Abstract
Human DNA polymerase ι (polι) is a recently discovered enzyme that exhibits extremely low fidelity on undamaged DNA templates. Here, we show that polι is able to facilitate limited translesion replication of a thymine–thymine cyclobutane pyrimidine dimer (CPD). More importantly, however, the bypass event is highly erroneous. Gel kinetic assays reveal that polι misinserts T or G opposite the 3′ T of the CPD ∼1.5 times more frequently than the correct base, A. While polι is unable to extend the T·T mispair significantly, the G·T mispair is extended and the lesion completely bypassed, with the same efficiency as that of the correctly paired A·T base pair. By comparison, polι readily misinserts two bases opposite a 6-4 thymine–thymine pyrimidine–pyrimidone photoproduct (6-4PP), but complete lesion bypass is only a fraction of that observed with the CPD. Our data indicate, therefore, that polι possesses the ability to insert nucleotides opposite UV photoproducts as well as to perform unassisted translesion replication that is likely to be highly mutagenic.
Keywords: DNA polymerase η/DNA polymerase ζ/Rad30/Rad30B/xeroderma pigmentosum variant
Introduction
Exposure of cells to UV light results in the formation of a variety of lesions in their DNA, the two most common being cyclobutane pyrimidine dimers (CPDs) and 6-4 pyrimidine–pyrimidone photoproducts (6-4PP) at adjacent pyrimidines (Friedberg et al., 1995). These lesions present considerable impediments to replication and are normally removed from DNA by efficient repair processes (Friedberg et al., 1995). Nevertheless, situations arise where repair cannot occur and the only recourse is lesion bypass if the cell is to survive. Recent studies suggest that translesion synthesis past a CPD in humans is facilitated by polη (Masutani et al., 1999a,b, 2000; Washington et al., 2000). This process appears to be efficient and largely accurate, with adenines being inserted opposite both bases of a thymine–thymine CPD (Masutani et al., 1999b, 2000; Washington et al., 2000). In addition to T–T CPDs, polη can also replicate across certain other types of DNA damage, such as abasic sites, acetyl aminofluorene (AAF) guanine adducts and cisplatinated guanines (Masutani et al., 2000; Vaisman et al., 2000). While it appears that polη can readily misinsert opposite these lesions, extension and subsequent bypass is only achieved from the correctly paired base, explaining why polη-dependent translesion replication is generally error free (Masutani et al., 2000).
Analysis of the mutation spectra generated from UV-irradiated cultured human cell lines reveals that mutational events are targeted at di-pyrimidines (Friedberg et al., 1995). Is this simply the small fraction of error-prone translesion replication performed by human polη, or is it caused by the actions of another low fidelity DNA polymerase? In the yeast Saccharomyces cerevisiae, polζ (consisting of Rev3 and Rev7 proteins) has been shown to replicate across a T–T CPD (Nelson et al., 1996a). Human homologs of Rev3 and Rev7 have been identified (Gibbs et al., 1998; Morelli et al., 1998; Xiao et al., 1998; Lin et al., 1999a; Murakumo et al., 2000) and overexpression of Rev3 antisense mRNA leads to a dramatic drop in the extent of UV-induced mutagenesis (Gibbs et al., 1998), thereby implicating human polζ as having a pivotal role in error-prone translesion replication in normal cells. Presumably, both polη and polζ can compete for the same 3′ primer terminus at the site of a lesion, but it is not yet known why the error-free polη-dependent bypass appears to be much more efficient compared with that of error-prone polζ.
If there is competition between the error-free polη and error-prone polζ to perform translesion replication, then one would predict that there would be a dramatic increase in mutagenesis in UV-irradiated cells lacking polη. Humans with the Xeroderma pigmentosum variant (XP-V) phenotype have mutations in their POLH (XPV/RAD30A) gene that, in most cases, result in a severely truncated and inactive polη protein (Johnson et al., 1999a; Masutani et al., 1999b; Washington et al., 2000), and cultured XP-V cells are indeed hypermutable by UV light (Wang et al., 1993; Raha et al., 1996; McGregor et al., 1999). Interestingly, however, the spectrum of UV-induced mutations in the hypermutable XP-V cells is very different from that of wild-type cells (Wang et al., 1993; McGregor et al., 1999). If, as hypothesized above, mutagenesis in normal cells is dependent upon polζ out-competing polη, why would the spectra of mutations be different in cells completely lacking polη? An alternative possibility is that yet another DNA polymerase is, in fact, responsible for translesion replication in XP-V cells.
Obvious candidates for such a polymerase are paralogs of human polη. polη is a member of the recently described UmuC/DinB/Rev1/Rad30 family of DNA polymerases (Gerlach et al., 1999; McDonald et al., 1999; Woodgate, 1999), and phylogenetic analyses reveal that humans contain two Rad30 paralogs, polη and polι (McDonald et al., 1999), as well as a DinB homolog (polκ) (Gerlach et al., 1999; Ogi et al., 1999) and a Rev1 homolog (Lin et al., 1999b; Gibbs et al., 2000).
