Abstract
Double-stranded RNA induces a post-transcriptional gene silencing process, termed RNAi, in diverse organisms. It is shown here that transcriptional gene silencing accompanied by de novo methylation of a target promoter in plants can be triggered by a double-stranded RNA containing promoter sequences. Similar to the double-stranded RNA involved in RNAi, this promoter double-stranded RNA, which is synthesized in the nucleus, is partially cleaved into small RNAs ∼23 nucleotides in length. Both transcriptional and post-transcriptional gene silencing can thus be initiated by double-stranded RNAs that enter the same degradation pathway. The results also implicate double-stranded RNA in directing DNA methylation. Different constructs designed to produce double-stranded promoter RNA in various ways were evaluated for their ability to induce gene silencing in tobacco and Arabidopsis. RNA hairpins transcribed from inverted DNA repeats were the most effective trans-acting silencing signals. This strategy could be useful for transcriptionally downregulating genes in a variety of plants.
Keywords: DNA methylation/double-stranded RNA/RNAi/small RNAs/transcriptional gene silencing
Introduction
RNA interference (RNAi) is a post-transcriptional gene silencing (PTGS) process in which double-stranded (ds) RNA induces the degradation of homologous RNA sequences (Bass, 2000; Bosher and Labouesse, 2000; Sharp and Zamore, 2000). First discovered in Caenorhabditis elegans (Fire et al., 1998), RNAi is similar to PTGS phenomena in plants (Kooter et al., 1999) and to quelling in Neurospora (Cogoni and Macino, 1999a). The relationship between these various types of PTGS has been confirmed recently by the analysis of mutants, which has revealed that these processes require some of the same gene products in different organisms (Cogoni and Macino, 1999b,c; Ketting et al., 1999; Tabara et al., 1999; Dalmay et al., 2000; Mourrain et al., 2000; Smardon et al., 2000).
RNAi and other PTGS phenomena require sequence homology in protein-coding or transcribed regions. In plants, transcriptional gene silencing (TGS) resulting from sequence homology in promoter regions has also been observed and correlated with increased promoter methylation (Kooter et al., 1999). Methylation associated with PTGS in plants is acquired in transcribed or coding regions, where its role is still unclear. Because the DNA methylation changes seen in both types of silencing are confined largely to regions of sequence homology between interacting genes, sequence-specific methylation signals consisting of either DNA–DNA or RNA–DNA associations are believed to be involved.
Precedents for DNA–DNA pairing as a trigger for DNA modifications are provided by the RIP (repeat-induced point mutation) and MIP (methylation induced pre-meiotically) phenomena in the filamentous fungi Neurospora crassa and Ascobolus immersus, respectively. While RIP produces mutations in duplicated sequences, MIP is a purely epigenetic process involving pairing-dependent silencing and methylation of duplications longer than ∼400 bp (Faugeron, 2000). The pairwise modification of simple sequence duplications in these fungi contrasts to the situation in plants, where most of the evidence for somatic DNA pairing that modifies gene expression has been obtained with complex, repetitive loci (Luff et al., 1999). DNA pairing might nevertheless provide a signal for de novo methylation in plants considering the recent finding of an Arabidopsis Masc1 type of methyltransferase (Finnegan and Kovac, 2000), which is required for MIP in Ascobolus (Faugeron, 2000).
RNA-directed DNA methylation (RdDM) was first discovered with a viroid system in plants (Wassenegger, 2000). Viroids are plant pathogens consisting solely of non-coding, highly base-paired RNAs that are replicated in the nucleus by the host RNA polymerase II. Viroid cDNA copies integrated into the tobacco genome become methylated only during viroid replication (Wassenegger et al., 1994). These results suggested that viroids and/or their replication intermediates, which have extensive secondary structure, are directly involved in inducing methylation of the homologous cDNA copies. RdDM has also been observed with silencing systems based on cytoplasmically replicating RNA viruses (Jones et al., 1998, 1999). Following virus infection, nuclear transgenes homologous to viral sequences became methylated, suggesting that viral RNAs present in the cytoplasm entered the nucleus and triggered DNA methylation.
The first non-pathogenic system demonstrating RdDM involved a TGS phenomenon in which nopaline synthase promoter (NOSpro) sequences were intentionally transcribed by the cauliflower mosaic virus 35S promoter to produce NOSpro RNAs. In one transgenic tobacco line, a non-polyadenylated NOSpro RNA that deviated from the expected size was able to induce methylation and transcriptional inactivation of homologous NOSpro copies in trans (Mette et al., 1999). A requirement for this aberrant NOSpro RNA was established by suppressing its synthesis by introducing the 271 locus, a general repressor of 35S promoter-driven genes (Vaucheret, 1993), which alleviated silencing and reduced methylation of target NOSpros (Mette et al., 1999). Because the aberrant RNA was synthesized from an inverted DNA repeat (IR) containing NOSpro sequences, it was hypothesized that it might be double stranded. The involvement of dsRNA would be consistent with RdDM in viroid and virus systems, where dsRNAs either constitute the genome and/or are produced during genome replication. Moreover, the involvement of dsRNA in directing DNA methylation would indicate that this molecule acts not only in the cytoplasm to initiate the RNA degradation step of PTGS (Kooter et al., 1999) but also at the genome level to induce epigenetic modifications. dsRNA might thus be a common molecular component of the mechanisms of PTGS and at least some cases of TGS. Here, these hypotheses are tested by examining whether the NOSpro RNA involved in trans-silencing and methylation is indeed double stranded. We have also assessed additional strategies for generating nuclear NOSpro dsRNA that could potentially induce silencing of NOSpro-driven target genes in tobacco and Arabidopsis.
