Abstract
Given its significant role in the maintenance of genomic stability, histone methylation has been postulated to regulate DNA repair. Histone methylation mediates localization of 53BP1 to a DNA double-strand break (DSB) during homologous recombination repair, but a role in DSB repair by nonhomologous end-joining (NHEJ) has not been defined. By screening for histone methylation after DSB induction by ionizing radiation we found that generation of dimethyl histone H3 lysine 36 (H3K36me2) was the major event. Using a novel human cell system that rapidly generates a single defined DSB in the vast majority of cells, we found that the DNA repair protein Metnase (also SETMAR), which has a SET histone methylase domain, localized to an induced DSB and directly mediated the formation of H3K36me2 near the induced DSB. This dimethylation of H3K36 improved the association of early DNA repair components, including NBS1 and Ku70, with the induced DSB, and enhanced DSB repair. In addition, expression of JHDM1a (an H3K36me2 demethylase) or histone H3 in which K36 was mutated to A36 or R36 to prevent H3K36me2 formation decreased the association of early NHEJ repair components with an induced DSB and decreased DSB repair. Thus, these experiments define a histone methylation event that enhances DNA DSB repair by NHEJ.
Keywords: double-strand break, I-Sce-I, chromatin immunoprecipitation, MRN complex, mathematical modeling
Histone methylation is highly regulated by a family of proteins termed histone methylases, which usually share a SET domain (1–3). Histone methylation plays a key role in chromatin remodeling and as such regulates transcription, replication, cell differentiation, genome stability, and apoptosis (1–3). Because of its role in replication and genome stability, histone methylation has been hypothesized to play an important role in DNA repair. DNA double-strand breaks (DSBs) are a cytotoxic form of DNA damage that disrupts many of the cellular functions regulated by histone methylation described above (4–6). Previous reports indicate that histone methylation may be important in DNA DSB repair by homologous recombination: The DSB repair component 53BP1, which is required for proper homologous recombination, is recruited to sites of damage by methylated histone H3 lysine 79 (H3K79) and histone H4 lysine 20 (H4K20) (7–9). However, neither H3K79 nor H4K20 methylation is induced by DNA damage (9), so other histone methylation events at sites of DNA damage have been sought. In addition, a mechanism by which histone methylation might regulate NHEJ DSB repair has yet to be defined. In this study, a survey of histone methylation events after DSB induction revealed that the major immediate H3 methylation event is H3K36me2.
Metnase is a DNA DSB repair component that is a fusion of a SET histone methylase domain with a nuclease domain and a domain from a member of the transposase/integrase family (10–14). We showed previously that Metnase enhances nonhomologous end-joining (NHEJ) repair of, and survival after, DNA DSBs, and that its SET domain was essential for this activity (10). We also found that Metnase directly dimethylated H3K36 in vitro (10). Because H3K36me2 is the major histone dimethylation event induced by the formation of DSBs, and Metnase enhances DSB repair, we hypothesized that the SET domain of Metnase mediated this histone methylation event. Because the SET domain of Metnase was required for its ability to enhance NHEJ DSB repair, we investigated whether Metnase directly methylated H3K36 at a DSB and how this histone methylation event functioned in DSB repair by NHEJ. To answer these questions, we generated a human cell system in which a single DSB is rapidly and synchronously induced in a defined sequence and this DSB is preferentially repaired by NHEJ. This unique cell system provided the template for analysis of repair protein recruitment to the DSB, and histone modification at that site, using chromatin immunoprecipitation. We found that (i) Metnase directly mediated the formation of H3K36me2 at the induced DSB, (ii) H3K36me2 recruited and stabilized other DNA repair components at the DSB, and (iii) H3K36me2 levels were proportional to DSB repair efficiency.
Results
Histone H3 Lysine 36 is Dimethylated at DSBs.
