Abstract
Even as genetic studies identify alleles that influence human disease susceptibility, it remains challenging to understand their functional significance and how they contribute to disease phenotypes. Here, we describe an approach to translate discoveries from human genetics into functional and therapeutic hypotheses by relating human genetic variation to small-molecule sensitivities. We use small-molecule probes modulating a breadth of targets and processes to reveal disease allele-dependent sensitivities, using cells from multiple individuals with an extreme form of diabetes (maturity onset diabetes of the young type 1, caused by mutation in the orphan nuclear receptor HNF4α). This approach enabled the discovery of small molecules that show mechanistically revealing and therapeutically relevant interactions with HNF4α in both lymphoblasts and pancreatic β-cells, including compounds that physically interact with HNF4α. Compounds including US Food and Drug Administration–approved drugs were identified that favorably modulate a critical disease phenotype, insulin secretion from β-cells. This method may suggest therapeutic hypotheses for other nonblood disorders.
Keywords: synthetic interactions, chemical screen
Genetic variants that influence susceptibility to disease have been identified for many diseases. Reproducing human genetic lesions in cellular or animal models for functional studies can be challenging, particularly when the causal variants are not known or fall outside coding regions. Furthermore, for genetically complex diseases such as type-2 diabetes, multiple genes influence an individual’s risk of disease. The use of patient-derived cells allows disease alleles to be studied in their native genetic background but is often hampered by limited access to traditional disease-related cell types (such as pancreatic β-cells for diabetes). In genetically tractable model organisms, genetic interaction screens provide a powerful, systematic method to reveal genes and pathways that interact with a given mutation (1–3); similarly, in genetically defined cell lines, the concept of synthetic lethality has been used to identify compounds that selectively kill cells expressing an oncogene (4).
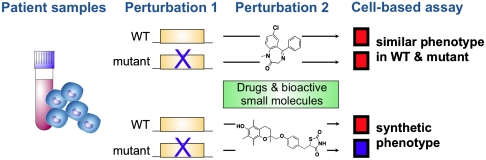
Here, we apply the logic of genetic interaction screens to study human disease alleles in cells from individual patients, using small-molecule probes as a source of systematic perturbation (particularly marketed drugs and bioactives with characterized activity). To discover small-molecule–gene interactions, we screen for small molecules that induce distinct cellular phenotypes depending on the presence or absence of a disease mutation (Fig. 1). This implies that the small molecules (or their protein targets) functionally interact with the disease mutation and identifies nodes where cellular pathways intersect with a disease gene. Both directions of small-molecule–induced effects could potentially be informative (i.e., small-molecule–induced assay measurements greater in either mutant or wild-type cells). Small-molecule–gene interactions can potentially be identified using cell-based measurements that reflect specific biological pathways or broader measures of cell function or viability; different measurements would likely identify partially overlapping subsets of interactions.
Fig. 1.
Overview of synthetic genetic interaction approach in patient-derived cells. Small molecules are identified that induce distinct cellular ATP measurements in the presence vs. absence of a disease mutation.
Type-2 diabetes is a genetically heterogenous disease; monogenic forms comprise approximately 2–5% of cases but may shed light on the pathogenesis or treatment of the common form of type-2 diabetes. Maturity onset diabetes of the young type 1 (MODY1) is an extreme form of diabetes characterized by early onset diabetes (often before age 25) due to impaired pancreatic insulin secretion (5–7). The causal mutation is in the orphan nuclear hormone receptor HNF4α, a transcription factor that plays wide-ranging roles in embryonic development, and the function of liver, pancreas, and other tissues. HNF4α binds to upstream sequences of hundreds of genes involved in glucose, lipid, cholesterol, amino acid, and drug metabolism (8–10). Two different conditional knockouts of HNF4α in β-cells yielded conflicting phenotypes: although both knockout models showed impaired insulin secretion in response to glucose challenge (11, 12), one model also unexpectedly showed elevated resting insulin levels with associated hypoglycemia (12), highlighting our incomplete understanding of the physiologic effects of HNF4α mutation.