Results
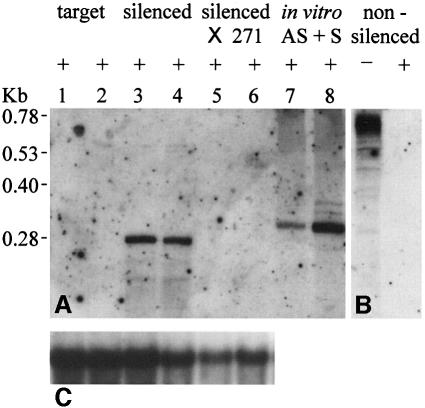
In previous work, aberrant RNA transcribed by the 35S promoter from an IR containing NOSpro sequences was able to trigger methylation and transcriptional silencing of unlinked copies of the NOSpro in trans (Figure 1B; Mette et al., 1999). To test whether this RNA was double stranded, which would result from uninterrupted transcription through the entire IR, RNA prepared from silenced and non-silenced plants was treated with RNase One, an enzyme that degrades single-stranded RNA (ssRNA) while leaving dsRNA intact. In the non-silenced line, NOSpro RNA was rapidly degraded by this enzyme (Figure 2B). In contrast, an RNase One-resistant dsRNA of the expected size was present in plants from the silenced line (Figure 2A, lanes 3 and 4). These clear bands representing dsRNA were in contrast to the smears of NOSpro RNA observed previously in untreated preparations (Mette et al., 1999), suggesting that untreated NOSpro dsRNAs contained variable lengths of overhanging single-stranded regions. The NOSpro dsRNA was not detectable in the original target line (Figure 2A, lanes 1 and 2) or in the presence of the 271 locus, which transcriptionally represses the 35S promoter (Figure 2A, lanes 5 and 6). This result is consistent with the loss of methylation and reactivation of the target NOSpro in the presence of the 271 locus (see Mette et al., 1999).
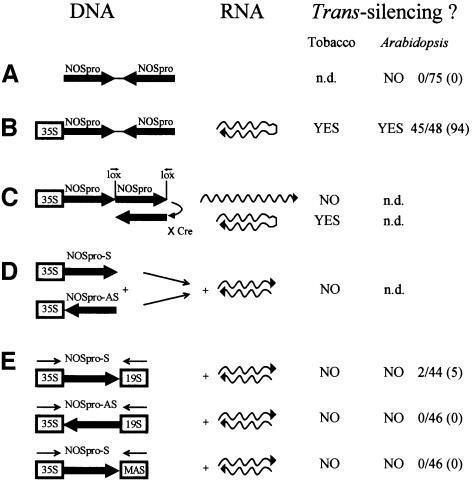
Fig. 1. DNA constructs designed to produce NOSpro dsRNA and their effectiveness in different plants. NOSpro IR without (A) and with (B) a transcribing 35Spro, which should produce a NOSpro hairpin RNA. (C) Conversion of a transcribed NOSpro DR into an IR via Cre recombinase activity (Odell and Russell, 1994) results in a switch from synthesis of a single-stranded NOSpro RNA to an RNA hairpin. (D) Synthesis of separate sense and antisense NOSpro RNAs from different loci. (E) Production of overlapping sense and antisense NOSpro transcripts at one locus. Three constructs based on this strategy were tested. The 35S and MAS promoters are approximately the same strength; the 19Spro is somewhat weaker. The Arabidopsis column shows the number of silenced plants over the total number of independent transformants tested. The percentage silencing is shown in parentheses. The target gene tested for construct A was a NOSpro-NPTII gene and for constructs B and E an intact nopaline synthase gene. S, sense orientation; AS, antisense orientation; MAS, mannopine synthase promoter. n.d., not determined.
Fig. 2. Detection of NOSpro dsRNA in silenced plant lines. RNA preparations (50 µg/lane) were treated with RNase One (‘+’ lanes), which digests ssRNA and leaves dsRNA intact. (A) Silenced plants contained a resistant NOSpro dsRNA of the expected size (∼280 bp; Mette et al., 1999) (lanes 3 and 4). This NOSpro dsRNA was not detected after introduction of the 271 locus (lanes 5 and 6), which represses the 35S promoter transcribing NOSpro sequences, or in the original target line (lanes 1 and 2). Lanes 7 and 8 show positive controls [0.1 pg (left) and 1 pg (right)] consisting of NOSpro dsRNA (∼300 bp) formed by annealing antisense (AS) and sense (S) transcripts synthesized in vitro. (B) In a non-silenced line, an ∼0.6 kb poly adenylated NOSpro RNA is present (‘–’ lane) but readily degraded by RNase One. (C) Actin mRNA controls for the same RNA preparations used in (A). Size markers are shown to the left.
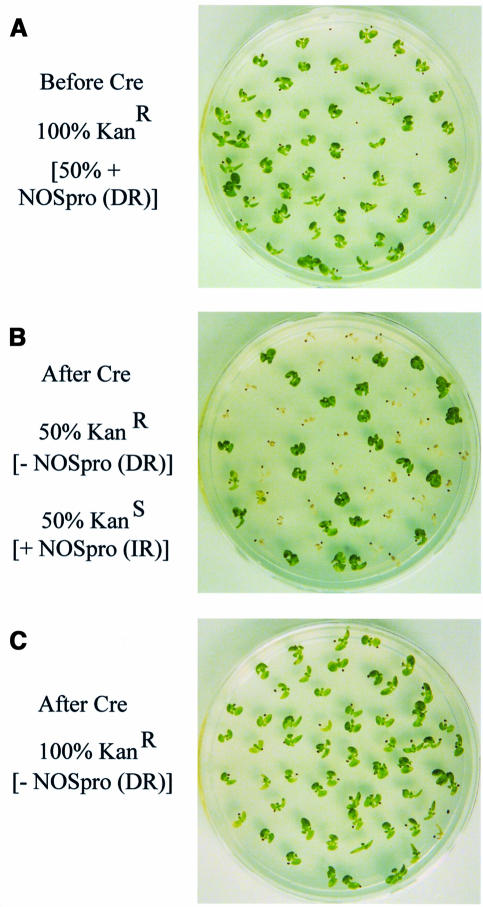
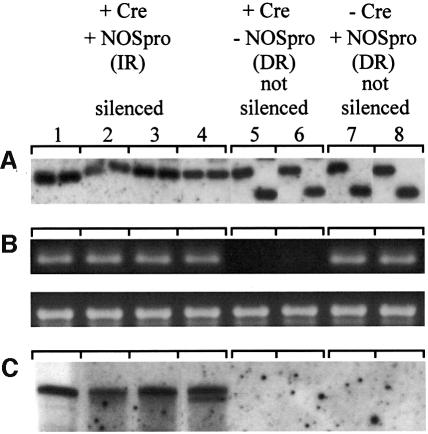
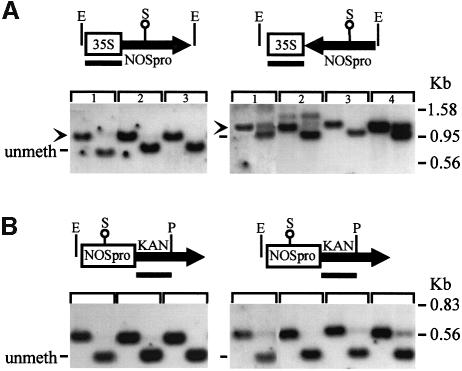
