Abstract
The two major functions of human natural killer (NK) cells are conventionally associated with distinct cell subsets. Thus, cytolytic activity is mostly confined to the CD56dimCD16+ subset, whereas cytokine production is generally assigned to CD56brightCD16+/− cells. In this study, we reevaluated the functional capabilities of these NK subsets with regard to the production of IFN-γ at different time points after cell triggering via NKp46 and NKp30 activating receptors. Different from previous studies, cytokine production was also assessed at early intervals. We show that CD56dim NK cells produce IFN-γ already at 2 to 4 h, whereas no cytokine production is detected beyond 16 h. In contrast, CD56bright cells release IFN-γ only at late time intervals (>16 h after stimulation). The rapid IFN-γ production by CD56dim NK cells is in line with the presence of IFN-γ mRNA in freshly isolated cells. Rapid IFN-γ production was also induced by combinations of IL-2, IL-12, and IL-15. Our data indicate that not only cytolytic activity but also early IFN-γ production is a functional property of CD56dim NK cells. Thus, this subset can assure a rapid and comprehensive NK cell intervention during the early phases of innate responses.
Natural killer (NK) cells are a major player in innate immune responses, in which they exert cytolytic activity against virus-infected or tumor cells and produce various proinflammatory cytokines and chemokines (1, 2). NK cell activation is finely tuned by a number of activating and inhibitory receptors (3). In humans, the major activating receptors include NKp46, NKp30, and NKp44 [collectively called natural cytotoxicity receptors (NCRs)], NKG2D, CD16, 2B4, NKp80, and DNAM-1 (4, 5). The major inhibitory receptors are specific for HLA class I molecules and prevent the NK-mediated attack to most normal (HLA class I+) autologous cells (4–6). NK cell triggering can also be induced by cytokines, including IL-2, IL-12, IL-15, and IL-18 (2, 7, 8).
Although NK cells are abundant in peripheral blood, they exert their function primarily in tissues and secondary lymphoid organs, where they migrate early in response to inflammation caused primarily by invading pathogens (9). In addition, a unique NK cell subset is largely represented during pregnancy in the decidua and is thought to play a relevant role in tissue remodeling, neoangiogenesis, and immunoregulation (10, 11).
Two main NK cell functions, i.e., cytolytic activity and cytokine production, have been primarily assigned to distinct NK cell subsets defined on the basis of the cell surface density of CD56 and on the expression of CD16 (1, 7, 12, 13). Namely, CD56dimCD16+ NK cells, predominant in peripheral blood, display potent cytolytic activity, whereas the poorly cytolytic CD56brightCD16+/− NK cells that predominate in tissues and secondary lymphoid organs would be responsible for cytokine production, primarily IFN-γ and TNF-α, in addition to other cytokines and chemokines [including GM-CSF, MCP-1, CCL3 (MIP1α), CCL4 (MIP1β), CCL5 (RANTES)] (1, 2, 14, 15). The fact that these NK cell subsets have distinct functional capabilities and different localization raises the question of how NK cells can coordinate and harmonize their defensive mechanisms. An efficient defense would require not only the rapid killing of “abnormal cells” (i.e. infected or neoplastic), but also a prompt production of cytokines and chemokines to promote/amplify inflammatory responses, recruit/activate other cell types, and favor polarization of downstream adaptive responses toward Th1 cells, which is particularly efficacious against various infectious agents and tumor cells. It should be emphasized that cytokine production by NK cells has been routinely analyzed at late intervals (≥16 h after cell stimulation) but no suitable experimental setting has been adopted to explore early cytokine production. Therefore, we investigated this issue and also analyzed early time points after NK cell triggering.
We show that CD56dim NK cells do release high amounts of IFN-γ very early after activation, i.e., in a time interval that overlaps with the process of target cell killing. Our data provide clear evidence that the same NK cell subset can simultaneously exert two important effector functions and provides a rational picture for the NK cell intervention in the framework of early innate immune responses.
Results
Early IFN-γ Production by Fresh CD56dim NK Cells upon Receptor-Mediated Stimulation.