Recently, we have shown that polι replicates undamaged DNA with very low fidelity, exhibiting misinsertion frequencies averaging 1 × 10–2 (Tissier et al., 2000). Interestingly, most errors occurred at template T, where misinsertion of the ‘wobble’ base G, opposite T, was in fact favored ∼3:1 over the correct base A, and misinsertion of T opposite T occurred with roughly the same efficiency as A opposite T. As polη exhibits the same misincorporation frequency at undamaged and CPD thymines (Washington et al., 2000), we were interested in determining if the same might be true for the related polι enzyme. We find that polι can bypass a CPD, but with a reduced efficiency compared with polη. More importantly, however, the bypass is likely to be error prone, as polι is able to extend both correctly paired and mispaired bases opposite the thymine–thymine CPD. In contrast to polη, which usually inserts just one base opposite the 3′ T of a 6-4PP, polι readily inserts two bases opposite the 6-4PP, and, under certain conditions, can completely bypass the lesion. Our data suggest, therefore, that polι has the capacity to facilitate unassisted translesion replication of certain types of DNA damage with a relaxed nucleotide incorporation specificity. We discuss these findings in the context of UV-induced mutagenesis in wild-type and XP-V cells.
Results
Ability of polι to facilitate translesion replication of a thymine–thymine CPD and 6-4PP
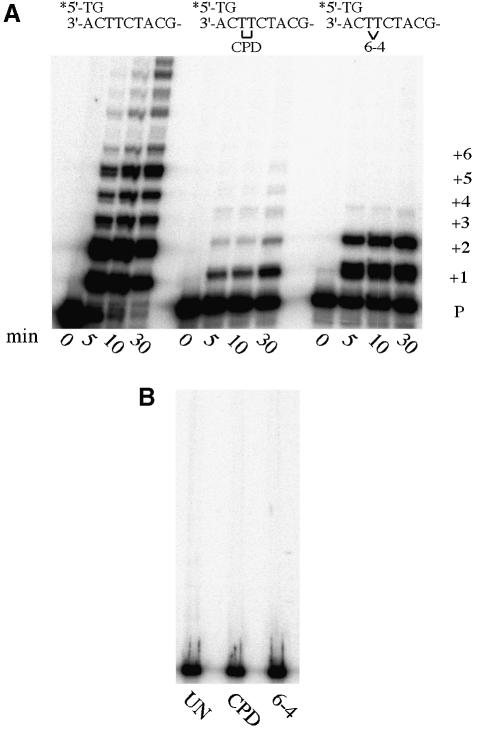
Given the phylogenetic relationship between polι and polη, we were interested in determining the ability of polι to perform translesion replication of UV-induced DNA lesions. To do so, we utilized a 30mer oligonucleotide containing a single CPD or 6-4PP at a defined site (Masutani et al., 1999a,b). The radiolabeled primer in these experiments is adjacent to the lesion, and is therefore considered a ‘standing-start’ reaction. The reason for using such a primer, rather than one with a short distance between the primer and the lesion (a ‘running-start’ reaction) is that, due to its high level of misincorporation, polι is a highly distributive enzyme that only replicates 1–2 bases before dissociating from the template (Tissier et al., 2000). The distributive nature of polι can be seen from the very same primer annealed to an undamaged template, where the replication products are predominantly 1–2 bases longer than the primer alone (Figure 1). At longer time intervals, larger sized replication products are observed, but these clearly arise from multiple binding/extension events as all of the primer has been utilized. By comparison, replication of the CPD was rather inefficient under the same assay conditions. However, after 30 min a small but significant amount of bypass product (P+5) was clearly detectable, indicating that polι-dependent translesion replication had occurred (Figure 1). It should be mentioned that the limited ability of polι to bypass a CPD is in complete agreement with previous in vitro studies with polη (Cordeiro-Stone et al., 1997; Ensch-Simon et al., 1998; Cordonnier et al., 1999; Masutani et al., 1999a). polη is clearly the most efficient enzyme to perform translesion replication past CPDs in cell extracts, and in its absence (such as in XP-V extracts) translesion replication only occurs with ∼10% or less of the efficiency observed in its presence (Cordonnier et al., 1999; Masutani et al., 1999a). As a consequence, we expected that polι-dependent bypass would not be as robust as polη-dependent bypass.
Fig. 1. polι-dependent replication of undamaged, CPD-containing and 6-4PP-containing DNA templates. Reactions were performed as described in Materials and methods for the times noted below each track. The template sequence is indicated at the top of the figure, and replication products (P+1, P+2, etc.) are indicated at the right of (A). (A) Replication promoted by wild-type GST–polι. (B) Replication in the presence of a mutant (D126A/E127A) GST–polι devoid of polymerase activity (Tissier et al., 2000). In these experiments, reactions were performed for 30 min in the presence of an undamaged template (UN), a CPD template (CPD) or a 6-4PP-containing template (6-4).
In contrast to the bypass seen with the CPD, considerable polι-dependent misinsertion opposite both bases of the 6-4PP (Figure 1; P+1 and P+2) was seen, as was a faint bypass product (P+3) after a short 5 min reaction. However, the extent of misincorporation and bypass of the 6-4PP did not increase even after a 30 min reaction (Figure 1).