We screened H3 for the induction of dimethylation events after DSB induction with IR or etoposide using Western analysis. We found that of the methylation events assessed, H3K36me2 was the major event, and was rapidly induced after IR (Fig. 1A and Fig. S1). This histone modification is a general response to DSB formation because H3K36me2 was induced by IR to some extent in eight of eight cell lines tested (Fig. S2). However, we did not see any increase in H3K36 trimethylation after IR. To test whether H3K36me2 was associated with DSB formation and repair, and whether Metnase could mediate this methylation event, we generated a model human cell system in which a single DSB could be induced rapidly and efficiently within a defined unique sequence, where this DSB would preferentially be repaired by NHEJ. The human sarcoma cell line HT1080 was engineered to contain an I-SceI site in a single-copy puromycin acetyltransferase (puro) gene sequence (Fig. 1 and Fig. S3). By using adenoviral-mediated transduction of I-SceI endonuclease (15–17), DSBs were produced in 90% of cells within 60 min (Fig. 1 B and C and Figs. S3 and S4). Given that the puro sequence is integrated as a single copy, cells experience either a single DSB in one chromosome or two DSBs in sister chromatids, and these DSBs are likely to be preferentially repaired by NHEJ. This engineered model cell system was termed HT1904. These cells were further manipulated to over- or underexpress H3 methylase and demethylase activities.
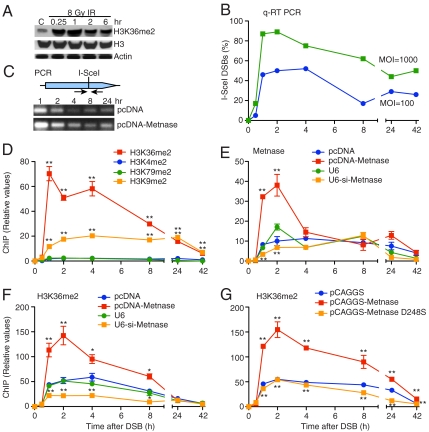
Fig. 1.
Metnase dimethylates H3K36 at DSBs. (A) Western blot of H3K36me2 after γ-radiation. (B) Quantitative real-time PCR analysis using PCR primers spanning the I-SceI site. Plotted are relative values calculated as the inverse of the total amount of amplified puro DNA compared to input GAPDH DNA for I-SceI adenovirus MOIs of 100 and 1,000. (C) Schematic of the HT1904 I-SceI DSB cell system (see also Figs. S3 and S4). (D) ChIP time course of methylated H3 species adjacent to a single induced DSB quantified by real-time PCR. Methylated H3K36 was not detected prior to DSB induction. For all ChIP and DSB repair experiments, each time point is the average of three quantitative real-time PCR measurements normalized to input DNA. All H3K36me2 ChIP data were also normalized to the presence of total H3 assessed by ChIP at the DSB. All data points include errors bars (SEM), but in many cases the error bars are smaller than data point symbols. Statistics were calculated for each time point vs. controls. In this and all subsequent figures, * indicates P ≤ 0.05 and ** indicates P ≤ 0.01. (E and F) ChIP analysis of Metnase and H3K36me2 adjacent to a single induced DSB. pcDNA-Metnase indicates Metnase overexpression, U6-si-Metnase indicates repression; pcDNA and U6 are empty vector controls. There was no detectable Metnase or H3K36me2 prior to DSB induction. (G) The Metnase D248S SET domain mutant prevents H3K36me2 formation at the DSB.
The presence of H3K36me2, Metnase, and other DNA repair components at the induced DSB in HT1904 cells can be quantified using ChIP analysis followed by real-time PCR. Functional ChIP primer targets were located within one nucleosome of the I-SceI DSB site (152 nt from the I-SceI site) in order to analyze events immediately adjacent to the DSB. We first examined whether H3K36me2 was induced at the single I-SceI DSB over a 42 h time course (Fig. 1D and Fig. S4). H3K36me2 was not present adjacent to the I-SceI site before DSB induction but was markedly induced within 1 h of DSB induction. Consistent with the Western blot results after IR (Fig. 1A and Fig. S1), other H3 methylation events were detected at the I-SceI DSB to a far lesser extent than H3K36me2 (Fig. 1D). Mathematical modeling the rate (dy/dt) of H3K36me2 association with the DSB region demonstrated that H3K36me2 appeared significantly faster than any of the other H3 methylation events (Figs. S6). H3K36me2 (and other H3 methylation events) were also assessed by ChIP analysis at a site 604 nt from the DSB (Fig. S3) but were nearly undetectable. Thus, H3K36 appears to be dimethylated only in the immediate vicinity of DSBs. We did find a decrease in total H3 present at the induced DSB over time, consistent with histone eviction, and therefore all H3K36me2 ChIP data is also normalized to total H3 at the DSB at each time point as well as input DNA. There was no detectable colocalization of H3K36me2 and γ-H2AX foci by confocal immunofluorescence microscopy, perhaps because of the limited region of H3K36 dimethylation at DSBs, especially when compared to the presence of H3K36me2 in chromatin generally.