While these experiments supported the involvement of NOSpro dsRNA in trans-silencing, it remained possible that some unique feature of the original silencing locus (H9NP) was responsible for its silencing ability. If NOSpro dsRNA were sufficient to induce silencing and methylation of the target NOSpro, then any NOSpro IR that is transcribed, regardless of its location in the genome, should act as a trans-silencer. To test this, NOSpro IRs were created in planta at different genomic locations by site-specific recombination. A construct that contained a 35Spro-driven NOSpro direct repeat (DR), in which the second copy was flanked by two lox sites in inverse orientation (Figure 1C), was introduced into a tobacco line homozygous for the target NOSpro-nptII gene (K81; Jakowitsch et al., 1999). DNA blot analysis of three independent lines demonstrated the presence of the intact NOSpro DR and the absence of methylation at a SacII site in the first NOSpro of the DR (Figure 3A). The full NOSpro DR was transcribed, as demonstrated by an RNase protection assay, which produced two bands corresponding to the slightly different sizes of the two NOSpro copies in the DR (Figure 3B). The presence of the transcribed NOSpro DR induced little or no methylation of the target NOSpro (Figure 3C) and did not affect kanamycin resistance in seedlings (Figure 4A). After crossing in the Cre locus, however, 50% of the progeny seedlings were sensitive to kanamycin, indicating silencing of the NOSpro-nptII target gene after conversion of the NOSpro DR into a NOSpro IR (Figure 4B). Consistent with the acquired kanamycin-sensitive phenotype, the target NOSpro driving nptII gene expression became methylated (Figure 5A, panels 1–4) in plants containing the newly formed NOSpro IR locus (Figure 5B). Moreover, NOSpro dsRNA was detected only in this population of plants (Figure 5C, panels 1–4). Identical results were obtained with all three lines originally containing the NOSpro DR, demonstrating the reliability of this approach.
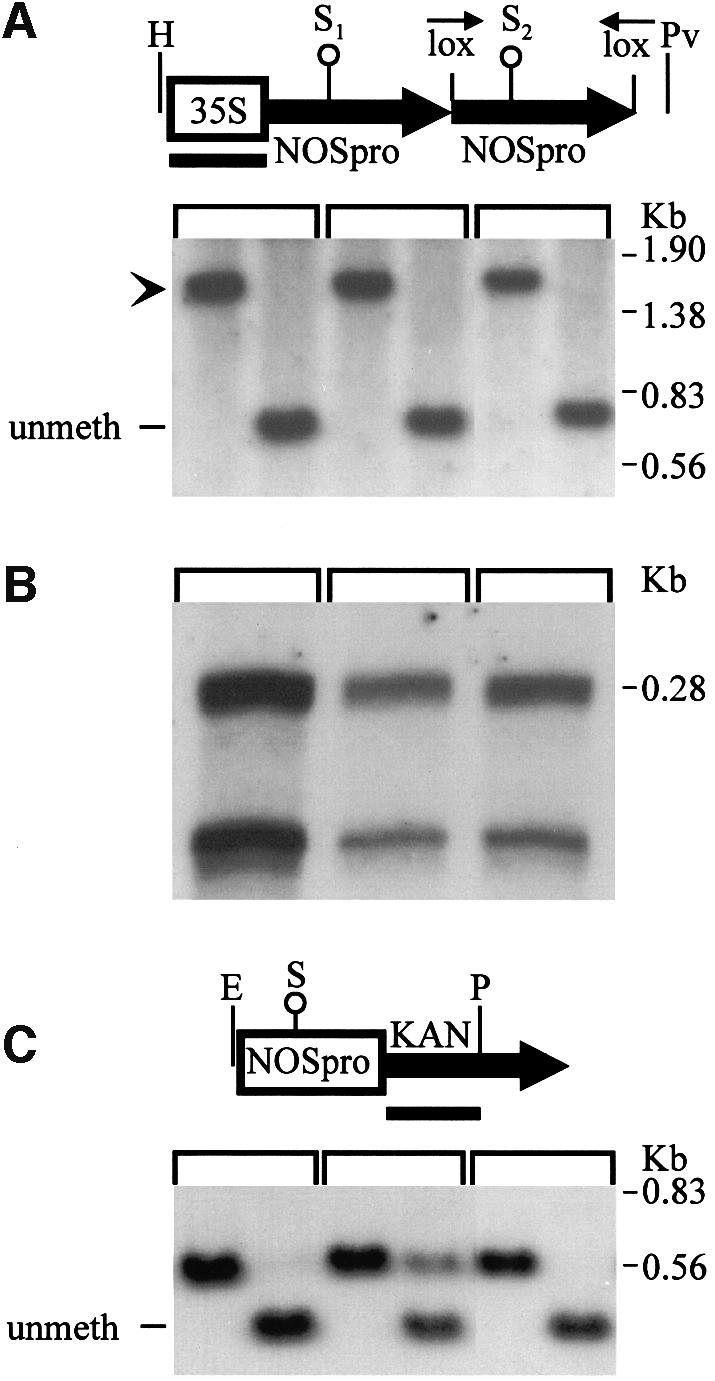
Fig. 3. A transcribed NOSpro direct repeat (DR) has a negligible effect on methylation of the target NOSpro. (A) Southern analysis demonstrating intact copies of the 35Spro-NOSpro DR construct integrated in three independent tobacco transformants. The first lane of each panel shows a HindIII (H)–PvuII (Pv) double digest probed with a 35Spro probe; in all three plant lines, a band of the expected size was obtained (arrowhead). The second lane of each panel shows triple digests with HPv plus SacII (S), which is methylation sensitive (open circle). Methylation at this S site in the NOSpro has been tightly correlated with silencing of this promoter. The appearance of the unmethylated fragment demonstrates that at least the S1 site in the first NOSpro copy of the DR is unmethylated in all three lines. (B) RNase protection demonstrating that the entire NOSpro DR is transcribed in all three lines. Because the two NOSpro copies in the DR differ somewhat in size (263 versus 197 bp; see T-DNA constructs, Materials and methods), two sizes of protected fragment were obtained. (C) Methylation analysis of the target NOSpro, which is the same in all lines, in the presence of the transcribed NOSpro DR. EcoRI (E)–PstI (P) digests of either minus (first lanes) or plus (second lanes) methylation-sensitive S were performed. In all cases, little or no methylation is present, as indicated by the complete or nearly complete shift to the smaller fragment after addition of S. The partial methylation seen in the middle panel could result from spontaneous partial methylation of the NOSpro that occurs occasionally in the target plant line (Matzke et al., 1993) or from additional complexity or binary vector sequences in the NOSpro DR locus. Probes are indicated by black bars beneath the maps. Size markers are shown to the right.