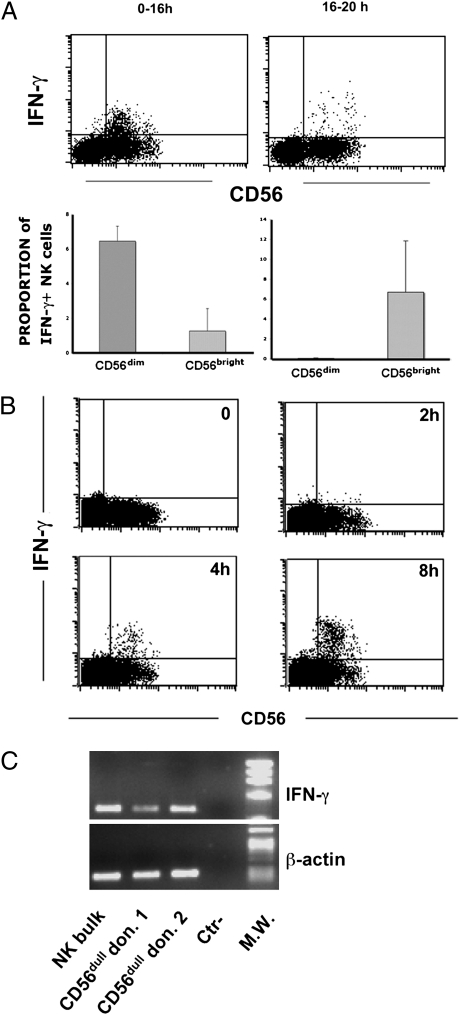
We reevaluated the ability of CD56dim and CD56bright NK cell subsets to produce IFN-γ, with particular focus on early time intervals after stimulation. In the first set of experiments, NK cell triggering was induced on peripheral blood mononuclear cells (PBMCs) by anti-NKp30 and anti-NKp46 mAbs together with FcγR+ P815 murine mastocytoma cells. To allow detection of cytokines produced at short intervals, GolgiPlug (a protein transport inhibitor) was added together with the stimulating mAb. For analysis, PBMCs were gated by forward and side scatter and on exclusion of CD3+CD14+CD19+ cells. Flow cytometric analysis was performed 16 h after stimulation. By this procedure, it is possible to detect all NK cells producing cytokines within the time interval analyzed (i.e., between 0 and 16 h; Fig. S1). Data were compared with those obtained by adding GolgiPlug at 16 h after NK cell stimulation (i.e., the approach routinely used to assess cytokine production by NK cells). By using this conventional method, mostly CD56bright NK cells were found to produce IFN-γ, in agreement with data obtained in previous studies (Fig. 1A). On the contrary, analysis of IFN-γ released during the first 16 h after NK cell triggering revealed that its production was predominantly confined to CD56dim NK cells. As IFN-γ production by CD56dim NK cells was confined to the first 16 h after stimulation, we further analyzed its production at earlier time intervals. As shown in Fig. 1B, some IFN-γ–producing CD56dim NK cells were detectable 2 h after receptor-mediated triggering, whereas their numbers markedly increased at 4 h and 8 h. NK cells were also analyzed for their cytolytic function as evaluated by the surface expression of CD107a in a 4-h assay. As expected, CD107a was fully detectable in CD56dim NK cells (Fig. S2), thus paralleling the early IFN-γ production.
Fig. 1.
IFN-γ production by peripheral blood CD56dim and CD56bright NK cell subset. (A) Flow cytometric analysis. PBMCs were stimulated in a redirected triggering assay (reverse antibody-dependent cell-mediated cytotoxicity) using FcγR + P815 cells and anti-NKp30/anti-NKp46 mAbs (γ isotype). GolgiPlug was added immediately at the time of stimulation until processed (Upper, 0–16 h) or after 16 h with analysis of cells after 4 h (Lower, 16–20 h; Fig. S1). CD3−19−14− cells were gated by flow cytometric analysis and analyzed for CD56 expression and intracellular IFN-γ production. Data are representative of 12 experiments. (B) Analysis of IFN-γ production at early time points. Cell populations enriched in NK cells by depletion of CD3+, CD19+, and CD14+ cells were analyzed for CD56 expression and intracellular IFN-γ production by flow cytometry at given sequential times. Data are representative of five experiments. Numbers and bars are referred to the proportions of IFN-γ+ cells within the specific CD56dim and CD56bright subpopulations. (C) IFN-γ mRNA expression. Sorted CD56dim freshly isolated from different donors were analyzed by RT-PCR for IFN-γ mRNA expression. Polyclonal IL-2–activated NK cells were used as control. PCR products were run on a 0.8% agarose gel and visualized by ethidium bromide staining. As positive control, RT-PCR was also performed with primers specific for β-actin.
IFN-γ Transcripts Are Detectable in Freshly Isolated CD56dim NK Cells.