In this study, we develop an approach to identify small-molecule–gene interactions in MODY1 diabetes, using patient-derived lymphoblast cell lines (LCLs, derived from B-lymphocytes) as a surrogate cell model and a luminescence-based assay for cellular ATP as the cell measurement. LCLs (and other cells derived from the peripheral circulation) are easily accessible from patient cohorts, but their utility as cell models for functional studies of nonblood disorders is not widely established. Gene expression data from LCLs have been used to study DNA variants that influence gene expression (13) as well as the mechanism of genetic variants implicated in asthma (14) and systemic lupus erythematosus (15). We identify multiple small molecules that functionally interact with HNF4α in LCLs, including fatty acids that physically interact with HNF4α. Several of these small-molecule–gene interactions are preserved in murine pancreatic β-cells, a canonical cell model for diabetes studies. Furthermore, our approach identifies compounds that modulate a key disease phenotype, insulin secretion from β-cells. These results suggest that systematic identification of synthetic interactions between small molecules and disease alleles can provide a functional context for disease-causing alleles and suggest testable therapeutic hypotheses.
Results
Small-Molecule–Disease Allele Interaction Screen in Patient LCLs.
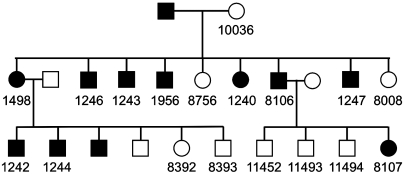
We studied LCLs from 18 members of a MODY1 family (10 with diabetes and 8 without; Fig. 2) in which affected members possess a Q268X nonsense mutation (16). This mutation truncates the ligand-binding domain and leads to impaired DNA binding, abnormal cellular trafficking, and loss of transcriptional activity (8, 17). Sequencing and RT-PCR confirmed the presence of a heterozygous C-to-T substitution in codon 268 exclusively in the diabetic subjects, and low levels of HNF4α expression. LCLs were treated with 3,973 clinically used drugs and small molecules whose activities have been previously characterized. The effects of small-molecule treatments were measured using a luminescence assay for cellular ATP content, chosen as a viability assay, and also for reasons related to diabetic metabolism: Type-2 diabetes has been associated with impairments in oxidative phosphorylation (18), and ATP concentrations in pancreatic islets are a key sensor during insulin secretion (19). ATP assay values were converted to a Z score (multiples of the standard deviation of the distribution of control wells in the same cell line) (20). The assay was highly reproducible with a coefficient of variation routinely < 10% (Table S1). Each small-molecule treatment was performed in duplicate at a single concentration.
Fig. 2.
Pedigree showing relations among MODY1 individuals screened in this study. Individuals whose lymphoblasts were screened have a number below their symbol. (Numbers correspond to the cell line’s designation in the Coriell Repository.) The above is a partial pedigree of the previously described MODY1 family R–W (16).
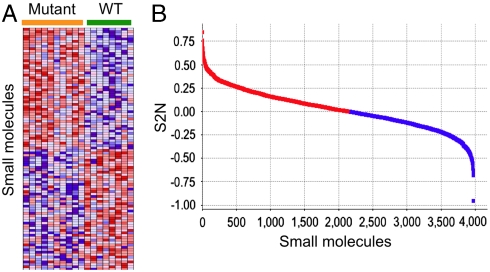
To assess the extent to which each small molecule induced different assay Z scores in mutant vs. wild-type LCLs, we calculated a signal-to-noise statistic (S2N): S2N = (μMutant - μWT)/(σMutant + σWT) (21). (μMutant and σMutant are the mean and standard deviation, respectively, of the ATP assay Z scores averaged across all mutant cell lines; μWT and σWT are the corresponding values across wild-type cell lines.) The magnitude of S2N is greatest (either positive or negative in sign) for small molecules that best discriminate between mutant vs. wild-type LCLs in our assay (Fig. 3 A and B and Dataset S1); this suggests a small-molecule–gene interaction, such as a physical association or functional interaction between HNF4α and the small molecule (or its target protein).
Fig. 3.
Identification of disease allele-dependent chemical sensitivities, using cells derived from individual patients. (A) Heatmap of small molecules that caused the most distinct effects on cellular ATP in mutant vs. wild-type LCLs; i.e., with the most positive and most negative S2N from the entire dataset of 3,973 screened small molecules. Each row represents a different small molecule, and each column is an individual patient-derived LCL sample; heatmap cells reflect the Z score of the ATP assay. (B) Distribution of S2N for all 3,973 screened compounds.
Compound Set Enrichment Analysis Identifies Small Molecules with Disease Allele-Dependent Effects.