Fig. 4. Kanamycin resistance before and after Cre-mediated conversion of a transcribed NOSpro DR into an IR. Silencing of the homozygous target NOSpro-NPTII gene was assessed by analyzing the kanamycin resistance of seedlings in the presence of the hemizygous 35Spro-NOSpro locus. (A) Before introducing the Cre locus, 100% of the seedlings were resistant, even though 50% contained the NOSpro DR. (B) Following the cross with a plant homozygous for the Cre locus, 50% of the seedlings were sensitive to kanamycin; these were ones that inherited the NOSpro IR (resistant seedlings did not inherit the 355pro-NOSpro locus). (C) Control cross with the target locus showing that the Cre locus alone did not weaken kanamycin resistance.
Fig. 5. Methylation of the target NOSpro and production of NOSpro dsRNA following conversion of a transcribed NOSpro DR into an IR by Cre recombinase. (A) Target NOSpro methylation (digests as in Figure 3C). Before introduction of the Cre locus and in the presence of the NOSpro DR [–Cre, +NP(DR)], the target NOSpro was not methylated, as indicated by the conversion to the smaller fragment after addition of SacII (second lane of panels 7 and 8). After the Cre cross [+Cre, +NP(IR)], target NOSpros became methylated, as indicated by lack of conversion to the smaller fragment (panels 1–4). This methylation was not induced by the Cre locus alone [+Cre, –NP(DR)] (panels 5 and 6). (B) PCR detection of the 19S promoter unique to the NOSpro DR/IR locus. The NOSpro IR was confirmed by cloning and sequencing a PCR fragment (data not shown). The bottom panels show a PCR control for the HPT gene that was present in all plants examined. (C) RNase One-resistant NOSpro dsRNA was only detected in plants containing a NOSpro IR and a methylated target NOSpro (panels 1–4).
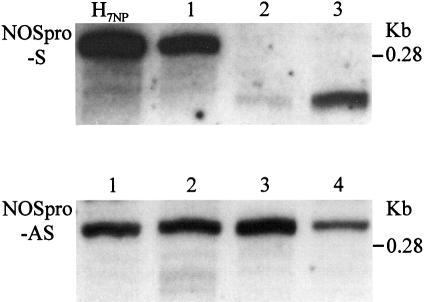
Transcription through an IR produces a NOSpro RNA hairpin. To test whether open dsRNA would act as a trans-silencer, constructs designed to synthesize separate NOSpro sense and antisense RNAs (Figure 1D) were introduced into plants homozygous for the NOSpro-nptII K81 target locus. Three sense (Figure 6A, left) and four antisense lines (Figure 6A, right) were chosen and the presence of the respective NOSpro RNA in each line was confirmed by RNase protection (Figure 7). Individually, these sense and antisense NOSpro RNAs did not trigger substantial silencing or methylation of the target NOSpro (Figure 6B, left and right). Seeds obtained from intercrossing the lines were sown on kanamycin-containing medium to assess silencing of the NOSpro-nptII target gene. No silencing was observed as indicated by the expected number of kanamycin-resistant seedlings and the undiminished strength of kanamycin resistance (data not shown).
Fig. 6. Presence of 35Spro-NOSpro-Sense or -Antisense constructs does not trigger methylation of the target NOSpro. The intact sense construct was integrated in three independent tobacco transformants (A, left) and the antisense construct in four (A, right) as indicated by the presence of EcoRI (E) fragments of the expected sizes (arrowheads; first lanes of each panel). The SacII (S) site in the NOSpro was largely unmethylated in the sense plants (A, left), as indicated by the conversion to the smaller fragment after an ES double digest (second lanes of each panel). The SacII site was slightly methylated in several antisense plants (A, right), as indicated by the less than complete conversion to the smaller fragment following an ES double digest (second lanes of each panel). The significance of this methylation is not known. (B) The target NOSpro, which was the same in all lines, remained unmethylated in the presence of the transcribed sense (B, left) and antisense (B, right) constructs. Digests and explanations as in Figure 3C. Size markers are shown to the right.
Fig. 7. Production of NOSpro sense (S) and antisense (AS) RNAs. The three sense and four NOSpro antisense lines shown in Figure 6 were analyzed by RNase protection for the respective NOSpro RNAs. A fourth sense line described previously (H7NP; Mette et al., 1999) was included in the analysis. The size of the sense transcripts varies depending on the presence (longer) or absence (shorter) of leader sequences (see T-DNA constructs, Materials and methods).
The failure of separate sense and antisense NOSpro RNAs to trigger silencing might have been due to their inability to locate each other in the nucleus and form dsRNA. To attempt to minimize this problem, constructs were designed to synthesize overlapping NOSpro sense and antisense RNAs from the same locus (Figure 1E). These constructs contained a 35Spro adjacent to a NOSpro in either a sense or antisense orientation followed by either the 19S promoter of cauliflower mosaic virus or mannopine synthase (MAS) promoter (Weigel et al., 2000) arranged to transcribe NOSpro sequences from the opposite strand. Following transformation of tobacco lines homozygous for the K81 target NOSpro-nptII gene with these constructs, no appreciable trans-silencing was observed (data not shown).
