomain structure of p53 allows it to distribute the roles of these two states between the two DNA-binding domains. We propose that, in tetrameric p53, the search state corresponds to a conformation in which the C termini are bound to DNA and the core domains are undocked, thus allowing for nonspecific binding and fast translocation. The recognition state, in turn, involves docking of the core domains to DNA and specific recognition (Fig. 4D). Conformational switching between the two states allows both sufficiently fast translocation and specific binding. In agreement with such a two-state mechanism, the rate of translocation of the full-length proteins is a factor of five lower than that of the C terminus construct because the protein spends a certain fraction of time in the immobile recognition conformation. Using the structure of p53 revealed by electron microscopy (16), the ratio of the corresponding rotational diffusion coefficients, controlled for increased size of the full-length p53 as compared to C terminus construct, can be estimated. This approximation results in an estimate of 40–50% for the fraction of time spent in the search state (see SI Text). Because analysis of the full-length single-molecule trajectories was focused on mobile particles, this estimate provides an upper bound.
It is also possible to estimate the minimal rate of the conformational transition in p53 required for fast search and specific binding. Using the measured diffusion coefficient for full-length p53 and in vivo time on DNA (22, 23), we obtain a rate constant of about 103 s-1 (see SI Text). Thus, if the conformational transition happens on a submillisecond time scale, the protein can efficiently search for its cognate site. This prediction may be tested by H/D-exchange or similar techniques (24).
Finally, we are able to relate our single-molecule measurements to published in vivo fluorescence recovery studies (22, 23) and p53 copy-number measurements (25) to calculate the time it takes p53 to find a specific site (e.g., p21) on DNA (see SI Text). Assuming that only about 5% of genomic DNA is accessible due to chromatinization and that about 1,000 copies of p53 are activated, we obtain a search time in the range of 3–30 min. This estimate is consistent with about an hour for initial expression of downstream genes (25). Moreover this reasoning suggests that the latent p53 with a lifetime of about 20 min and its slow oligomerization kinetics (26, 27) is unlikely to yield significant occupancy in tetrameric form at hundreds of target promoters. The search, however, is fast enough to allow a long-lived activated form (lifetime of ∼200 min) (28, 29) to bind most of target promoters.
Our framework of a one-dimensional/three-dimensional search process and our single-molecule data allow a reconciliation of seemingly contradictory studies of the role of the C terminus in p53 recognition. From a thermodynamic point of view, the C-terminal domain functions as a negative regulator for p53 by sequestering it onto nonspecific DNA. From a kinetic perspective, the C-terminal region functions as positive regulator for p53 by facilitating the search process. An optimal affinity is required for fast search and a stable cognate complex. This model explains how experimental alterations of the C-terminal domain have both positive and negative effects on p53 function. Truncation of the C terminus or binding by specific antibodies eliminates sequestration and leads to better binding to the cognate sites on short DNA fragments (5), while making binding to long DNA molecules kinetically inefficient. Sequestration to nonspecific DNA also explains the fact that long nonspecific DNA molecules inhibit binding of the full-length p53 to short cognate DNA molecules, but have no effect on C terminally truncated form (5). Moreover, modulation of affinity for nonspecific DNA can serve as a regulatory mechanism. For example, activation of p53 by acetylation of the C-terminal domain reduces its affinity for nonspecific DNA several fold, and thus can activate p53 by allowing rapid search for target sites.
In summary, we used single-molecule experiments to visualize and quantitatively characterize diffusive motion of individual p53 proteins along DNA molecules. We demonstrated that the C-terminal domain is nonspecifically bound to DNA and is capable of sliding very rapidly along DNA, while the full-length protein moves on DNA at a much slower rate. We demonstrated that single-molecule measurements are consistent with the theory of sliding, and the two-state mechanism of sliding/recognition (9, 20, 30) and proposed that while on DNA p53 rapidly interconverts between two conformations. This rapid switching allows the protein to sample sequences for specific, core-domain mediated binding, while enabling rapid search through the interaction between the C-terminal domain and DNA.
Materials and Methods
DNA Preparation and Flow Stretching.