Replication products were not observed with any of the three templates when wild-type polι was substituted with a mutant protein that is devoid of polymerase activity (Tissier et al., 2000), indicating that the misinsertion and bypass of the two UV lesions is entirely polι dependent (Figure 1).
Specificity of polι-dependent nucleotide incorporation opposite the CPD and 6-4PP
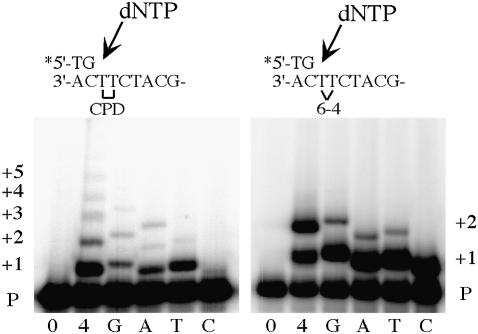
To determine the specificity of nucleotide incorporation opposite both lesions, we assayed the extent of polι-dependent (mis)incorporation in the presence of all four dNTPs and each individual dNTP (Figure 2). After a 30 min reaction, it appears that T is most efficiently incorporated opposite the 3′ T of the CPD. By comparison, G and A appeared to be incorporated slightly less efficiently, but this may be misleading because both the misincorporated G, and the correctly incorporated A, are clearly extended to the 5′ T of the dimer and beyond. In the case of G, this represents misincorporation of G opposite the two thymines of the dimer followed by the correct incorporation of G opposite an undamaged C immediately after the lesion. In the case of A, this represents the correct incorporation of two As opposite the dimer followed by misincorporation opposite C. Under these conditions very little misincorporation of C opposite the 3′ T of the CPD was observed.
Fig. 2. Incorporation of nucleotides opposite a CPD or 6-4PP. The sequence of each template is indicated above each panel. Each primer/template was incubated for 30 min with polι in the absence of dNTPs (0), all four dNTPs (4), or each dNTP individually (G, A, T, C). The size of replication products (P+1, P+2, etc.) is indicated at the right and left of the figure.
A very different picture emerged with the 6-4PP, with polι inserting all four dNTPs with roughly equal efficiency at the 3′ T of the lesion and limited insertion of G, A or T at the 5′ site (Figure 2). Interestingly, the intensity of incorporation at P+2 was much greater in the presence of all four dNTPs suggesting that polι favors different nucleotides at the 3′ and 5′ sites of the 6-4PP (Figure 2).
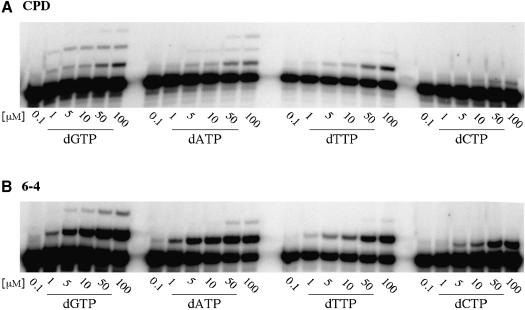
Using a gel kinetic assay (Creighton et al., 1995), we have quantitatively determined which dNTP polι inserts opposite the 3′ T of the CPD and 6-4PP. Unlike the experiments shown in Figure 2, these experiments are performed under steady-state ‘single-hit’ kinetics. This was achieved by varying the concentration of incoming dNTP from 0.1–100 µM and reducing the reaction time from 30 to 10 min for the CPD, and to 2 min for the 6-4PP. Under these conditions, <20% of the primer is extended and, when determining the misincorporation frequency, we take into account that any bands observed at P+2 and P+3 could only have arisen from extension of the initial insertion at P+1 (opposite the 3′ T of each dimer) (Figure 3A). These quantitative experiments revealed that T is misincorporated ∼1.7-fold better than the correct base, A. Similarly, G is misincorporated ∼1.4-fold better than A. These misincorporation frequencies are in dramatic contrast to those reported for polη, where polη-dependent misinsertion of T occurs with a frequency of 1.1 × 10–2 and misinsertion of G with a frequency of 4.2 × 10–3 relative to A (Johnson et al., 2000). Thus, polι is ∼150- to 300-fold more likely to misincorporate at the 3′ T of the CPD than polη.
Fig. 3. Steady-state kinetic analysis of polι-dependent nucleotide incorporation opposite the 3′ T of a CPD or 6-4PP. (A) CPD. (B) 6-4PP. The concentration of each nucleotide (in µM) added to the reaction is indicated below each track. Experiments illustrating incorporation opposite the CPD were performed for 10 min, while those showing polι-dependent incorporation opposite the 6-4PP were for 2 min.
The qualitative misincorporation experiments with the 6-4PP suggested that polι inserts all four dNTPs opposite the 3′T of the lesion with roughly equal efficiency. However, quantitative analysis of the incorporation (Figure 3B) revealed that incorporation of A was, in fact, slightly favored over the three misincorporated bases. The estimated misincorporation frequency of T, G or C insertion opposite the 3′ T of the 6-4PP was 6.4 × 10–1, 3.6 × 10–1 and 1.0 × 10–1, respectively.