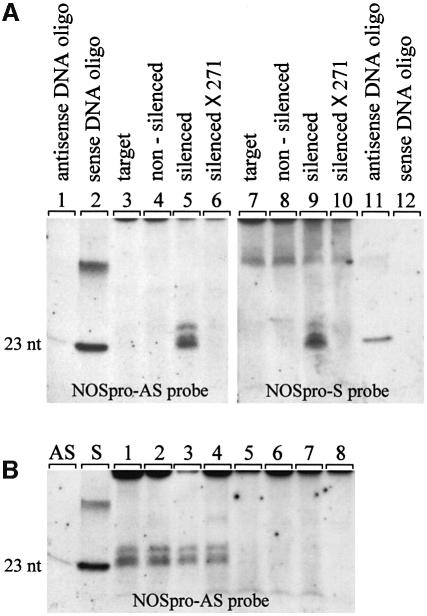
dsRNA involved in PTGS in plants (Hamilton and Baulcombe, 1999) and RNAi in animal systems (Zamore et al., 2000) is degraded to small RNAs that are 21–25 nucleotides (nt) in length. To test whether the NOSpro dsRNA was similarly degraded, a procedure to enrich for these small molecules (A.Hamilton, personal communication) was used to isolate RNAs from silenced and non-silenced plants. Indeed, both sense and antisense NOSpro RNAs ranging in length from 23 to 25 nt were detected in silenced plants (Figure 8A, lanes 5 and 9), but not in the original target line (Figure 8A, lanes 3 and 7) or in non-silenced plants (Figure 8A, lanes 4 and 8), which contained a single-stranded NOSpro RNA (Figure 2A). These small RNAs were no longer detectable after crossing in the 271 locus (Figure 8A, lanes 6 and 10), which also repressed synthesis of the NOSpro dsRNA (Figure 2A, lanes 5 and 6). NOSpro small RNAs were also detected in all silenced plants containing a transcribed NOSpro IR that had been created in planta by Cre recombinase (Figure 8B, lanes 1–4) but not in non-silenced plants harboring a transcribed NOSpro DR before Cre-mediated conversion (Figure 8B, lanes 7 and 8). These results indicate that silencing and methylation of the NOSpro target promoter depend on synthesis of a NOSpro dsRNA that can be degraded to small RNAs in a manner similar to dsRNAs that induce PTGS.
Fig. 8. Detection of small RNAs in silenced plants. Total RNA from the indicated plants was prepared using a procedure to enrich for small RNAs (Hamilton and Baulcombe, 1999). This RNA was electrophoresed on 15% acrylamide gels, electroblotted onto a nylon membrane, and probed with NOSpro sense (S) and antisense (AS) RNA probes. (A) Small NOSpro sense (lane 5) and antisense (lane 9) RNAs ranging in size from 23 to 25 nt were detected only in silenced plants. These small RNAs were not present in the original NOSpro target line (lanes 3 and 7), in non-silenced plants that synthesized polyadenylated single-stranded NOSpro RNA (lanes 4 and 8), or in silenced plants following the introduction of the 271 locus (lanes 6 and 10), which disrupted synthesis of NOSpro dsRNA (Figure 2). As size and polarity controls, NOSpro sense (lanes 2 and 12) and antisense (lanes 1 and 11) DNA 23mers were used. The larger band visible in lane 2 is probably a multimer of the oligonucleotide present in the commercial preparation. Larger bands in lanes 7–10 are due to non-specific hybridization of the RNA probe. (B) Detection of NOSpro small RNAs in plants containing a transcribed NOSpro IR created in planta by Cre recombinase (lanes 1–4). NOSpro small RNAs were not detected in plants containing a transcribed NOSpro DR before conversion by Cre recombinase (lanes 5–8). Identical results were obtained with a NOSpro-S probe (not shown). The eight plants shown here are the same as those shown in Figure 5, panels 1–8. AS and S lanes show control antisense and sense DNA 23mers, respectively.
To establish whether transcriptional silencing by NOSpro dsRNA would work in another plant species, the constructs shown in Figure 1A, B and D were introduced into two Arabidopsis lines containing different NOSpro-nptII target genes. While neither a non- transcribed NOSpro IR (Figure 1A) nor constructs intended to produce overlapping sense and antisense NOSpro transcripts (Figure 1D) induced significant silencing, a NOSpro IR adjacent to a transcribing 35S promoter, which would produce a NOSpro hairpin RNA (Figure 1B), resulted in a high frequency of NOSpro-nptII silencing in both Arabidopsis target lines.
Discussion
We have demonstrated that NOSpro dsRNA can trigger methylation and transcriptional inactivation of homologous copies of the NOSpro in trans. In addition, this NOSpro dsRNA is degraded into small RNAs ∼23 nt in length, similar to the dsRNA degradation products observed with PTGS systems in plants (Hamilton and Baulcombe, 1999) and an RNAi in vitro system in Drosophila (Zamore et al., 2000). The production of these small RNAs indicates that at least some of the NOSpro dsRNA, which is synthesized in the nucleus, enters the same pathway as cytoplasmic dsRNAs involved in PTGS/RNAi. Therefore, the degradation process is not restricted to dsRNAs containing sequences that are normally transcribed (untranslated and coding regions) and targeted to mRNA. The similar fate of these dsRNAs strengthens the link between RNA-mediated TGS and PTGS. dsRNA can trigger either the sequence-specific RNA degradation step of PTGS/RNAi or DNA methylation, which can lead to TGS if promoter sequences are involved. dsRNA generated in the cytoplasm can potentially play a dual role by initiating PTGS and by entering the nucleus to direct methylation of homologous DNA sequences. This provides a means by which PTGS/RNAi events could be imprinted at the genome level in organisms that methylate their DNA. Whether dsRNA can also stimulate chromatin structural changes and whether promoter dsRNAs synthesized in the nucleus induce TGS in other organisms remain to be determined.