Purified DNA from λ phage (New England Biolabs) was linearized and biotinylated at one end by annealing a 3′ biotin-modified oligo (5′AGGTCGCCGCCC3′-biotin; Integrated DNA Technologies) to the complementary λ-phage 5′ overhang. Flow cells (0.1 mm height, 2.0 mm width) with a streptavidin-coated surface were prepared as described previously (17, 31, 32). The streptavidin-coated flow-cell surfaces were blocked by incubation with blocking buffer (Tris 20 mM, EDTA 2 mM, NaCl 50 mM, BSA 0.2 mg/mL, Tween 20 0.005%; pH 7.5) for 20 min. Biotin-modified DNA constructs were introduced into the flow cell at a rate of 0.1 mL/ min at a concentration of 10 pM for 20 min. These conditions resulted in an average density of ∼100 surface-tethered DNA molecules per field of view (∼50 × 50 μm2).
The single-molecule imaging experiments were performed in an imaging buffer, containing 20 mM Hepes, 0.5 mM EDTA, 2 mM MgCl2, 0.5 mM DTT, 0.05 mg/mL BSA (pH 7.9), and varying amounts of KCl. Imaging buffer was drawn into the channel by a syringe pump at a flow rate of 0.1 mL/ min, creating shear flow near the coverslip surface (11). Single-molecule imaging was done with 30–100 pM TC (Tetramerization + C-terminal) p53 and 10–50 pM NCT (N-terminal + Core domain + Tetramerization) in imaging buffer. The proteins were kept at low-micromolar concentration, and were diluted right before the single-molecule experiment. The single-molecule experiments were done within less than 1 h from the time of dilution. Due to the slow kinetics of the tetramer-dimer transition (26), all constructs are assumed to be in the tetrameric form during the single-molecule experiment.
Protein Preparation and Labeling.
Expression and purification of TC.
P53 Tet + C (293–393) with an N-terminal cysteine was cloned in PET 24-HLTev using BamHI and EcoRI sites. The resulting plasmid encodes a fusion protein with an N-terminal 6xHis tag, followed by a lipoyl domain, a TeV protease cleavage site and the p53 Tet + C (293–393) sequence of interest. The proteins were expressed in Escherichia coli strain BL21 and purified by a Ni-affinity column followed by cleavage with TeV overnight. Subsequent purification by cation exchange chromatography on SP Sepharose and gel filtration on Superdex 75 yielded a purity of > 99% (33). To measure the oligomerization state of the TC domain, we measured the lifetime of different TC domains as well as only the C-terminal domain on DNA. The average lifetime of the C-terminal domain on DNA is 0.88 ± 0.05 seconds, whereas the TC domain has the lifetime of 2.41 ± 0.08 seconds on DNA (Fig. S5). Both experiments are done in 25 mM total salt concentration. Because the tetramerization domain does not interact with DNA, we conclude than the TC domain in our single-molecule experiment conditions must be a dimer or tetramer.
Labeling of TC.
The labeling was carried out in phosphate buffer (20 mM sodium phosphate, 150 mM NaCl, pH 7.0) with a protein concentration of 100 μM on ice. 10-fold excess Alexa Fluor 555 maleimide was added in the presence of 1 mM of tris(2-carboxyethyl) phosphine (TCEP). The labeling progress was followed by matrix assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The reaction was quenched with 10 mM β-mercaptoethenol after ∼1 h. The mixture was then loaded onto a G-25 desalting column to separate excess dye.
Purification and labeling of NCT.
The superstable mutant of NCT p53 (N-terminal + core domain + Tetramerization domain, residues 1–363) with mutations M133L, V203A, N239Y, and N268D (34) was used. The protein was expressed in Escherichia coli and purified as described (33, 35).
The NCT was labeled with Alexa Fluor 555 carboxylic acid succinimidyl ester (Invitrogen) through the N terminus amine. The labeling was carried out in phosphate buffer (50 mM sodium phosphate, 150 mM NaCl, pH 6.4). Alexa Fluor 555 of equal molarity was added to 1 mL of NCT solution (30 μM). The labeling progress was followed by MALDI-TOF MS. The reaction was quenched after about 1 h with 0.2 mL of 1 M Tris (hydroxymethyl) aminomethane-HCl (pH 7.4) and the labeled protein was separated from the free dye on a G-25 desalting column.
Supplementary Material
Acknowledgments.
A.T. thanks Dr. J. J. Loparo for his help with the quantum-dot experiments, and J.S. Leith for helpful discussions. A.M.v.O. acknowledges support from the National Science Foundation and the National Institutes of Health. L.A.M. acknowledges support by the National Cancer Institute through Physical Sciences in Oncology Center at MIT.