The finding that IFN-γ was produced as early as 2 to 4 h after stimulation suggested that CD56dim NK cells could contain preformed intracytoplasmic IFN-γ or IFN-γ–encoding mRNA, which could be rapidly translated upon cell triggering. However, the first possibility was excluded by the finding that no intracytoplasmic IFN-γ was detected in CD56dim NK cells before stimulation (Fig. 1B, time 0). To evaluate the presence of IFN-γ mRNA, CD56dim NK cells were separated by flow cytometric cell sorting and analyzed by RT-PCR. PBMCs were first enriched in NK cells by using commercial magnetic bead separation assays. Negatively selected NK cell-enriched preparations were then further purified by flow cytometric cell sorting after anti-CD56 and anti-CD3 labeling. Highly purified CD3−CD56dim and CD3−CD56bright populations were thus obtained. As shown in Fig. 1C, CD56dim NK cells freshly isolated from different donors expressed IFN-γ transcripts.
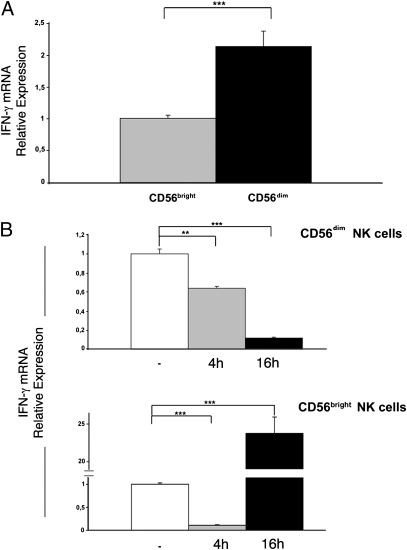
The possibility of cell contamination during cell sorting should be considered. Sorting purity was 98% for CD56dim NK cells, whereas CD56bright NK cells represented fewer than 5% of PBMCs. Contamination of the CD56dim subset with CD56bright NK cells was no greater than 0.1% (i.e., 5% of 2%). This would result in an amount of mRNA barely detectable by PCR. However, to completely rule out the possible effect of contamination, we developed a real-time PCR assay for IFN-γ mRNA quantification. In these experiments, NK cell subsets were isolated from six additional donors. As shown in Fig. 2A, the relative expression of IFN-γ mRNA in freshly sorted resting CD56dim NK cells was 2.14 times that of CD56bright NK cells (P = 0.0001, unpaired t test). These data unequivocally indicate that a minor contamination of CD56dim with CD56bright NK cells during cell sorting would not alter the interpretation of the results.
Fig. 2.
IFN-γ mRNA expression in CD56bright and CD56dim NK cell subsets. (A) IFN-γ mRNA was analyzed by real-time PCR in sorted CD56bright and CD56dim NK cells freshly isolated from six donors. Gene expression levels were normalized to GAPDH mRNA and relative quantification was performed using the ΔΔCT method. The levels of IFN-γ mRNA in CD56bright NK cells were chosen as reference and were arbitrarily normalized to 1. Data are representative of six independent experiments with six different donors (mean ± SE; ***P = 0.0001). (B) Time course: levels of IFN-γ mRNA were assessed at time 0 (white bars) or following stimulation with anti-NKp46/NKp30 mAbs for 4 h (gray bars) or 16 h (black bars). For each NK cell subset, unstimulated cells were chosen as reference and levels of IFN-γ mRNA in these cells were arbitrarily normalized to 1. All samples were analyzed in three independent experiments, processing cells obtained from two donors at a time. For each bar, the SE is indicated (**P = 0.002; ***P = 0.0001).
We further assessed the kinetics of mRNA transcription by purified NK cell subsets by real-time PCR. Cells were triggered by NCR crosslinking using mAb-coated plastic plates and harvested before triggering or after 4 h and 16 h, respectively. As shown in Fig. 2B, in CD56dim cells, the relative expression of IFN-γ mRNA progressively decreased whereas in CD56bright cells a 24-fold increase was detected after 16 h (Fig. 2C).
IFN-γ Production by Fresh CD56dim NK Cells upon Cytokine Stimulation Occurs Early and Remains Sustained Over Time.