Metnase Dimethylates H3K36 at DSBs.
Because Metnase dimethylates H3K36 in vitro (10), we used ChIP analysis to test whether Metnase was recruited to the induced DSB in HT1904 cells. Metnase indeed appeared at the DSB, and with similar kinetics as the appearance of H3K36me2. Overexpression of Metnase increased Metnase recruitment to the DSB region, while decreasing Metnase reduced its presence (Fig. 1E and Fig. S5). Like H3K36me2, Metnase was not detected adjacent to the DSB site before expression of I-SceI. Because both H3K36me2 and Metnase were present at the induced DSB, we tested whether altering Metnase levels could alter H3K36me2 levels at the DSB site. As shown in Fig. 1F, increasing Metnase levels enhanced the peak appearance of H3K36me2 at the DSB region, while decreasing Metnase levels decreased its appearance, implying that Metnase was responsible for H3K36 dimethylation at the DSB. Mathematical modeling of the ChIP data revealed a marked increase in the rate of Metnase and H3K36me2 association with the DSB 30–60 min after DSB induction, and increasing Metnase levels further increased the rate of Metnase and H3K36me2 appearance near the DSB (Fig. S7).
It was possible that H3K36 dimethylation was coincidental with Metnase association at DSBs, rather than being directly caused by Metnase. To test this we next determined whether the Metnase D248S SET domain mutant, which lacks the ability to promote NHEJ (10 and below) could still increase H3K36me2 at the DSB in HT1904 cells. Unlike overexpression of wild-type Metnase, overexpression of the D248S mutant blocked accumulation of H3K36me2 at the DSB (Fig. 1G), indicating that H3K36 dimethylation is directly catalyzed by Metnase near DSBs.
Metnase-Dependent Methylation of H3K36 at DSBs Enhances NHEJ Repair Component Recruitment and DSB Repair.
H3K36me2 has previously established roles in regulating gene transcription (18, 19), but potential roles for this histone modification in DSB repair by NHEJ were unknown. Because there is evidence that 53BP1 is recruited to DNA DSBs by methylated histones (7–9), we postulated that H3K36me2 might similarly recruit repair components to the DSB. We again induced the formation of H3K36me2 with IR, immunoprecipitated H2K36me2, and analyzed the immunoprecipitate for the presence of DNA DSB repair components (Fig. 2). We found that early acting NHEJ factors, such as NBS1 and Ku70, were present in the H3K36me2 immunoprecipitate, and their presence was induced by IR. In contrast, the homologous recombination repair component 53BP1 did not coimmunoprecipitate with H3K36me2 after IR, consistent with its recruitment by other methylated histones during DSB repair (7–9). The slight presence of NBS1 and Ku70 within the H3K36me2 immunoprecipitate before exposure to IR may be due to endogenous DSBs arising from internal sources, such as collapsed replication forks or oxidative damage (20, 21).
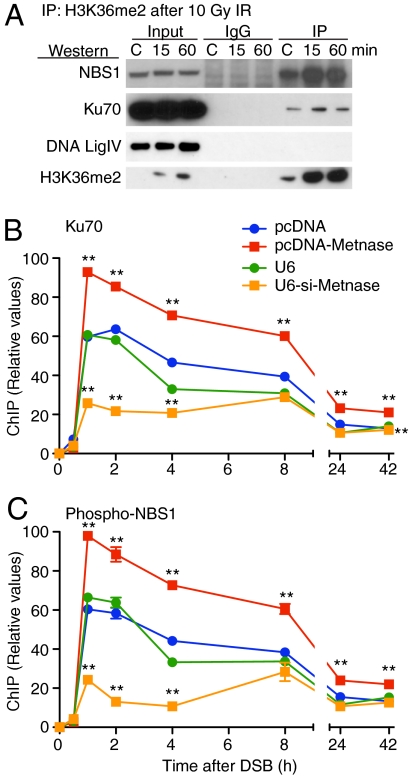
Fig. 2.