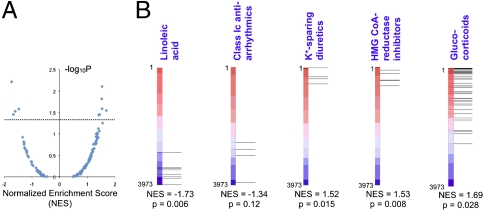
Although many small molecules with the most positive or most negative S2N had P values < 0.05 (calculated by permutation of mutant and wild-type class labels; Dataset S1), after correcting for multiple hypothesis testing, individual compounds did not meet statistical significance. To better identify patterns in these data, we tested whether sets of related compounds were statistically enriched among the most discriminating compounds, even if the discriminating effect of individual compounds was subtle. We curated the screened compounds into 141 compound sets (each containing from 3–60 compounds) based on drug class or activity in a common biologic pathway (Table S2). A weighted Kolmogorov–Smirnov-like statistic [normalized enrichment score (NES)] was calculated for each compound set that reflects the extent to which set members are statistically enriched among compounds with the most positive or negative S2N; P values were calculated by randomly permuting mutant vs. wild-type class labels 1,000 times (Fig. 4 A and B). This approach is analytically analogous to gene set enrichment analysis (18, 22) for gene expression data, and has been used to robustly identify groups of genes (e.g., related by virtue of belonging to a common biological process or pathway) that are differentially expressed in two cell types.
Fig. 4.
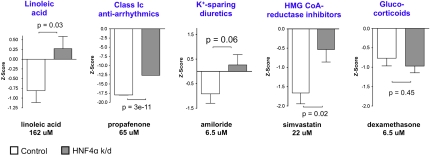
Compound set enrichment analysis identifies classes of small molecules that interact with an HNF4α mutation. (A) NES from compound set enrichment analysis, plotted against - log 10P (where P values are determined by randomly permuting wild-type vs. mutant class labels 1,000 times). Each data point corresponds to a compound set; compound sets are listed in Table S2. Dotted line corresponds to P = 0.05. (B) Display of output of compound set enrichment analysis. The “linoleic acid,” “class Ic antiarrhythmics,” “K+-sparing diuretics,” ‘HMG CoA-reductase inhibitors,” and “glucocorticoids” compound sets are among those that showed an interaction with HNF4α in LCLs. The verticle red-blue bar represents the ranked list of 3,973 screened compounds, ranked according to S2N (compounds with most positive S2N at the top); horizontal lines indicate where each member of the indicated compound set falls within this ranked list. Corresponding NES and permutation P value for the indicated compound set are shown below each bar.
The most highly ranked compound sets from this approach are listed in Table 1 (compound set members are listed in Table S3). These include several classes of drugs approved for conditions other than diabetes, as well as a subtype of fatty acids (Fig. 4B). Of note, these compound sets scored narrowly better than a set comprised of the most widely used therapy for MODY1, the insulin secretagogues belonging to the sulfonylurea class (such as glimepiride and tolbutamide; NES = 1.27). We verified that previously reported confounders of small-molecule treatment in LCLs such as LCL growth rate and EBV copy number (23) do not differ in mutant vs. wild-type LCLs (P = 0.59 and 0.26, respectively) (Fig. S1). We also confirmed that the observed variation in small-molecule–induced ATP measurements did not correlate with the growth rate of individual LCLs (Fig. S2).
Table 1.
Compound sets that best discriminate between HNF4α mutant vs. wild-type LCLs, based on a cellular ATP assay and compound set enrichment analysis
| Compound set | NES | P value | Representative compound | Interaction in β-cells |
| Linoleic acid | −1.73 | 6.1 × 10-3 | linoleic acid | + |
| ω-6 fatty acids | −1.61 | 0.03 | linoleic acid | + |
| Oxicam antiinflammatories | −1.48 | 0.026 | meloxicam | − |
| Class Ic antiarrhythmics | −1.34 | 0.12 | propafenone | + |
| Coxib antiinflammatories | 1.44 | 0.035 | rofecoxib | − |
| Potassium-sparing diuretics | 1.52 | 0.015 | amiloride | + |
| Acetic acid antiinflammatories | 1.53 | 0.026 | diclofenac | − |
| HMG CoA-reductase inhibitors | 1.53 | 7.9 × 10-3 | simvastatin | + |
| Imidazole/triazole antiinfectives | 1.55 | 0.056 | fluconazole | − |
| Glucocorticoids | 1.69 | 0.028 | dexamethasone | − |
A Significant Fraction of Small-Molecule–Gene Interactions Are Preserved from LCLs to Pancreatic β-Cells.