The requirement for a dsRNA to provoke methylation in our system is consistent with other examples of RdDM, which involve RNA pathogens that have a dsRNA genome (viroids) or replicate via dsRNA intermediates (viruses). It is not yet clear how a NOSpro dsRNA triggers methylation and silencing of homologous promoter sequences. One possibility is a direct RNA–DNA interaction based on sequence homology. In this case, an RNA helicase to unwind the dsRNA would presumably be required, since it is likely that RNA–DNA pairing requires an ssRNA that is complementary to target DNA (Pélissier and Wassenegger, 2000). An alternative mechanism that does not rely on DNA–RNA sequence interactions involves titration of transcription factors by the NOSpro dsRNA, which is easily detectable in total RNA preparations and thus relatively abundant. RNA aptamers containing core binding domains for transcription factors have been used as decoys to sequester the corresponding proteins (Lebruska and Maher, 1999). However, titration of a transcription factor would possibly produce a pleiotropic phenotype in the plants and this was not observed. A further possibility is that the small RNAs resulting from dsRNA degradation are responsible for directing homologous DNA methylation. Their size approaches the lower limit of DNA target length for RdDM (∼30 bp; Pélissier and Wassenegger, 2000). Small RNAs produced during PTGS/RNAi appear to guide a dsRNA endonuclease to the homologous RNA and target it for degradation (Bass, 2000). It is conceivable that the small RNAs also guide DNA methyltransferase to homologous sequences in the genome. Future analysis of DNA methylation in mutants defective in small RNA production will help to address this question.
By testing various strategies to potentially produce dsRNA in the nucleus, we have shown that an RNA hairpin transcribed from an IR of NOSpro sequences provides the most effective means to trigger trans-silencing. In tobacco, the most reliable and predictable means proved to be conversion in planta of a NOSpro DR into an IR by the activity of Cre recombinase, which avoids manipulating unstable IRs in bacteria during plasmid cloning steps. In all three tobacco lines tested, this conversion resulted in silencing and methylation of the target NOSpro in all progeny inheriting the IR. These experiments, which showed that NOSpro dsRNAs synthesized at different genomic locations can induce trans-silencing and methylation, eliminated position effects as an influence on silencing ability. These experiments also established that neither two copies of the NOSpro in direct orientation nor transcription through a DR was sufficient for trans-silencing and methylation, thus arguing against a simple copy number-dependent mechanism.
The inability of separate sense and antisense NOSpro transcripts to trigger silencing of NOSpro-driven target genes contrasts to several cases of PTGS, where enhanced silencing and virus resistance were observed when sense- and antisense-expressing lines were crossed (Waterhouse et al., 1998). This disparity might reflect differences between RNA-mediated TGS and PTGS, which take place in separate cellular compartments that could present distinct environments for RNA–RNA interactions. Attempts to produce trans-acting NOSpro dsRNA by synthesizing overlapping sense and antisense NOSpro transcripts also proved unsuccessful. It is possible, however, that this arrangement will trigger methylation in cis of the NOSpro positioned between the two transcribing promoters (Figure 1E). This possibility will be tested in the future by taking advantage of the 271 locus, which suppresses the 35S and 19S promoters (Vaucheret, 1993), but not the MAS promoter.
Long, perfect duplex RNAs are normally not abundant in cells, and when present they are often associated with pathogenic states that trigger profound physiological reactions (Kumar and Carmichael, 1998). dsRNAs in the cytoplasm are usually associated with virus infections and they can provoke strong antiviral defenses (Smith, 1999). In the nucleus, dsRNAs corresponding to repetitive sequences have been found in the hnRNA fraction in animal cells (Kumar and Carmichael, 1998). Some of these nuclear dsRNAs might correspond to transposable element transcripts, suggesting a connection between RdDM and genome defense (Matzke et al., 2000). Viroid pathogenicity, which remains mysterious after decades of study, might be based on dsRNA-directed methylation of plant sequences (Pélissier and Wassenegger, 2000). While the extent of viroid-homologous DNA sequences in host plant genomes is not yet known, some viroids and 7S RNA share sequence similarity, which is potentially a factor in pathogenesis (Symons, 1989).
A transcribed NOSpro IR efficiently silenced target NOSpros not only in tobacco but also in Arabidopsis. While not all promoters might be sensitive to this method of silencing, preliminary data suggest that at least some others, including those active in specific tissues, can also be silenced via promoter dsRNA in Arabidopsis (W.Aufsatz, T.Kanno and A.Matzke, unpublished results). If so, promoter hairpin RNA-mediated methylation and TGS provides an alternative to PTGS/RNAi (Chuang and Meyerowitz, 2000) for gene knockouts. Individual members of gene families or individual alleles that have distinct promoter regions can be targeted by promoter dsRNAs. TGS is potentially a more stable means to silence genes than PTGS because promoter methylation will tend to persist through the maintenance activity of methyltransferase even if the inducing signal is temporarily removed. The establishment of a promoter dsRNA- mediated silencing system in Arabidopsis will allow mutant screens to identify proteins involved in this process.
Materials and methods
T-DNA constructs
All NOSpro coordinates in the following description are relative to the transcriptional start site as given in DDBJ/EMBL/GenBank accession No. V00087. The structure of the NOSpro-NPTII-NOSpro-NOS target transgene locus in tobacco line K81 has been described by Jakowitsch et al. (1999). Target lines containing this pair of NOSpro-driven reporter genes were produced in Arabidopsis. These lines were retransformed with potential NOSpro silencing constructs using a 19Spro-HPT (hygromycin phosphotransferase) gene as a second selectable marker. The non-transcribed NOSpro IR (Figure 1A) harbored the same NOSpro portions as the silencing transgene locus H9NP (Mette et al., 1999), but without flanking 35S promoters. The 35Spro-driven NOSpro IR used in Arabidopsis (Figure 1B) comprised a –264 to +34 NOSpro fragment in sense orientation with respect to the 35S promoter and a –264 to +34 NOSpro fragment in antisense orientation. The two NOSpro fragments were separated by a 263 bp spacer containing a soybean promoter for the α′ subunit of β-conglycinin (–309 to –46 with regard to the translational start site as described in DDBJ/EMBL/GenBank accession No. M13759) in sense orientation relative to the 35Spro. A NOSpro DR under transcriptional control of the 35Spro (Figure 1C) harbored two NOSpro fragments in sense orientation: one from position –264 to –1 of the NOSpro sequence, followed by a second one from position –264 to –67. The second NOSpro copy was flanked by two lox sites in inverse orientation (Figure 1C), which allows the formation of a transcribed NOSpro IR in planta by crossing with a line expressing Cre recombinase. Transcribed NOSpro sense and antisense constructs (Figure 1D) contained either NOSpro fragments from position –264 to +34 (including the leader sequence) or –264 to –1 (without the leader sequence) in either polarity relative to the 35Spro. In both sense and antisense constructs, the NOSpro fragment was not followed by terminator sequences. All constructs containing the NOSpro sequence flanked by two other promoters in opposite orientation (Figure 1E) harbored a NOSpro fragment from position –264 to +34.