Coimmunoprecipitation of DNA repair components with H3K36me2 after IR. (A) Immunoprecipitates (IP) of H3K36me2 were collected from HT1904 cells before (C) and 15 or 60 min after 10 Gy γ-radiation and analyzed by Western blot for NBS1, Ku70, DNA ligase IV; input, nonspecific IgG, and H3K36me2 controls are also shown. ChIP analysis of early DNA repair components at a single induced DSB. Increasing Metnase levels enhances the presence of Ku70 (B) and phospho-NBS1 (C) at the single DSB, while repressing Metnase reduced the presence of these proteins. Neither Ku70 nor phospho-NBS1 were detected at the target DSB site prior to DSB induction (B and C).
Based on these findings, we used ChIP to examine whether Metnase levels regulated the recruitment of Ku70 and phosphorylated NBS1 to the region adjacent to an induced DSB in HT1904 cells. We found that increasing Metnase, which elevates H3K36me2 levels adjacent to the induced DSB (Fig. 1F), enhanced both the peak amount of these repair components at the induced DSB region (Fig. 2 B and C) and their rate of association with the DSB (Fig. S6). Repressing Metnase levels decreased both the peak amount and the rate of association of Ku70 and phospho-NBS1 with the region adjacent to the I-SceI-induced DSB (Fig. 2 and Fig. S6). In addition, mathematical analysis of the ChIP data revealed that repressing Metnase increased the rate of disassociation of Ku70 and NBS1 from the DSB (Fig. S7).
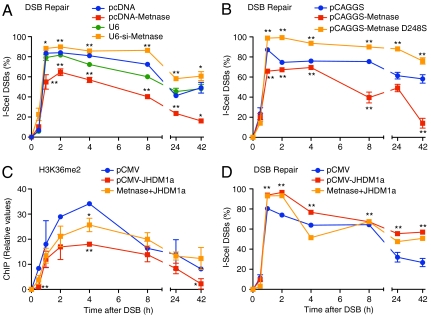
These changes in H3K36 dimethylation and NHEJ repair factor recruitment correlate with DSB repair efficiency. Overexpression of Metnase increased DSB repair, and reducing Metnase with siRNA decreased DSB repair (Fig. 3A) as analyzed using real-time PCR with primers spanning the I-SceI site (Fig. 1 B and C). Importantly, DSB repair was also decreased in cells expressing the SET domain mutant Metnase D248S (Fig. 3B), implying that Metnase-dependent dimethylation of H3K36 regulates DSB repair efficiency. There is less DSB induction when high levels of Metnase are present, likely from more rapid repair of the DSB before the first time point is measured.
Fig. 3.
Dimethylation of H3K36 enhances DSB repair. (A) Repair of a single DSB measured by real-time quantitative PCR in cells over- or underexpressing Metnase; values are inverse percentages of input (uncut) DNA prior to DSB induction and normalized to GAPDH (input) controls. Statistics calculated at each time point for over- or underexpression vs. cognate empty vector controls. (B) As in A but with cells expressing wild-type or D248S (SET-defective) Metnase. (C and D) Overexpression of JHDM1a demethylase reduces H3K36me2 presence at an induced DSB (C) and reduces DSB repair (D).
Recently, a group of histone demethylases has been described that share a Jumonji domain (22). The Jumonji domain protein JHDM1a is a conserved, H3K36me2-specific demethylase (23). We therefore examined whether overexpression of JHDM1a (Fig. S8) could alter the presence of H3K36me2 at the induced DSB in HT1904 cells. As shown in Fig. 3C, overexpression of JHDM1a reduced formation of H3K36me2 at the induced DSB site. In addition, JHDM1a overexpression slowed the ability of Metnase to induce H3K36me2 at the induced DSB site (Fig. 3C) and inhibited the ability of Metnase to enhance DSB repair (Fig. 3D). Thus, JHDM1a reduced the effect Metnase had on histone methylation and repair at the induced DSB.