Efficiency of extending paired/mispaired primers opposite the 3′ T of the CPD or 6-4PP
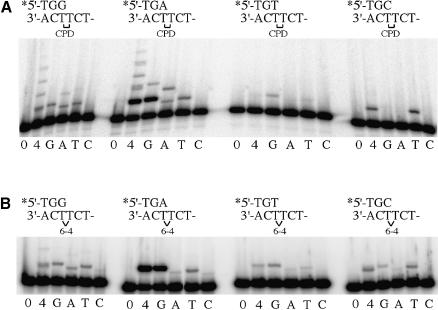
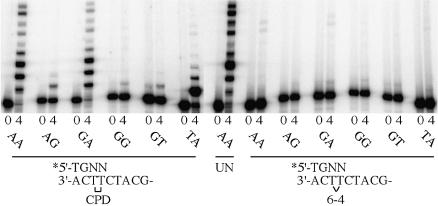
While it is clearly important to determine the misincorporation frequency at the 3′ site of both lesions, a question as to the biological significance of the data is raised if the (mis)incorporated base is not efficiently extended. Primers were therefore synthesized in which each of the four bases would be correctly or incorrectly paired with the 3′ T of the CPD or 6-4PP (Figure 4). polι-dependent extension of these annealed primers was assayed after a 2 min reaction for the 6-4PP template and 10 min for the CPD template, in the presence of all four dNTPs or each individual dNTP (100 µM). Surprisingly, the most efficient extension of a primer annealed to the CPD-containing template occurred with the correctly paired 3′ A, but in the presence of dGTP rather than dATP (Figure 4A). Indeed, there was significant misincorporation of G opposite the 5′ T of the CPD, and the lesion bypassed by a further incorporation opposite the adjacent undamaged C in the template (Figure 4A). The A·T primer was also extended in the presence of A and T, but complete lesion bypass only occurred in the presence of dATP (Figure 4A). By comparison, extension of the G·T mispair was less efficient in the presence of dGTP, but was similar with A and T, and the extent of complete lesion bypass in the presence of all four dNTPs was only slightly less than that seen with the correctly paired A·T primer. In contrast, only a small amount of polι-dependent extension of the T·T mispair was observed in the presence of dGTP. The C·T mispair was extended, but only in the presence of dTTP.
Fig. 4. polι-dependent extension of primers opposite the 3′ T of the CPD or the 6-4PP. The 3′ sequence of each primer is shown above each group of experiments. (A) CPD. (B) 6-4PP. 0, 4, G, A, T and C indicate reactions in the absence of nucleotides, all four nucleotides, or G, A, T or C, respectively.
Extension of primers annealed to the 6-4PP-containing template was varied. Similar to the results obtained with the CPD lesion, the best extension was seen with the correctly paired A·T primer in the presence of dGTP (Figure 4B). This result also explains why we observed efficient incorporation of two nucleotides opposite the 6-4PP in Figures 1 and 2. Although four dNTPs can be inserted at the 3′ T, only the A·T primer is extended significantly in the presence of dGTP. In contrast to the CPD, however, no complete bypass products were detected. Extension of the G·T mispair was qualitatively similar with both the CPD and 6-4PP. polι was able to insert G, A or T opposite the 5′ T of the 6-4PP, and in the presence of all four dNTPs, a faint but detectable bypass product was observed (Figure 4B). While virtually no extension of the T·T mispair was observed with the CPD, extension in the presence of G or T was observed with the 6-4PP, as was extension of the C·T mispair (Figure 4B). Thus, in these reactions, significant incorporation of the correct nucleotide, A, opposite the 5′ T of the 6-4PP only occurs when the 3′ base is a misinserted G.
polι-dependent extension of primers with two bases opposite the CPD or 6-4PP
The data presented in Figures 2, 3 and 4 indicate that polι can insert a variety of bases opposite the 3′ T of each lesion, but that the efficiency at which the (mis)pair is extended varies considerably. But what happens to the primers that are actually extended to the 5′ site of the lesion? Can these now be utilized to bypass the lesion completely? To determine if this might be the case, we synthesized primers in which the terminal 3′ bases would be located directly opposite the CPD or 6-4PP in the template DNA (Figure 5). Instead of analyzing all theoretical combinations of nucleotides at the 3′ and 5′ site of each lesion, we focused on a set of six primers that represented the best misincorporation and/or extension at the 3′ and 5′ sites of each dimer (Figure 5). Under these conditions, polι was able to extend efficiently a primer with two As opposite the CPD, and the extent of replication was only slightly less than that seen with the same primer annealed to an undamaged template (Figure 5). Interestingly, a virtually identical pattern was observed with the ‘GA’ primer, indicating that it too is a good substrate for polι-dependent extension and complete lesion bypass. Although the misincorporation/extension results presented in Figures 3 and 4 suggest that polι readily inserts an A opposite the 3′ T followed by a G at the 5′ T, only a small fraction of the ‘AG’ primers were extended beyond the lesion (Figure 5).