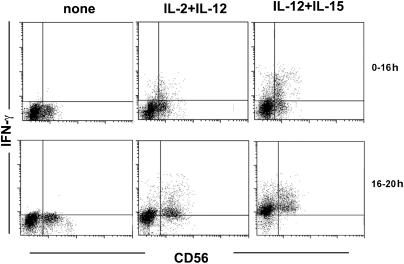
As IFN-γ production by NK cells can also be induced upon exposure to cytokines including IL-2, IL-12, and IL-15, we further investigated whether early production of IFN-γ could also be detected in this experimental setting. To this end, fresh NK cells were cultured with cytokine combinations (IL-2 plus IL-12 or IL-12 plus IL-15) for 16 h in the presence of GolgiPlug. As shown in Fig. 3, IFN-γ production was readily detected in CD56dim NK cells. Different from what occurs in NK cells stimulated via NKp46/NKp30, addition of GolgiPlug at 0–16 h revealed intense and accumulating IFN-γ production by both CD56dim and CD56bright NK subsets (as assessed after an additional 4 h). Thus, CD56dim NK cells show an early IFN-γ production upon cytokine stimulation as well. However, in this case, the response is not transient but increases and persists over time.
Fig. 3.
IFN-γ production by peripheral blood NK cell subsets following exposure to IL-2, IL-12, and IL-15. Flow cytometric analysis of peripheral blood NK cells triggered by combinations of cytokines (IL-2 plus IL-12, IL-12 plus IL-15). GolgiPlug was added immediately at the time of stimulation until processed (0–16 h) or after 16 h with analysis of cells after 4 h (16–20 h). Data are representative of four experiments.
Discussion
In the present study, we revisited the functional capabilities of two major NK cell subsets as defined by the expression of CD56 and CD16 surface antigens. At variance with the common knowledge, we show that the cytolytic CD56dimCD16+ NK cells can rapidly produce IFN-γ in a time span (2–4 h) that overlaps with their cytolytic activity. The rapid cytokine response is related to the presence of preformed IFN-γ mRNA in these cells.
The two major NK-mediated functions, i.e., cytolytic activity and cytokine production, have been attributed to CD56dim and to CD56bright cells, respectively. Notably, whereas cytolytic activity takes place rapidly upon NK cell triggering (within 4 h), cytokine production was reported to occur predominantly at later time intervals (over 16 h). The association of the two major NK cell functions with distinct cell subsets residing in different compartments (13) together with marked differences in the kinetics of these effector functions made it difficult to understand how these defense mechanisms could be effectively coordinated. In addition, the substantial delay in the production of proinflammatory cytokines was not in line with the concept that NK cells provide a rapid and effective first line of defense (1, 9). This delay would also have consequences on the cytokine-mediated crosstalk of NK cells with other cell types of the innate immunity [e.g., dendritic cells (DCs) and macrophages] that are responsible for the rapid amplification of innate responses and for the polarization of downstream adaptive immune responses (16, 17). The present study reconciles this apparent functional gap. Indeed, it assigns the capability of a prompt and abundant IFN-γ production to the CD56dim cytolytic NK cell subset. Thus, in response to proinflammatory chemokines, this subset migrates to inflamed tissues, where it can kill virally infected (or tumor) cells and simultaneously release IFN-γ. In turn, IFN-γ potentiates macrophages and DC function, contributes to DC maturation and migration to lymph nodes, and favors Th1 polarization. Notably, the timeframe of IFN-γ production by CD56dim NK cells is short and virtually no IFN-γ production was detectable 16 h after cell triggering. Whereas the intervention of CD56dim subset can now fulfill the requirement of rapid (i.e., hours) responses typical of frontline innate responses, a more prolonged IFN-γ production would be provided by the CD56bright subset, which starts cytokine production at later intervals and could play a prominent immunoregulatory role.
Our data are substantially in agreement with a recent report showing a time gradient in the chemokine and cytokine production in NK cells stimulated via different triggering receptors including NKG2D, CD16, 2B4, and DNAM-1 (15). It is of note that cytokines active on NK cells, such as IL-2, IL-12, and IL-15, can also induce an early IFN-γ production in CD56dim NK cells, however the effect is amplified and sustained over time.