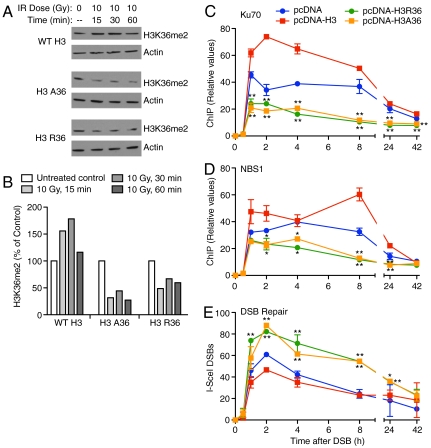
The formation of H3K36me2 at the DSB may only be correlated with the enhanced association of early NHEJ components with the DSB and not the direct cause of their association. Therefore, we next examined the effect of overexpressing mutant versions of histone H3 that lack the K36 target site for Metnase (R36 and A36) on the recruitment of NBS1 and Ku70 to the region adjacent to an induced DSB. We expected that cells expressing H3A36 and H3R36 species in competition with endogenous wild-type H3 will have decreased formation of H3K36me2 after DSB induction. Indeed, consistent with the original observation of increased H3K36me2 after IR (Fig. 1A and Fig. S1), cells overexpressing wild-type H3 showed IR-induced H3K36me2, but this was not the case in cells expressing either H3A36 or H3R36 (Fig. 4 A and B). Importantly, overexpression of either mutant H3 in HT1904 cells markedly decreased recruitment of Ku70 and NBS1 to the DSB (Fig. 4 C and D) and significantly decreased DSB repair (Fig. 4E). This is consistent with a requirement of H3K36me2 formation for maximal NHEJ repair of the DSB.
Fig. 4.
Expression of H3A36 and H3R36 mutant proteins limits Ku70 and NBS1 recruitment to DSBs and decreases DSB repair. (A) HT1904 cells transfected with vectors expressing WT H3, H3R36, or H3A36 were treated with 10 Gy IR or untreated and analyzed for H3K36me2 by Western blot. (B) Quantification of H3K36me2 signals in A normalized to actin loading controls. (C–E) ChIP analysis of Ku70 and NBS1, and DSB induction and repair in HT1904 cells expressing wild-type or mutant histone H3, or empty vector control.
Discussion
There are several advantages to the HT1904 cell system used here that might make it generally applicable to NHEJ DNA repair investigation. The DSB can be generated using adenoviral transduction of the I-SceI nuclease, rapidly inducing a single, defined DSB in the vast majority of cells (Fig. 1 B and C and Figs. S3 and S4). This allows ChIP analysis over time to monitor protein/DNA association, which can be mathematically modeled to provide information on cascades of repair components at the DSB. Repair in this system should occur preferentially by the NHEJ pathway, because when a sister chromatid template is present there is a high likelihood that both sister chromatids will suffer a DSB. However, the system can be modified for study of homologous recombination repair by addition of a second copy of puro lacking an I-SceI to serve as a donor locus during repair.
In this study, Western analysis showed that H3K36me2 was markedly induced after DSBs were induced by IR, and ChIP analysis showed that H3K36me2 is formed at a defined, nuclease-induced DSB. These data imply that H3K36me2 marks the local presence of a DSB. The finding that DSB-induced H3K36me2 levels correlate with Metnase expression levels and that the Metnase SET domain mutant (D248S) repressed generation of H3K36me2 indicates that Metnase is directly responsible for the induction of H3K36me2 at the DSB. We had previously shown that the D248S SET mutant of Metnase fails to promote NHEJ of a transfected plasmid substrate, and the data here indicate that Metnase promotes chromosomal DSB repair and that the D248S mutant suppresses chromosomal DSB repair due to its inability to methylate H3K36.
We had initially hypothesized that the formation of H3K36me2 at the DSB might improve histone eviction at the DSB and enhance access to the DSB by repair complexes. However, this was not the case; we did not observe alterations in histone H3 occupancy with increased or decreased Metnase levels. It is possible that we did not assay at time points early enough to see a difference in histone eviction from altered Metnase. We found that H3K36me2 enhances the presence of MRN complex components and Ku70 at the induced DSB. These DNA repair proteins show an increased interaction with H3K36me2 after IR, and their presence at an induced DSB also correlated with Metnase levels. In addition, mathematical modeling of the ChIP data revealed that H3K36me2 not only enhances the rate of association of these repair proteins with the DSB but decreases their disassociation rates as well. Because the MRN and Ku complexes can bind free DNA ends at a DSB in nonchromatinized DNA, the decreased rates of disassociation are likely the more important role of H3K36me2, as opposed to increased association rates. This implies that the main benefit of H3K36me2 in DSB repair is more likely to stabilize these repair components at the DSB than to enhance their recruitment.