Plant transformation and marker gene assay
Tobacco (‘Petit Havana SR1’) leaf disk transformation and seed germination assays were performed as described previously (Matzke et al., 1989). Arabidopsis plants (ecotype Columbia) were grown at 22°C in a 16 h/8 h day/night cycle. Transgenic Arabidopsis lines were obtained by the floral dip method (Clough and Bent, 1998). To study antibiotic resistance phenotypes of seedlings, the germination and growth medium contained 40 mg/l hygromycin and/or 50 mg/l kanamycin for tobacco, and 20 mg/l hygromycin and/or 50 mg/l kanamycin for Arabidopsis. Nopaline was detected using high-voltage paper electrophoresis and phenantherenequinone staining (Matzke et al., 1989).
DNA analysis
Plant genomic DNA was purified using a DNeasy Plant Maxi Kit (Qiagen, Hilden). DNA blot analysis using 32P-labeled RNA probes and methylation analysis of the SacII site in the NOSpro-NPTII gene were performed as described previously (Matzke et al., 1989).
dsRNA analysis
Total RNA was extracted from young, expanding tobacco leaves using the Hybaid-AGS RNAClean system (Chemomedica, Vienna) including the RNAClean Extension protocol according to the manufacturer’s instructions. As a quality control for RNA preparations, RNA blot analysis to detect tobacco actin mRNA was performed as described previously (Mette et al., 1999). For the detection of dsRNA, RNase One (Promega, Mannheim) was used to degrade selectively the ssRNA in these preparations. Using 0.1–0.2 U of RNase per 1 µg of RNA, 50 or 100 µg of total RNA were digested in a volume of 100 µl for 10 min at 37°C in 10 mM Tris–HCl pH 7.5, 5 mM EDTA, 200 mM sodium acetate. After phenol extraction and ethanol precipitation, the remaining, RNase-resistant RNA was separated by electrophoresis on a 5% polyacrylamide–7 M urea gel and transfered to Zeta-Probe GT nylon membrane (Bio-Rad, Vienna) by electroblotting using a Bio-Rad Trans-Blot SD semidry transfer cell. 32P-labeled probes were synthesized from an isolated DNA fragment containing the complete NOSpro sequence using a Multiprime DNA labeling system (Amersham-Pharmacia, Vienna) and were hybridized to the membrane-bound RNA at 42°C in 125 mM sodium phosphate pH 7.2, 250 mM sodium chloride, 7% SDS, 50% deionized formamide. Prior to exposure, membranes were washed once in 2× SSC, 0.1% SDS at room temperature, once in 0.5× SSC, 0.1% SDS at ambient temperature and twice in 0.1× SSC, 0.1% SDS at 64°C. Annealed sense and antisense NOSpro in vitro transcripts were used as positive controls in these experiments. To detect single-stranded sense or antisense NOSpro transcripts in plants, 50 µg of total RNA were first hybridized to 5 ng of unlabeled antisense or sense NOSpro in vitro transcripts, respectively (42°C; 40 mM PIPES–NaOH pH 6.4, 400 mM sodium acetate, 1 mM EDTA, 80% formamide; 100 µl volume) and then subjected to RNase One treatment (500 µl volume) and gel-blot analysis as described above.
Small RNA analysis
For the detection of small RNA fragments with NOSpro homology, the procedure described by Hamilton and Baulcombe (1999) was used. Young tobacco leaves were frozen in liquid nitrogen, ground to fine powder and resuspended in extraction buffer (50 mM Tris–HCl pH 9.0, 10 mM EDTA, 100 mM sodium chloride, 2% SDS; 5 ml per 1 g of tissue). After extracting the homogenate twice with equal volumes of buffer-saturated phenol–chloroform, nucleic acids were precipitated from the solution by adding 1/10 vol. of 3 M sodium acetate pH 5.0 and 3 vols of absolute ethanol. After incubation for at least 2 h at –20°C, the precipitate was collected by centrifugation, washed with 70% ethanol, dried and redissolved in bidistilled water. High molecular weight nucleic acids were precipitated from this solution by adding polyethylene glycol (MW 8000) and sodium chloride to final concentrations of 5% and 500 mM, respectively, followed by incubation on ice for 30 min. After removing the precipitate by centrifugation, the low molecular weight nucleic acids remaining in the supernatant were recovered by sodium acetate–ethanol precipitation as described above. From this fraction, 50 µg per lane were separated on a 15% polyacrylamide–7 M urea gel and transferred to Zeta-Probe GT nylon membrane as described above. As size and polarity controls, the DNA 23mers 5′-AATTTATGGAACGTCAGTGGAGC-3′ (antisense) and 5′-CCACTGACGTTCCATAAATTCCC-3′ (sense) were included on the same gels (10 pmol per lane). Using T7 or T3 RNA polymerase (Roche, Vienna) and [α-32P]UTP (125 µCi per 20 µl reaction volume; specific activity 3000 Ci per mmol; New England Nuclear, Vienna), internally labeled in vitro transcripts of the complete NOSpro sequence were synthesized in both antisense and sense orientation. The template DNA was removed from the reaction by treatment with RNase-free DNase I (Roche, Vienna). To shear the labeled transcripts to an average size of 50 nt, 300 µl of alkaline buffer (80 mM sodium bicarbonate, 120 mM sodium carbonate) per 20 µl of reaction volume were added and the solution was incubated at 60°C for 2.5 h [the incubation time was calculated according to the formula t = (Li – Lf)/(k × Li × Lf); where t is time in min, Li is the initial length of probe in kb, Lf is the final length of probe in kb and k is a rate constant of 0.11 kb–1 min–1]. The solution was neutralized by adding 20 µl of 3 M sodium acetate pH 5.0 and then immediately used as a hybridization probe. Hybridization and washes were carried out at low stringency, in 125 mM sodium phosphate pH 7.2, 250 mM sodium chloride, 7% SDS, 50% deionized formamide at 42°C and twice in 2× SSC, 0.2% SDS at 42°C for 30 min, respectively. Unspecific background on the hybridized membranes could be efficiently removed by RNase A treatment (20 mM Tris–HCl pH 7.5, 5 mM EDTA, 60 mM sodium chloride, 10 µg/ml RNase A; incubation for 1 h at 37°C).