Fig. 5. polι-dependent extension of primers with various 3′ dinucleotides opposite the CPD or 6-4PP. Each reaction was performed for 15 min in the absence of nucleotides (0), or all four nucleotides (4). The 3′ dinucleotide sequence of each primer is given below each panel. The left panel shows extension from the CPD-containing template, while on the right are extensions from the 6-4PP-containing template. The track in the middle, labeled UN, demonstrates replication from an ‘AA’ primer and an undamaged template. Note that any extension from these primers represents complete bypass of the respective lesion.
The most frequent misincorporation opposite the 3′ T of the CPD was T (Figure 3A). However, virtually no polι-dependent extension of this mispair was observed (Figure 4A). But what if the T·T mispair is, perhaps, extended by a different polymerase? We reasoned that the most likely base inserted at the 5′ T would be A. Therefore, we synthesized a ‘TA’ primer in which the T is mispaired with the 3′ T of the CPD, but the base opposite the 5′ T is the correct base, A. Interestingly, this primer was efficiently extended in the presence of all four dNTPs, but generally only by 1–2 bp (Figure 5). Longer replication products were detectable, but these were much less intense than those observed with the ‘AA’ or ‘GA’ primers, suggesting that the ‘TA’ primer/terminus may still be somewhat distorted. By comparison, there was very little extension of the ‘GG’ or ‘GT’ primer annealed to the CPD-containing template. The results with the ‘GG’ primer contrast with our results shown in Figures 2 and 3, where we clearly observed a small amount of bypass in the presence of dGTP alone. This suggests that polι is unable to extend a ‘standing-start’ primer containing two Gs opposite the lesion, but can do so (on a limited scale) if the initial primer is located before the lesion. The bypass data shown in Figures 2 and 3 presumably occur as a result of a single binding/extension event prior to the lesion, rather than from misinsertion, dissociation and subsequent elongation.
None of the aforementioned primers was significantly extended by polι when annealed to the 6-4PP-containing template. Similar to the results shown in Figures 1 and 4B, faint bypass products are detectable in the ‘AA’ and ‘GA’ tracks, but it is difficult to determine if these levels are of biological significance. We conclude, therefore, that while polι can readily misinsert nucleotides opposite both bases of the 6-4PP, and may be able to bypass the lesion completely under certain conditions, the efficiency at which it bypasses a 6-4PP is greatly reduced compared with that of the CPD.
Discussion
To date, 12 eukaryotic DNA polymerases have been identified (for a recent review see Hubscher et al., 2000). Of these polymerases, only polζ and polη have been shown to facilitate translesion replication of a CPD in vitro (Nelson et al., 1996a; Johnson et al., 1999b, 2000; Masutani et al., 1999a,b). The studies reported here indicate that polι can now be added to the list of enzymes that can perform unassisted lesion bypass. polι-dependent translesion replication differs from that of the related polη in the efficiency and the accuracy of the reaction. The limited ability of polι to bypass a CPD is not surprising given the fact that XP-V cell extracts, in which polη is inactive, exhibit only ∼10% of the lesion bypass activity of normal cell extracts, indicating that the majority of translesion replication is normally polη dependent (Cordonnier et al., 1999; Masutani et al., 1999a). Perhaps the most striking aspect of these studies is the fact that the polι-dependent incorporation of nucleotides opposite the lesion and subsequent bypass is highly error prone. Both T and G were incorporated opposite the 3′ T of the CPD ∼1.5-fold better than A. At first glance, this might not seem much, but consider the fact that polη-dependent misinsertion of T and G occurs with a frequency of 10–2–10–3 less than A! Interestingly, most of the bases, whether they be correctly or incorrectly inserted, were not efficiently extended by polι. Extension from the misinserted T was barely detectable, and extension of the mispaired G or correctly paired A was limited. It should be noted, however, that any primers which were extended by the incorporation of an A opposite the 5′ T of the CPD (AA, GA or TA; Figure 5) served as excellent substrates for subsequent replication and complete lesion bypass by polι.
In contrast to the CPDs, which are clearly bypassed by polι in our in vitro assays, only very limited bypass of 6-4PP occurs and, as noted above, at the present time we cannot ascertain whether the extent of polι-dependent bypass will ultimately turn out to be of biological significance. The inability of polι to bypass the 6-4PP with great efficiency is similar to that of the related polη, which is also unable to traverse a 6-4PP (Masutani et al., 1999a). However, in contrast to polη, which usually inserts one nucleotide opposite the 3′ T (Masutani et al., 1999a) and a second nucleotide opposite the 5′ T at high enzyme concentrations (Masutani et al., 2000), polι readily inserts two bases opposite the lesion. Given the fact that neither polη nor polι can perform significant levels of unassisted bypass of a 6-4PP in vitro, one has to hypothesize that both enzymes lack a factor(s) that normally allows for lesion bypass in vivo, or that it is perhaps facilitated by another enzyme.