The possibility existed that the induction of H3K36me2 at DSBs was an epiphenomenon and was not responsible for enhanced localization of early DSB repair components. However, when H3K36 was mutated to H3R36 or A36, there was a marked decrease in both the recruitment of NBS1 and Ku70 to the DSB (Fig. 4 C and D) and in DSB repair (Fig. 4E), indicating that reduced substrate (H3K36) availability suppresses repair factor recruitment and DSB repair, and demonstrating that H3K36me2 is required for efficient assembly/retention of repair components at DSBs and for optimum DSB repair. The identification of dimethylated H3K36 as a chromatin modification that enhances DSB repair by NHEJ places this modification alongside phosphorylated H2Ax and ubiquitylated H2A as DNA damage-induced histone modifications that recruit repair components to DSBs and enhance repair (24–28). In this regard, H3K36 methylation by Metnase and demethylation by JHDM1a is consistent with an NHEJ histone code, as defined in the original histone code hypothesis for transcriptional regulation as histone modifications, acting in a combinatorial fashion on histones, which specify unique downstream functions (29). We believe that H3K36me2 is reserved for NHEJ, because Ku70 and Metnase are involved in DSB repair by NHEJ rather than homologous recombination and because the latter requires complete histone eviction adjacent to the DSB.
Finally, this study defines a specific mechanism by which Metnase enhances DSB repair. We have found that human cancer cells that express Metnase at high levels display enhanced resistance to treatment with radiation or chemotherapy. Thus targeting Metnase may improve the response to these modalities (30, 31). The resistance mediated by Metnase could reflect improved stabilization of the assembly of DSB repair components at DSB sites due to the generation of H3K36me2 at these sites. Specific targeting of Metnase methylase activity may improve the efficacy of common cancer therapies based on DNA damaging agents.
Materials and Methods
Cell Line Construction.
HT1904 cells were derived from the human sarcoma cell line HT1080 by stable transfection of a linearized vector containing the puromycin acetyltransferase (puro) gene with a phosphogylcerol kinase promoter and containing a single I-SceI site (TGGTTCCTGGATTACCCTGTTATCCCTACGCGCCGGGG, where the I-SceI site is shown in bold italics flanked by puro sequence). This vector contains a blasticidin resistance cassette for selection (Fig. S3).
I-SceI Adenovirus Generation and Quantification.
The adenovirus expressing I-SceI was a generous gift from K. Valerie (17). The virus was propagated using AD293 cells as previously described (15–17). Unless otherwise specified, HT1904 cells were exposed to the I-SceI adenovirus at an MOI of 1,000 for 3 h and then washed three times in media to remove the adenovirus (Figs. S3, S4, and S5). For all experiments using adenoviral I-SceI, time 0 was before adenoviral infection, and subsequent time points were after adenovirus was removed by washing. Real-time PCR analysis measuring DSB repair was performed using primers spanning the I-SceI site (Table S1). Percent DSBs were calculated at each time point after adenoviral I-SceI infection relative to uninfected control and normalized to real-time PCR values for input GAPDH DNA (Fig. 1C).
Manipulation of Metnase Expression.
Cells overexpressing V5-tagged Metnase were generated by electroporation with pcDNA-Metnase, and cells underexpressing Metnase were generated by electroporation with U6-siRNA Metnase as described (10). For each experiment Western blot analysis was performed to ensure correct up- or down-regulation of Metnase protein (Fig. S5).
Coimmunoprecipitation.
HT1904 cells were treated with 10 Gy of γ-radiation from a 137Cs source and allowed to recover for 15 or 60 min. Total protein lysates were prepared from cells at each of these time points, as well as from untreated HT1904 cells. Proteins were analyzed by Western blot using the following primary antibodies: anti-H3K36me2 (Abcam), anti-phospho-NBS1(Ser343), and anti-NBS1 (Cell Signaling); anti-Ku70 (BD Biosciences); and anti-DNA Ligase IV (Genway).