Representative members of the compound sets in Table 1 were validated by two methods. First, through dose-ranging experiments, we identified concentrations at which representative members of a compound set induce distinct ATP measurements in mutant vs. wild-type LCLs. Second, we mimicked the HNF4α mutation in MIN6 cells (murine insulinoma cells composed of pancreatic β-cells) by stably knocking down HNF4α by lentiviral shRNA infection and tested whether discriminating compounds also cause distinct ATP measurements in HNF4α knockdown vs. control β-cells.
Representative individual compounds from compound sets in Table 1 were confirmed to induce distinct responses in mutant vs. wild-type LCLs (Fig. S3). In pancreatic β-cells, 44% (four out of nine) of the representative compounds induced distinct ATP measurements in HNF4α knockdown vs. control cells (Table 1; Fig. 5 shows results for these four compounds, as well as for dexamethasone, a glucocorticoid that did not show an interaction with HNF4α in β-cells). These include the ω-6 fatty acid linoleic acid and several US Food and Drug Administration (FDA)-approved drugs: amiloride (a potassium-sparing diuretic that inhibits the epithelial sodium channel), propafenone (an antiarrhythmic drug that inhibits sodium channels as well as other ion channels), and simvastatin [a hydroxymethylglutaryl (HMG) CoA-reductase inhibitor used to treat hyperlipidemia] (Fig. 5). These experiments demonstrate that a significant portion of small-molecule–gene interactions that are elucidated in individual patient LCLs are also preserved in pancreatic β-cells, a canonical cell model for diabetes.
Fig. 5.
Many small-molecule–disease allele interactions identified in LCLs are also preserved in β-cells. For each of the compound sets in Fig. 4B, a representative compound’s induced ATP measurement in β-cells is shown; dexamethasone, a representative compound from the glucocorticoids set, did not interact with HNF4α in β-cells. White bars, ATP Z scores in β-cells subjected to control knockdown; gray bars, Z scores in cells subjected to HNF4α knockdown. Data are plotted as mean ± standard error of the mean (SEM) and are representative results from 2–3 independent experiments.
Small Molecules that Functionally Interact with HNF4α Modulate Insulin Secretion.
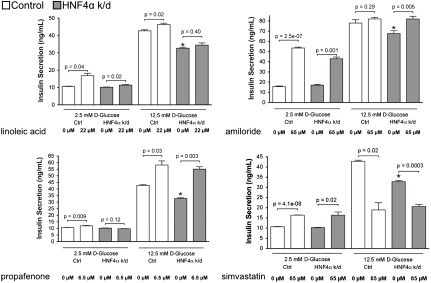
Small molecules that emerge from the screen or their protein targets presumably participate in the same or related biological processes as those involving HNF4α. We hypothesized that some of these proteins (and their associated pathways) may help mediate the cellular effects of disease mutations in HNF4α and that modulation of these proteins may favorably modify disease phenotypes. Because MODY1 patients display impaired insulin secretion (24), we tested the small molecules identified in our screen for their effects on insulin secretion from pancreatic β-cells. Several compounds that showed an interaction with HNF4α status in β-cells also modulated insulin secretion, and compounds from different classes exerted distinct patterns of effect (Fig. 6 and Table S4). In β-cells wild-type for HNF4α, linoleic acid, amiloride, propafenone, and simvastatin all increased insulin secretion at basal glucose concentrations (2.5 mM); linoleic acid and propafenone also increased insulin secretion at elevated glucose (12.5 mM). Notably, in HNF4α knockdown cells, linoleic acid, propafenone, and amiloride also increased insulin secretion in the presence of low glucose, high glucose, or both, respectively; at high glucose concentration, amiloride and propafenone restored insulin secretion in HNF4α knockdown cells to wild-type levels (Fig. 6). Simvastatin caused a complex phenotype, stimulating insulin secretion at 2.5 mM glucose but inhibiting insulin secretion at 12.5 mM glucose. The distinct effects these compounds exert on insulin secretion (e.g., as a function of glucose concentration and mutation status; Table S4) likely reflect both the complexity of insulin secretion physiology and the different mechanisms of different compounds. Overall, these data illustrate that small-molecule–disease allele interactions can identify compounds that modulate disease-modifying pathways and exert therapeutically beneficial or adverse effects on disease phenotypes.