Possible mechanisms of translesion replication in wild-type and XP-V cells
Although the experiments reported here were performed entirely in vitro, we believe that they shed light on the mutagenic process in vivo. Our in vitro studies indicate that the predominant product of polι-dependent misincorporation and extension at both the CPD and 6-4PP is the correct insertion of A opposite the 3′ T followed by misinsertion of G opposite the 5′ T (Figure 4). If similar events occurred in vivo, they would lead to 5′ T→C mutations. However, analysis of the mutagenic spectra observed after exposing human cells to UV light suggests that most mutations are, in fact, targeted to the 3′ site of a di-pyrimidine. Our data indicate, therefore, that polι probably does not participate in translesion replication in wild-type cells in vivo. Indeed, several lines of evidence suggest that the predominant lesion bypass pathway in humans is polη dependent. But what happens if polη is absent, such as in XP-V cells? Interestingly, the spectra of mutations found in XP-V cells are very different from those observed in normal cells and parallel the frequency of polι-dependent misincorporations at the 3′ T of the CPD (Figure 3A), i.e. T→C (misincorporation of G opposite T) and T→A (misincorporation of T opposite T) occur with roughly similar frequency (Wang et al., 1993; McGregor et al., 1999). However, as shown in Figure 4, the T·T mispair is poorly extended by polι in vitro. This apparent incongruity can be reconciled if one postulates that the T·T mispair is, in fact, extended by a different enzyme in vivo. An obvious candidate is human polζ. Saccharomyces cerevisiae polζ is able to extend mispaired bases (Lawrence et al., 2000), as well as Rev1- (Nelson et al., 1996b) and polη-dependent misinsertions (Yuan et al., 2000), so if the human polζ shares similar properties, it should be able to extend any polι-dependent misinsertions opposite the 3′ T of the CPD. Indeed, such a propensity of polζ to extend mispaired bases opposite lesions is probably not restricted to polι-dependent misinsertions in XP-V cells, but is also likely to occur with polη-dependent misinsertions in normal cells as well.
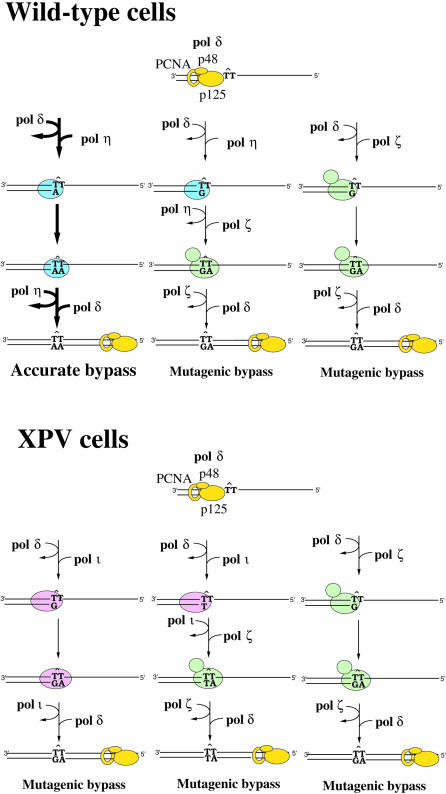
As a consequence, we would like to propose a model for translesion replication in human cells that takes into account the known biochemical properties of polζ, polη and polι, as well as incorporating physiological data gathered from a variety of in vivo experiments (Figure 6). It is clear that the predominant error-free/accurate mode of translesion DNA synthesis in wild-type cells is dependent upon polη (Johnson et al., 1999a; Masutani et al., 1999a,b). However, polη also frequently misincorporates bases opposite the 3′ T of a CPD (Johnson et al., 2000; Masutani et al., 2000). These are not extended efficiently by polη, but may be substrates for another enzyme, such as polζ. Alternatively, human polζ, like its S.cerevisiae counterpart, may also perform unassisted error-prone translesion replication. Although wild-type cells express polι, our in vitro data suggest that polι has a limited capacity to replicate damaged DNA, and/or extend any misinserted bases. As a consequence, we believe that polι may play only a minor role in translesion replication in normal cells. However, the situation is very different in the absence of polη, such as in XP-V cells. Although polζ could conceivably perform all of the bypass, since the mutational spectra observed in XP-V cells are strikingly similar to polι-dependent misinsertion opposite the 3′ T of the CPD (Wang et al., 1993; McGregor et al., 1999), we suggest that most of the mutagenic events scored in XP-V cells are, in fact, probably polι dependent. Some of these will be unassisted polι-dependent misinsertion and bypass, while others will be polι-dependent misincorporations that are subsequently extended by polζ. Thus, we propose that most error-free bypass of lesions (at least CPDs) is likely to represent unassisted polη-dependent translesion replication, while the error-prone component possibly results from unassisted polι or polζ translesion replication and a combination of polη–polζ, or polι–polζ bypass. Experiments are currently in progress to test various aspects of this model.