Chromatin Immunoprecipitation (ChIP).
ChIP was performed using primers targeting sites 152 and 650 bp from the DSB (Table S1) before I-SceI adenovirus infection and at 0.5, 1, 2, 4, 8, 24, and 42 h after removal of the I-SceI adenovirus. Triplicate plates of 107 exponentially growing HT1904 cells per experimental condition per time point were washed with PBS and incubated for 10 min with 1% formaldehyde. After quenching reactions with 0.125 M glycine, cells were harvested by centrifugation, cells were resuspended in 2 mL lysis buffer (0.1 M PIPES pH 8, 1 M KCl, 10% NP-40), incubated on ice for 30 min, and disrupted by douncing 10 times. Nuclei were centrifuged for 10 min at 4 °C and resuspended in 1 mL of nuclear lysis buffer without EDTA (50 mM Tris–HCl, pH 8, 0.5% deoxycholic acid). DNA digestion was performed with 40 U of micrococcal nuclease (MNase I, New England Biolabs) at room temperature for 15 min. Digestion was stopped by placing the reaction at 4 °C and adding EDTA to a final concentration of 20 mM. ChIP assays were performed with 3 μg of the following antibodies: anti-Metnase (10), anti-H3K36me2 (Abcam), anti-Ku70 (Cell Signaling), anti-phospho-NBS1 (ser343, Cell Signaling), anti-NBS1 (Cell Signaling, for coimmunoprecipitation), antiphospho H2Ax (Millipore), anti-H3K79me2 (Millipore), anti-H3K9me2 (Cell Signaling), anti-H3K4me2 (Cell Signaling), and anti-H3K27me2 (Cell Signaling). Formaldehyde cross links were reversed by adding NaCl to a final concentration of 0.3 M followed by incubation at 65 °C overnight with RNase A (10 μg/μL), then at 50 °C for 3 h with Proteinase K at 1 μg/μL. DNA was purified using Qiagen purification kits and electrophoresed on a 2% agarose gel to ensure product. DNA associated with immunoprecipitated protein was then quantified using real-time PCR. There was no detectable Metnase, H3K36me2, phospho-NBS1, Ku70, or DNA Ligase 4 adjacent to the I-SceI site prior to DSB induction. For all ChIP experiments, each time point is the average of three distinct measurements repeated at least twice, normalized to input DNA GAPDH. Data for H3K36me2 were also normalized to total H3 assessed by ChIP at the same site.
Real-time PCR.
Quantitative real-time PCR was performed using SYBR green reagent (Applied Biosystems) with the ABI PRISM 7000 Sequence Detection System (Table S1). All experimental values were normalized to the input DNA using amplification of GAPDH.
Mathematical Modeling.
In order to compare the rates of appearance or disappearance of repair proteins at the DSB site for the different experimental conditions, we constructed a FORTRAN computer program that used piecewise cubic interpolating polynomials to fit the experimental data (32). To avoid oscillation in the resulting fits, we used a cubic Hermite formulation designed to preserve monotonicity in each interval (32–34). FORTRAN routines from the Netlib collection of mathematical software (www.netlib.org) (34) were used to generate the monotone piecewise cubic Hermite interpolant to the experimental data. Using these routines we computed the Hermite function and its temporal derivative at 1 min intervals over the entire experimental period of 42 h.
Supplementary Material
Acknowledgments.
The authors acknowledge K. Valerie for the kind gift of the adenovirus I-SceI system, the support of National Institutes of Health (NIH) Grant R01 GM084020 and NIH Grant R01 CA100862 (J. N.), APRC Supplement CA100862 (J.N. and R.H.), NIH Grant R01 CA102283 (R.H.), NIH Grant R01 HL075783 (R.H.), NIH Grant R01 CA139429 (R.H.), and Leukemia and Lymphoma Society Grant SCOR 7388-06 (R.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013571108/-/DCSupplemental.