Fig. 6.
Several small molecules that interact with HNF4α also enhance insulin secretion. Glucose-stimulated insulin secretion assays were performed on either control- (white bars) or HNF4α-knockdown (gray bars) β-cells in the presence of the indicated compounds; compound treatments were performed in the presence of either 2.5 mM or 12.5 mM glucose. ∗= P < 0.05 for comparison of glucose-stimulated insulin secretion in control- vs. HNF4α-knockdown β-cells (at 12.5 mM glucose) in absence of compound treatment. Data are plotted as mean ± SEM and are representative results from 2–3 independent experiments.
Discussion
By studying blood-derived cells from individuals, we have identified functional connections between HNF4α, several drugs, cellular ATP, and insulin secretion. These connections add a functional framework to the existing vast catalog of HNF4α transcriptional targets, and suggest several testable mechanistic and therapeutic hypotheses. None of the drugs identified in our study have been studied in association with MODY1, and none were approved for diabetes-related indications.
One of the compounds identified in our screen, linoleic acid, was the predominant species bound to HNF4α (based upon gas chromatography/mass spectrometry) in livers from fed mice or when rat HNF4α was expressed in human cells (25). Furthermore, crystal structures of HNF4α show a mixture of fatty acids spontaneously bound to the ligand-binding pocket (26, 27). These reports suggest that the emergence of linoleic acid and related fatty acids from our screen likely arises from their physical association with HNF4α, which strongly supports the ability of our method to identify physiologically relevant small-molecule–gene interactions in cells from individual patients. A physical interaction has also been reported between HNF4α and the glucocorticoid receptor (the target of dexamethasone); the glucocorticoid receptor immunoprecipitates with HNF4α in HeLa cells and human hepatocytes, and dexamethasone increases HNF4α-mediated transcriptional activation (28).
Amiloride inhibits the epithelial sodium channel (SCNN1A/B) and also shows activity against Na+/H+ exchangers (29). Derivatives of amiloride have been reported to increase insulin secretion from β-cells in vitro and in vivo (30, 31). In contrast, no association between propafenone and insulin secretion has been previously reported. Propafenone inhibits several channels, especially the voltage-gated sodium channel SCN5A and the voltage-gated potassium channel KCNH2 (29). Propafenone also inhibits ATP-sensitive potassium channel (KATP) currents in myocytes (32) and vascular smooth muscle (33), and the delayed rectifier channel Kv2.1 in Xenopus oocytes (34). Decreased activity of KATP or Kv2.1 in islets increases insulin secretion (19), suggesting that these channels may mediate propafenone’s effects on insulin secretion. Although amiloride and propafenone have not been previously studied in the context of HNF4α, SCNN1A (encoding the target of amiloride) and KCNJ11 (encoding Kir6.2, a subunit of KATP) are among the hundreds of reported transcriptional targets of HNF4α (though data on the dependence of KCNJ11 expression on HNF4α in vivo are conflicting) (10–12).
Our data suggest that functional connections relevant to β-cell physiology can be elucidated from small-molecule–gene interactions in a collection of patient-derived LCLs; the observation that some compounds showed an interaction with HNF4α in LCLs but not β-cells may reflect tissue- or species-specific interactions. LCLs have also been used as a model cell system for congenital hyperinsulinism/hyperammonemia (35), pharmacogenomic studies (36–38), ion channel disorders (39), and Huntington’s disease (40). Our approach may be extended to other diseases and patient-derived cells, including primary circulating blood cells, or even induced pluripotent stem cells; the choice of cell model for each disease study will reflect a balance between access to cells and disease-relevant physiology.