Fig. 6. Possible mechanisms of translesion replication in wild-type and XP-V cells. In both cases, a CPD is encountered by the cell’s main replicase, polδ (shown in yellow). In a normal cell, translesion replication is accurately performed by polη (shown in light blue). This is clearly the main replicative pathway utilized by the cell to bypass a CPD and we have indicated this fact by larger sized arrows and labeling. On occasions, however, polη misinserts a base opposite the 3′ T of the CPD (Johnson et al., 2000; Masutani et al., 2000). These mispairs cannot be extended by polη, but are probably extended by polζ (shown in light green). Alternatively, polζ may perform unassisted translesion replication, which is error prone. This model incorporates a central role in error-prone lesion bypass attributed to polζ and explains why overproduction of antisense to hRev3 (the catalytic cores of polζ) results in a dramatic reduction of UV light-induced mutagenesis in human cells (Gibbs et al., 1998). In XP-V cells devoid of polη, lesion bypass is highly error prone and probably occurs as a consequence of three discrete pathways: unassisted polι-dependent bypass (polι is shown in pink); polι-misincorporation followed by polζ extension of the mispair; or unassisted polζ-dependent bypass. If the two polι-dependent pathways predominate over the unassisted polζ-bypass pathway, it would explain why the spectra of mutations are very different in wild-type and XP-V cell lines (Wang et al., 1993; McGregor et al., 1999). In all cases, polη, polι or polζ only replicates a few nucleotides surrounding the lesion before dissociating from the primer/template and is rapidly replaced by polδ, which completes chromosome duplication.
Materials and methods
Human DNA polymerase ι
Wild-type or mutant glutathione S-transferase (GST)-tagged polι were purified by GST affinity chromatography and hydroxylapatite ion-exchange chromatography as previously described (Tissier et al., 2000).
DNA templates
The templates for the replication assays were synthetic oligonucleotide 30mers with the sequence 5′-CTCGTCAGCATCTTCATCATACAGTCAGTG-3′. The two thymines underlined indicate the position of the CPD or 6-4PP. Lesion-containing oligonucleotides were synthesized as previously described (Masutani et al., 1999a,b). The undamaged template and all oligonucleotide primers were synthesized by Loftstrand Laboratories (Gaithersburg, MD) using standard techniques and were gel purified prior to use. The initial primer used in these studies is a 16mer, 5′-CACTGACTGTATGATG-3′. This primer anneals to the template such that the first base replicated is the 3′ T of the T–T dimer. 17mer primers were subsequently synthesized that were identical to the 16mer, but contained an extra terminal 3′ A, T, G or C. When annealed, the 3′ base of each primer would be located opposite the 3′ T (in the template) of the lesion and the first base replicated would be the 5′ T of the dimer. 18mer oligonucleotides were also synthesized that had identical 5′ sequences to the 16mer, but contained different terminal 3′ dinucleotides (either AA; AG; GA; GG; GT; TA). When annealed, these dinucleotides are located directly opposite the lesion in the template. The first template base replicated from each primer is an undamaged C. Each primer was 5′-labeled with [γ-32P]ATP (5000 Ci/mmol; 1 Ci = 37 GBq) (Amersham Pharmacia Biotech, Piscataway, NJ) using T4 polynucleotide kinase (Life Technologies, Gaithersburg, MD).
Replication reactions
Radiolabeled primer-template DNAs were prepared by annealing the 5′-32P-labeled primer to the unlabeled template DNA at a molar ratio of 1:1.5. Standard reactions of 10 µl contained 40 mM Tris–HCl pH 8.0, 5 mM MgCl2, 100 µM of each ultrapure dNTP (Amersham Pharmacia Biotech, NJ), 10 mM dithiothreitol, 250 µg/ml bovine serum albumin, 60 mM KCl, 2.5% glycerol, 10 nM 5′-[32P]primer-template DNA and 10 nM GST–polι. After incubation at 37°C for 30 min, reactions were terminated by the addition of 10 µl of 95% formamide/10 mM EDTA and the samples heated to 100°C for 5 min. Five microliters of the reaction mixture were subjected to 20% polyacrylamide–7 M urea gel electrophoresis and replication products visualized by autoradiography or PhosphorImage analysis.
Kinetic analysis of replication products
Time course reactions using the standard replication conditions were initially performed so as to ensure that the reaction was in the linear range (usually <20% of primer utilization) (Boosalis et al., 1987; Creighton et al., 1995). Subsequent reactions measuring the incorporation of nucleotides opposite the 3′ T of the CPD were for 10 min, while those measuring incorporation opposite the 3′ T of the 6-4PP were for 2 min. In all cases, the concentration of each nucleotide in the reaction mixture was varied from 0.1 to 100 µM. All of the reactions were initiated by the addition of the appropriate dNTP. Reaction products were separated in a 20% polyacrylamide gel containing 7 M urea, and gels dried prior to quantitative PhosphorImage analysis using the ImageQuant software (Molecular Dynamics, CA). Nucleotide misincorporation frequencies were subsequently calculated as previously described and the data presented are the average of 2–3 separate experiments (Boosalis et al., 1987; Creighton et al., 1995; Tissier et al., 2000).
Acknowledgments
Acknowledgements
We would like to thank Alan Lehmann for his constructive comments on the manuscript, and Martín Gonzalez, Antonio Rodriguez, Mary McLenigan, Dominique Vandewiele and François Boudsocq for their helpful comments and suggestions during the course of this work. This work was supported in part by the NIH Intramural research program and by a CREST grant to F.H.