References
- 1.Trievel RC. Structure and function of histone methyltransferases. Crit Rev Eukar Gene. 2004;14:147–169. doi: 10.1615/critreveukaryotgeneexpr.v14.i3.10. [DOI] [PubMed] [Google Scholar]
- 2.Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18:159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng JC, Karpen GH. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 2009;5:e1000435. doi: 10.1371/journal.pgen.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munoz IM, Rouse J. Control of histone methylation and genome stability by PTIP. EMBO Rep. 2009;10:239–245. doi: 10.1038/embor.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oda H, et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 10.Lee SH, et al. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci USA. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaheen M, Williamson EA, Nickoloff JA, Lee S-H, Hromas R. Metnase/SETMAR: A domesticated primate transposase that enhances DNA repair, replication, and decatenation. Genetica. 2010;138:559–566. doi: 10.1007/s10709-010-9452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hromas R, et al. The human set and transposase domain protein Metnase interacts with DNALigase IV and enhances the efficiency and accuracy of non-homologous end-joining. DNA Repair. 2008;7:1927–1937. doi: 10.1016/j.dnarep.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roman Y, et al. Biochemical characterization of a SET and transposase fusion protein, Metnase (SETMAR) for its DNA binding and DNA cleavage activity. Biochemistry. 2007;46:11369–11376. doi: 10.1021/bi7005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck BD, et al. Human PSO4 is a Metnase (SETMAR) binding partner that regulates Metnase function in DNA repair. J Biol Chem. 2008;283:9023–9030. doi: 10.1074/jbc.M800150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golding SE, et al. Double strand break repair by homologous recombination is regulated by cell cycle-independent signaling via ATM in human glioma cells. J Biol Chem. 2004;279:15402–15410. doi: 10.1074/jbc.M314191200. [DOI] [PubMed] [Google Scholar]
- 16.Anglana M, Bacchetti S. Construction of a recombinant adenovirus for efficient delivery of the I-SceI yeast endonuclease to human cells and its application in the in vivo cleavage of chromosomes to expose new potential telomeres. Nucleic Acids Res. 1999;27:4276–4281. doi: 10.1093/nar/27.21.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valerie K, et al. Improved radiosensitization of rat glioma cells with adenovirus-expressed mutant herpes simplex virus-thymidine kinase in combination with acyclovir. Cancer Gene Ther. 2000;7:879–884. doi: 10.1038/sj.cgt.7700185. [DOI] [PubMed] [Google Scholar]
- 18.Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris SA, et al. Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryot Cell. 2005;4:1446–1454. doi: 10.1128/EC.4.8.1446-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: How to fix a broken relationship. Cell Mol Life Sci. 2009;66:1039–1056. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartlerode AJ, Scully R. Mechanisms of double-strand break repair in somatic mammalian cells. Biochem J. 2009;423:157–168. doi: 10.1042/BJ20090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anand R, Marmorstein R. Structure and mechanism of lysine-specific demethylase enzymes. J Biol Chem. 2007;282:35425–35429. doi: 10.1074/jbc.R700027200. [DOI] [PubMed] [Google Scholar]
- 23.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 24.Escargueil AE, Soares DG, Salvador M, Larsen AK, Henriques JA. What histone code for DNA repair? Mutat Res. 2008;658:259–270. doi: 10.1016/j.mrrev.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Sedelnikova OA, Pilch DR, Redon C, Bonner WM. Histone H2AX in DNA damage and repair. Cancer Biol Ther. 2003;2:233–235. doi: 10.4161/cbt.2.3.373. [DOI] [PubMed] [Google Scholar]
- 26.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 29.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 30.Wray J, et al. Metnase mediates resistance to topoisomerase II inhibitors in breast cancer cells. PLoS ONE. 2009;4:e5323. doi: 10.1371/journal.pone.0005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wray J, et al. Metnase mediates chromosome decatenation in acute leukemia cells. Blood. 2009;114:1852–1858. doi: 10.1182/blood-2008-08-175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes 3rd Edition: The Art of Scientific Computing. New York: Cambridge Univ Press; 2007. [Google Scholar]
- 33.Fritsch FN, Carlson RE. Monotone piecewise cubic interpolation. SIAM J Numer Anal. 1980;17:238–246. [Google Scholar]
- 34.Fritsch FN, Butland J. A method for constructing local monotone piecewise cubic interpolants. SIAM J Sci Stat Comp. 1984;5:300–304. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.