We have identified small-molecule sensitivities that are dependent on the presence of disease alleles in the native context of cells from individual patients. This approach can therefore be particularly useful to study disease alleles that are not easily studied by traditional genetic methods. By using small molecules as a source of targeted perturbations in patient-derived cells, the approach brings the logic of model organism genetics to genetically complex human diseases. Chemical–genetic interaction screens in patient samples can help discover proteins that participate in disease-relevant processes (thus placing the disease mutation in a biological context), and equally important, identify small molecules, FDA-approved drugs, or genes that act as disease modifiers. Biological discovery and clinical translation may be accelerated because a small molecule can be used both to elicit the interaction with the disease mutation and to ameliorate disease phenotypes. Small-molecule–based genetic interaction screens using patient-derived blood cells are useful for exploring a highly heritable form of diabetes and may prove useful for functional studies and clinical translation of human disease susceptibility alleles discovered through genome-wide association and other genetic studies.
Methods
LCL Culture, Small-Molecule Screen, and ATP Assay.
LCLs were purchased from Coriell Cell Repositories (Fig. 2), and cultured and maintained as previously described (23). For screening, LCLs were thawed from frozen stocks and maintained by diluting them to 100,000–300,000 cells/mL daily for ∼20 d prior to screening. LCLs were plated in 40 μL media at a density of 300,000 cells/mL; 100 nL was then pin transferred from compound libraries using a CyBi-Well Vario robot (CyBio US), and plates were incubated at 37 °C and 5% CO2 for 40 h. The CellTiter-Glo (Promega Corporation) assay for cellular ATP was performed as described (23); luminescence data were collected on an Analyst HT plate reader (LJL Biosystems, Molecular Devices). Each library member was screened in each LCL line in duplicate. For the small-molecule screen, LCLs from different individuals were screened in random order. When individual compounds were subsequently retested at various doses, LCLs were incubated with compounds for 20 h; each condition (compound dose and cell line) was tested in at least six replicates.
Small-Molecule Libraries.
Compounds screened included the following commercially available collections: Prestwick Chemical library of marketed drugs (Prestwick Chemical, 1120 compounds, 2 mg/mL stock concentration); Spectrum Collection of known bioactives, including drugs, tool compounds, and natural products (MicroSource Discovery Systems, 2000 compounds, 10 mM stock concentration); Institute of Chemistry and Cell Biology Bioactives collection (Enzo Life Sciences, 480 compounds, variable concentrations); Biomol-NT (Neurotransmitter) collection of neurotransmitter drugs and bioactives (Enzo Life Sciences, subset of 287 compounds, 10 mM stock concentration); and 86 discretes (various sources, 10 mM stock concentration).
Analysis of Interaction Screen.
Replicate raw luminescence values for each compound library member in each cell line were converted to a Z score using a standard analytic pipeline (20). Calculation of S2N for each compound across all LCL lines, and rank-ordering of compounds for their ability to discriminate LCLs that are mutant vs. wild-type at HNF4α, was performed using the Comparative Marker Selection module of GenePattern (41, 42), a data analysis platform for genomics and other systematically acquired datasets (http://www.broadinstitute.org/cancer/software/genepattern/); we used compound Z scores instead of gene expression values as inputs, and otherwise we used standard settings. P values were determined by randomly permuting mutant and wild-type class labels. For compound set enrichment analysis, we manually curated our screened compound list into 141 compound sets of at least 3 compounds (Table S2 for all 141 set names; individual compounds making up the top scoring sets are listed in Table S3). We used the Gene Set Enrichment Analysis module of GenePattern, which calculates a weighted Kolmogorov–Smirnov-like statistic (NES) for each compound set; this score reflects the extent to which set members cause statistically significant, concordant differences in ATP Z scores between mutant vs. wild-type (at HNF4α) LCLs (18, 22). (See SI Methods for more details.) P values for each compound set’s NES were calculated by randomly permuting mutant vs. wild-type HNF4α class assignments 1,000 times.
The majority of compound sets were curated based on the World Health Organization Anatomical Therapeutic Chemical Classification System (http://www.whocc.no/atc_ddd_index/), which classifies drugs according to therapeutic use and chemical properties; additional sets were curated based on membership in pathways based on the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) and Ingenuity Pathways Analysis (Ingenuity Systems).
β-Cell ATP and Insulin Assays.
MIN6 cells were cultured in DMEM containing: 25 mM glucose, supplemented with 15% FBS (ATCC), penicillin (50 international units/mL), streptomycin (50 μg/mL), and 27.5 μM β-mercaptoethanol under humidified conditions of 5% CO2 and 95% air at 37 °C. Upon stable infection with shRNA lentiviral infection particles, DMEM was supplemented with 1.25 μg/mL puromycin.