PCR
Tobacco genomic DNA for PCR was prepared using a DNeasy Plant Mini Kit (Qiagen, Hilden). In all cases, 40 amplification cycles were performed using AmpliTaq Gold DNA Polymerase (Perkin Elmer, Vienna) with 2.0 mM MgCl2 in the reaction mixture. NOSpro-IR-specific primers were 5′-TTTCTGACGTATGTGCTTAGCTC-3′ and 5′-GAGTTAGCTCAC TCATTAGGCA-3′; the annealing temperature was 66°C. 19Spro-specific primers were 5′-AAAGAATTACCACAGCAATGACAA AGAG-3′ and 5′-TCTCCAGAGATGTGTTTAAATAGGCAG-3′, HPT-specific primers were 5′-GTCCTGCGGGTAAATAGCTGC-3′ and 5′-CGTCTGCTGCTCCATACAAGC-3′; in both cases the annealing temperature was 57°C.
Acknowledgments
Acknowledgements
We thank Joan Odell for providing the Cre-expressing tobacco line, Hervé Vaucheret for the 271 plant line, Detlef Weigel for pSKI15 containing the MAS promoter, and Andrew Hamilton for providing a detailed protocol to detect small RNAs. This work was supported by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (Grant No. Z21-MED).
References
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. [DOI] [PubMed] [Google Scholar]
- Bosher J.M. and Labouesse,M. (2000) RNA interference: genetic wand and genetic watchdog. Nature Cell Biol., 2, E31–E36. [DOI] [PubMed] [Google Scholar]
- Chuang C.-F. and Meyerowitz,E.M. (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cogoni C. and Macino,G. (1999a) Homology-dependent gene silencing in plants and fungi: a number of variations on the same theme. Curr. Opin. Microbiol., 2, 657–662. [DOI] [PubMed] [Google Scholar]
- Cogoni C. and Macino,G. (1999b) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature, 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni C. and Macino,G. (1999c) Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science, 286, 2341–2344. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton,A., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Faugeron G. (2000) Diversity of homology-dependent gene silencing strategies in fungi. Curr. Opin. Microbiol., 3, 144–148. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J. and Kovac,K.A. (2000) Plant DNA methyltransferases. Plant Mol. Biol., 43, 189–210. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Jakowitsch J., Papp,I., Moscone,E.A., van der Winden,J., Matzke,M. and Matzke,A.J.M. (1999) Molecular and cytogenetic characterization of a transgene locus that induces silencing and methylation of homologous promoters in trans. Plant J., 17, 131–140. [DOI] [PubMed] [Google Scholar]
- Jones A.L., Thomas,C.L. and Maule,A.J. (1998) De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J., 17, 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Hamilton,A.J., Voinnet,O., Thomas,C.L., Maule,A.J. and Baulcombe,D.C. (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell, 11, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F., Haverkamp,T.H., van Luenun,H.G. and Plasterk,R.H.A. (1999) mut-7 of C.elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Kooter J., Matzke,M.A. and Meyer,P. (1999) Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci., 4, 340–346. [DOI] [PubMed] [Google Scholar]
- Kumar M. and Carmichael,G.G. (1998) Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev., 62, 1415–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebruska L.L. and Maher,L.J. (1999) Selection and characterization of an RNA decoy for transcription factor NF-κB. Biochemistry, 38, 3168–3174. [DOI] [PubMed] [Google Scholar]
- Luff B., Pawlowski,L. and Bender,J. (1999) An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol. Cell, 3, 505–511. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Primig,M., Trenovsky,J. and Matzke,A.J.M. (1989) Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J., 8, 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Neuhuber,F. and Matzke,A.J.M. (1993) A variety of epistatic interactions can occur between partially homologous transgene loci brought together by sexual crossing Mol. Gen. Genet., 236, 379–386. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Mette,M.F. and Matzke,A.J.M. (2000) Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol. Biol., 43, 401–415. [DOI] [PubMed] [Google Scholar]
- Mette M.F., van der Winden,J., Matzke,M.A. and Matzke,A.J.M. (1999) Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J., 18, 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Odell J.T. and Russell,S.H. (1994) Use of site-specific recombination systems in plants. In Paszkowski,J. (ed.), Homologous Recombination and Gene Silencing in Plants. Kluwer, Dordrecht, The Netherlands, pp. 219–270. [Google Scholar]
- Pélissier T. and Wassenegger,M. (2000) A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA, 6, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.A. and Zamore,P.D. (2000) RNA interference. Science, 287, 2431–2436. [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke,J.M., Stacey,S.C., Klein,M.E., Mackin,N. and Maine,E.M. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C.elegans. Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Smith H. (1999) Interfering with viral infection: plants do it too. Plant Cell, 11, 1191–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons B. (1989) Pathogenesis by antisense. Nature, 338, 542–543. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian,M., Kelly,W.G., Fleenor,J., Grishok,A., Timmons,L., Fire,A. and Mello,C.C. (1999) The rde-1 gene, RNA interference and transposon silencing in C.elegans. Cell, 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (1993) Identification of a general silencer for 19S and 35S promoters in a transgenic tobacco plant: 90 bp of homology in the promoter sequence are sufficient for trans-inactivation. C. R. Acad. Sci. Paris, 317, 1471–1483. [Google Scholar]
- Wassenegger M. (2000) RNA-directed DNA methylation. Plant Mol. Biol., 43, 203–220. [DOI] [PubMed] [Google Scholar]
- Wassenegger M., Heimes,S., Riedel,L. and Sänger,H.L. (1994) RNA-directed de novo methylation of genomic sequences in plants. Cell, 76, 567–576. [DOI] [PubMed] [Google Scholar]
- Waterhouse P., Graham,M.W. and Wang,M.-B. (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl Acad. Sci. USA, 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. et al. (2000) Activation tagging in Arabidopsis. Plant Physiol., 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]