Footnotes
The authors declare no conflict of interest.
See companion article on page 557.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016020107/-/DCSupplemental.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Wu WJ, et al. Allelic frequency of p53 gene codon 72 polymorphism in urologic cancers. Jpn J Cancer Res. 1995;86:730–736. doi: 10.1111/j.1349-7006.1995.tb02461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberosler P, Hloch P, Ramsperger U, Stahl H. p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halazonetis TD, Kandil AN. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993;12:5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 6.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 7.Muller-Tiemann BF, Halazonetis TD, Elting JJ. Identification of an additional negative regulatory region for p53 sequence-specific DNA binding. Proc Natl Acad Sci USA. 1998;95:6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKinney K, Mattia M, Gottifredi V, Prives C. p53 linear diffusion along DNA requires its C terminus. Mol Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 9.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Schwedes JF, Parks D, Mann K, Tegtmeyer P. Interaction of p53 with its consensus DNA-binding site. Mol Cell Biol. 1995;15:2157–2165. doi: 10.1128/mcb.15.4.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayaraman L, Prives C. Covalent and noncovalent modifiers of the p53 protein. Cell Mol Life Sci. 1999;55:76–87. doi: 10.1007/s000180050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitayner M, et al. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Huang F, et al. Multiple conformations of full-length p53 detected with single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2009;106:20758–20763. doi: 10.1073/pnas.0909644106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tidow H, et al. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc Natl Acad Sci USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tafvizi A, et al. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys J. 2008;95:L01–03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 19.Kalodimos CG, et al. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 20.Mirny LA, Wunderlich Z, Tafvizi A, Leith J, Kosmrlj A. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J Phys A-Math Theor. 2009;42:434013. [Google Scholar]
- 21.Viadiu H, Aggarwal AK. Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 22.Mueller F, Wach P, McNally JG. Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J. 2008;94:3323–3339. doi: 10.1529/biophysj.107.123182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinow P, et al. The DNA binding activity of p53 displays reaction-diffusion kinetics. Biophys J. 2006;91:330–342. doi: 10.1529/biophysj.105.078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalodimos CG, Boelens R, Kaptein R. A residue-specific view of the association and dissociation pathway in protein—DNA recognition. Nat Struct Biol. 2002;9:193–197. doi: 10.1038/nsb763. [DOI] [PubMed] [Google Scholar]
- 25.Wang YV, et al. Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc Natl Acad Sci USA. 2007;104:12365–12370. doi: 10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natan E, Hirschberg D, Morgner N, Robinson CV, Fersht AR. Ultraslow oligomerization equilibria of p53 and its implications. Proc Natl Acad Sci USA. 2009;106:14327–14332. doi: 10.1073/pnas.0907840106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedler A, Veprintsev DB, Hansson LO, Fersht AR. Kinetic instability of p53 core domain mutants: implications for rescue by small molecules. J Biol Chem. 2003;278:24108–24112. doi: 10.1074/jbc.M302458200. [DOI] [PubMed] [Google Scholar]
- 28.McVean M, Xiao H, Isobe K, Pelling JC. Increase in wild-type p53 stability and transactivational activity by the chemopreventive agent apigenin in keratinocytes. Carcinogenesis. 2000;21:633–639. doi: 10.1093/carcin/21.4.633. [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Dhanwada KR, Birt DF, Hecht S, Pelling JC. Increase in p53 protein half-life in mouse keratinocytes following UV-B irradiation. Carcinogenesis. 1994;15:1089–1092. doi: 10.1093/carcin/15.6.1089. [DOI] [PubMed] [Google Scholar]
- 30.Hu L, Grosberg AY, Bruinsma R. Are DNA transcription factor proteins maxwellian demons? Biophys J. 2008;95:1151–1156. doi: 10.1529/biophysj.108.129825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JB, et al. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 32.van Oijen AM, et al. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg RL, Freund SM, Veprintsev DB, Bycroft M, Fersht AR. Regulation of DNA binding of p53 by its C-terminal domain. J Mol Biol. 2004;342:801–811. doi: 10.1016/j.jmb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 34.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc Natl Acad Sci USA. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veprintsev DB, et al. Core domain interactions in full-length p53 in solution. Proc Natl Acad Sci USA. 2006;103:2115–2119. doi: 10.1073/pnas.0511130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.