All assays were performed in 384-well plates. For ATP assays, MIN6 cells were added to 384-well plates (10,000 cells in 30 μL) in white, solid-bottom 384-well plates (Corning 3570) and cultured for 20 h. Compound dilutions were pin transferred into cells in at least six replicates and incubated with cells for 20 h at 37 °C and 5% CO2. CellTiter-Glo (Promega) was then performed according to manufacturer instructions. Luminescent output was measured using an EnVision plate reader (PerkinElmer). The Z scores [Z = (meantreated - meanDMSO)/standard deviationDMSO) were calculated for each compound concentration (in each cell line) using the mean and standard deviation of the 42 control DMSO wells found on each plate.
For glucose-stimulated insulin secretion assays, MIN6 cells were seeded at 9,000 cells/well in black, clear bottom plates (Corning 3712) and cultured in DMEM for 24 h. DMEM was aspirated using an ELx405 microplate washer (Biotek). All further solutions were prepared in Krebs–Ringer buffer (KRB) containing 0.5 % BSA (128 mM NaCl, 5 mM KCl, 2.7 mM CaCl2, 1.2 mM MgSO4, 1 mM Na2HPO4, 1.2 mM KH2PO4, 5 mM NaHCO3, buffered with 10 mM Hepes, pH 7.4); 30 μL of 2.5 mM D-glucose was added to each well and incubated with cells for 30 min at 37 °C. The 2.5 mM glucose was aspirated, cells were washed once with KRB, and then 30 μL of either 2.5 or 12.5 mM D-glucose was added to each well. Small molecules were pin transferred using the CyBi-Well Vario robot, and incubated with cells for 2 h at 37 °C. At the end of the incubation period, cells were pelleted for 5 min at 4 °C, and 20 μL of the secretion medium was collected from each well and frozen at -20 °C until processing with an Insulin ELISA kit (Alpco Diagnostics).
Statistical Analysis.
When comparing the assay results of two different cell populations (e.g., individual compound treatments of mutant vs. wild-type LCLs, or of HNF4α- vs. control-knockdown β-cells), the two-sided, unpaired Student’s t test was used. For compound set enrichment analysis, P values for enrichment scores were calculated by randomly permuting wild-type vs. mutant class labels for the LCLs 1,000 times (see the Analysis of Interaction Screen section of Methods for details). Two-way ANOVAs were performed to examine the effect of compound treatment and HNF4α mutation status on insulin secretion assays.
Additional details on LCL characterization and shRNA knockdown of HNF4α in MIN6 cells are available in SI Methods.
Supplementary Material
Acknowledgments.
This work was supported by the National Institutes of Health (Grants K08 HL077186 to S.Y.S. and GM38627 to S.L.S.) and the de Gunzburg Family Foundation at Massachusetts General Hospital (S.Y.S.). S.L.S. is an Investigator at the Howard Hughes Medical Institute. The Chemical Biology screening facility at the Broad Institute is supported by the National Institutes of Health/National Cancer Institute under Contract N01-CO-12400.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016789108/-/DCSupplemental.