References
- Boosalis M.S., Petruska,J. and Goodman,M.F. (1987) DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem., 262, 14689–14696. [PubMed] [Google Scholar]
- Cordeiro-Stone M., Zaritskaya,L.S., Price,L.K. and Kaufmann,W.K. (1997) Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J. Biol. Chem., 272, 13945–13954. [DOI] [PubMed] [Google Scholar]
- Cordonnier A.M., Lehmann,A.R. and Fuchs,R.P.P. (1999) Impaired translesion synthesis in Xeroderma pigmentosum variant extracts. Mol. Cell. Biol., 19, 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton S., Bloom,L.B. and Goodman,M.F. (1995) Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading and lesion bypass efficiencies. Methods Enzymol., 262, 232–256. [DOI] [PubMed] [Google Scholar]
- Ensch-Simon I., Burgers,P.M. and Taylor,J.S. (1998) Bypass of a site-specific cis–syn thymine dimer in an SV40 vector during in vitro replication by HeLa and XPV cell-free extracts. Biochemistry, 37, 8218–8226. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology,Washington, DC. [Google Scholar]
- Gerlach V.L., Aravind,L., Gotway,G., Schultz,R.A., Koonin,E.V. and Friedberg,E.C. (1999) Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc. Natl Acad. Sci. USA, 96, 11922–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence,C.W. (1998) A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs P.E., Wang,X.D., Li,Z., McManus,T.P., McGregor,W.G., Lawrence,C.W. and Maher,V.M. (2000) The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc. Natl Acad. Sci. USA, 97, 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher U., Nasheuer,H.-P. and Syvaoja,J.E. (2000) Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci., 25, 143–147. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999a) hRAD30 mutations in the variant form of Xeroderma Pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1999b) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, polη. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (2000) Fidelity of human DNA polymerase η. J. Biol. Chem., 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- Lawrence C.W. et al. (2000) Roles of DNA polymerase ζ and Rev1 protein in eukaryotic mutagenesis and translesion replication. Cold Spring Harb. Symp. Quant. Biol., 65, in press. [DOI] [PubMed] [Google Scholar]
- Lin W., Wu,X. and Wang,Z. (1999a) A full-length cDNA of hREV3 is predicted to encode DNA polymerase ζ for damage-induced mutagenesis in humans. Mutat. Res., 433, 89–98. [DOI] [PubMed] [Google Scholar]
- Lin W., Xin,H., Zhang,Y., Wu,X., Yuan,F. and Wang,Z. (1999b) The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res., 27, 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999a) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C. et al. (1999b) The XPV (Xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.P., Rapic-Otrin,V., Epstein,J.A., Broughton,B.C., Wang,X., Lehmann,A.R., Wolgemuth,D.J. and Woodgate,R. (1999) Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics, 60, 20–30. [DOI] [PubMed] [Google Scholar]
- McGregor W.G., Wei,D., Maher,V.M. and McCormick,J.J. (1999) Abnormal, error-prone bypass of photoproducts by xeroderma pigmentosum variant cell extracts results in extreme strand bias for the kinds of mutations induced by UV light. Mol. Cell. Biol., 19, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli C., Mungall,A.J., Negrini,M., Barbanti-Brodano,G. and Croce,C.M. (1998) Alternative splicing, genomic structure and fine chromosome localization of REV3L. Cytogenet. Cell Genet., 83, 18–20. [DOI] [PubMed] [Google Scholar]
- Murakumo Y., Roth,T., Ishii,H., Rasio,D., Numata,S., Croce,C.M. and Fishel,R. (2000) A human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J. Biol. Chem., 275, 4391–4397. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996a) Thymine–thymine dimer bypass by yeast DNA polymerase ζ. Science, 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996b) Deoxycytidyl transferase activity of yeast REV1 protein. Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- Ogi T., Kato,T. and Ohmori,H. (1999) Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein DinB. Genes Cells, 4, 607–618. [DOI] [PubMed] [Google Scholar]
- Raha M., Wang,G., Seidman,M.M. and Glazer,P.M. (1996) Mutagenesis by third-strand-directed psoralen adducts in repair-deficient human cells: high frequency and altered spectrum in a Xeroderma pigmentosum variant. Proc. Natl Acad. Sci. USA, 93, 2941–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A., McDonald,J.P., Frank,E.G. and Woodgate,R. (2000) polι, a remarkably error-prone human DNA polymerase. Genes Dev., 14, 1642–1650. [PMC free article] [PubMed] [Google Scholar]
- Vaisman A., Masutani,C., Hanaoka,F. and Chaney,S.G. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry, 39, 4575–4580. [DOI] [PubMed] [Google Scholar]
- Wang Y.C., Maher,V.M., Mitchell,D.L. and McCormick,J.J. (1993) Evidence from mutation spectra that the UV hypermutability of Xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol. Cell. Biol., 13, 4276–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington M.T., Johnson,R.E., Prakash,S. and Prakash,L. (2000) Accuracy of thymine–thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl Acad. Sci. USA, 97, 3094–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R. (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev., 13, 2191–2195. [DOI] [PubMed] [Google Scholar]
- Xiao W., Lechler,T., Chow,B.L., Fontanie,T., Agustus,M., Carter,K.C. and Wei,Y.F. (1998) Identification, chromosomal mapping and tissue-specific expression of hREV3 encoding a putative human DNA polymerase ζ. Carcinogenesis, 19, 945–949. [DOI] [PubMed] [Google Scholar]
- Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.S. and Wang,Z. (2000) Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]