References
- 1.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 2.St Onge RP, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Systematic mapping of genetic interaction networks. Annu Rev Genet. 2009;43:601–625. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 4.Dixon SJ, Stockwell BR. Identifying druggable disease-modifying gene products. Curr Opin Chem Biol. 2009;13:549–555. doi: 10.1016/j.cbpa.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrne MM, et al. Altered insulin secretory responses to glucose in subjects with a mutation in the MODY1 gene on chromosome 20. Diabetes. 1995;44:699–704. doi: 10.2337/diab.44.6.699. [DOI] [PubMed] [Google Scholar]
- 6.Fajans SS. Scope and heterogeneous nature of MODY. Diabetes Care. 1990;13:49–64. doi: 10.2337/diacare.13.1.49. [DOI] [PubMed] [Google Scholar]
- 7.Vaxillaire M, Froguel P. Monogenic diabetes in the young, pharmacogenetics and relevance to multifactorial forms of type 2 diabetes. Endocr Rev. 2008;29:254–264. doi: 10.1210/er.2007-0024. [DOI] [PubMed] [Google Scholar]
- 8.Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci USA. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolotin E, et al. Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miura A, et al. Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J Biol Chem. 2006;281:5246–5257. doi: 10.1074/jbc.M507496200. [DOI] [PubMed] [Google Scholar]
- 12.Gupta RK, et al. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 15.Graham RR, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 16.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 17.Sladek FM, Dallas-Yang Q, Nepomuceno L. MODY1 mutation Q268X in hepatocyte nuclear factor 4alpha allows for dimerization in solution but causes abnormal subcellular localization. Diabetes. 1998;47:985–990. doi: 10.2337/diabetes.47.6.985. [DOI] [PubMed] [Google Scholar]
- 18.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 19.Remedi MS, Nichols CG. Hyperinsulinism and diabetes: Genetic dissection of β cell metabolism-excitation coupling in mice. Cell Metab. 2009;10:442–453. doi: 10.1016/j.cmet.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seiler KP, et al. ChemBank: A small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–D359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pomeroy SL, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choy E, et al. Genetic analysis of human traits in vitro: Drug response and gene expression in lymphoblastoid cell lines. PLOS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman WH, et al. Abnormal insulin secretion, not insulin resistance, is the genetic or primary defect of MODY in the RW pedigree. Diabetes. 1994;43:40–46. doi: 10.2337/diab.43.1.40. [DOI] [PubMed] [Google Scholar]
- 25.Yuan X, et al. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLOS One. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wisely GB, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002;10:1225–1234. doi: 10.1016/s0969-2126(02)00829-8. [DOI] [PubMed] [Google Scholar]
- 27.Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. 2002;277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- 28.Onica T, et al. Dexamethasone-mediated up-regulation of human CYP2A6 involves the glucocorticoid receptor and increased binding of hepatic nuclear factor 4 alpha to the proximal promoter. Mol Pharmacol. 2008;73:451–460. doi: 10.1124/mol.107.039354. [DOI] [PubMed] [Google Scholar]
- 29.Goodman LS, Gilman A, Brunton LL, Lazo JS, Parker KL. Goodman & Gilman’s The Pharmacological Basis of Therapeutics. New York: McGraw-Hill; 2006. [Google Scholar]
- 30.Gunawardana SC, Head WS, Piston DW. Amiloride derivatives enhance insulin release in pancreatic islets from diabetic mice. BMC Endocr Disord. 2005;5:9. doi: 10.1186/1472-6823-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunawardana SC, Head WS, Piston DW. Dimethyl amiloride improves glucose homeostasis in mouse models of type 2 diabetes. Am J Physiol Endoc M. 2008;294:E1097–E1108. doi: 10.1152/ajpendo.00748.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christe G, Tebbakh H, Simurdova M, Forrat R, Simurda J. Propafenone blocks ATP-sensitive K+ channels in rabbit atrial and ventricular cardiomyocytes. Eur J Pharmacol. 1999;373:223–232. doi: 10.1016/s0014-2999(99)00217-4. [DOI] [PubMed] [Google Scholar]
- 33.Cogolludo AL, et al. Propafenone modulates potassium channel activities of vascular smooth muscle from rat portal veins. J Pharmacol Exp Ther. 2001;299:801–810. [PubMed] [Google Scholar]
- 34.Madeja M, et al. Molecular site of action of the antiarrhythmic drug propafenone at the voltage-operated potassium channel Kv2.1. Mol Pharmacol. 2003;63:547–556. doi: 10.1124/mol.63.3.547. [DOI] [PubMed] [Google Scholar]
- 35.MacMullen C, et al. Hyperinsulinism/hyperammonemia syndrome in children with regulatory mutations in the inhibitory guanosine triphosphate-binding domain of glutamate dehydrogenase. J Clin Endocrinol Metab. 2001;86:1782–1787. doi: 10.1210/jcem.86.4.7414. [DOI] [PubMed] [Google Scholar]
- 36.Dolan ME, et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 37.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc Natl Acad Sci USA. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan S, et al. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui Y, et al. Loss of protein kinase C inhibition in the beta-T594M variant of the amiloride-sensitive Na+ channel. Proc Natl Acad Sci USA. 1997;94:9962–9966. doi: 10.1073/pnas.94.18.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seong IS, et al. HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum Mol Genet. 2005;14:2871–2880. doi: 10.1093/hmg/ddi319. [DOI] [PubMed] [Google Scholar]
- 41.Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics. 2006;22:1924–1925. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- 42.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.