Notably, the present data emphasize how different experimental settings may greatly influence the interpretation of NK cell functional capabilities. Thus, as CD56dim NK cells produce IFN-γ within a timeframe of few hours after cell triggering, the conventional addition of GolgiPlug (or brefeldin) after overnight stimulation led to the interpretation that these NK cells are poor cytokine producers. The rapid IFN-γ production by CD56dim NK cells could be explained by the presence of preformed IFN-γ mRNA. However, because IFN-γ mRNA was also detected in CD56bright NK cells, it is possible that differences in timing of IFN-γ release between the two subsets may reflect their different threshold of responsiveness via triggering receptors. Although this possible explanation requires further investigation, these data are reminiscent of earlier studies comparing responses of naive versus memory T cells, in which the number of T cell receptor/ligand interactions required for T cell activation was considerably higher for naive T cells (18, 19). Regarding the NK cell populations cultured in vitro in the presence of rIL-2, their functional behavior is similar to that of fresh CD56dim NK cells, as they rapidly produced cytokines and exerted cytolytic activity upon activation (Fig. S3). This functional property is important in view of their possible use in protocols of adoptive immunotherapy to treat leukemias or solid tumors (20, 21).
The present results suggest that analysis of early IFN-γ impairment may also be warranted in chronic infectious disease conditions in which NK cell impairments have been previously shown. Should an early IFN-γ impairment take place, for example, in patients with chronic hepatitis C virus or HIV infection, this would extend the concept of the relevance of the NK cell-mediated responses in these pathological conditions (22–25)
Taken together, our study provides a rational explanation for the NK cell intervention during the early phases of an innate response. Thus, CD56dim NK cells would play a predominant effector role coupled with a simultaneous, transient, cytokine-mediated immunoregulatory function. The immunoregulatory effect would be the predominant function of CD56bright subset. Our present data suggests the need of revisiting the relative contribution of CD56dim and CD56bright NK cell subsets in innate immune responses and their actual role at the interface between innate and adaptive immunity.
Materials and Methods
Cell Cultures.
PBMCs were obtained from 15 healthy uninfected donors and separated by gradient centrifugation (Ficoll-Hypaque) and cryopreserved until processed.
Culture medium was RPMI 1620 (BioWhittaker/Lonza) supplemented with 10% FCS, L-glutamine (2 mM), and 1% antibiotic mixture (penicillin–streptomycin 5 mg mL−1).
NK cells were enriched from PBMCs by negative selection by using an NK Cell Isolation Kit II (Miltenyi Biotec). To obtain in vitro purified NK cell cultures, 105 NK cells were cultured in 96-well round-bottom plates (Costar) in the presence of 100 IU/mL IL-2 (Roche). To obtain highly purified CD56bright and CD56dim NK cell subsets, PBMCs were first enriched in NK cells by using commercial magnetic bead separation assays. Negatively selected NK cell-enriched preparations were then further purified by flow cytometric cell sorting after anti-CD56 and anti-CD3 labeling. Highly purified CD3−CD56dim and CD3−CD56bright populations were thus obtained by using FACSAria (BD Biosciences) according to the lack of CD3 and the expression of CD56.
Antibodies.
The following panel of mouse anti-human mAbs was used: anti-CD3FITC, anti-CD19 FITC, and anti-CD14FITC (BD PharMingen). For intracellular staining, anti–CD107a-PE (BD PharMingen) and anti–IFN-γ APC (BD PharMingen) were used, and anti-CD56PC7 (Immunotech-Coulter) was used for surface labelling of NK cells. Anti-NKp46 (IgG1) BAB281 (26) and anti-NKp30 (IgG1) AZ20 were produced in our laboratory.
Intracellular Production of IFN-γ.
PBMCs were stimulated using FcγR+ mouse P815 target cells at 10:1 E:T ratio in complete medium in the absence or presence of anti-NKp30 and anti-NKp46 mAb mixture (0.1 μg mL−1), rIL-2 (100 U mL−1; Proleukin, Chiron), IL-12 (20 ng mL−1), and IL-15 (40 ng mL−1; PeproTech).
Phorbol myristate acetate (25 ng mL−1) plus ionomycin (1 μg mL−1; Sigma) were used for maximal IFN-γ production. GolgiPlug (BD Pharmingen) was added at 37 °C for 4 h after overnight incubation or from incubation start on for up to 16 h (Fig. S1). PBMCs were gated by forward and side scatter and on exclusion of CD3+CD14+CD19+ cells; staining was followed by permeabilization/fixation according to the Citofix/Citoperm protocol (BD Pharmingen) and subsequently intracellular staining for IFN-γ was performed in the presence of permeabilizing solution (0.1% saponin in PBS solution). Cells were analyzed on a FACSCalibur device (Becton Dickinson) using CellQuest Software after acquiring 10,000 gated events.
RT-PCR Analysis.
Total RNA was extracted using RNAeasy Micro Kit (Qiagen) from different cells: freshly isolated and sorted CD56dimCD16+ and CD56brightCD16+/− NK cells and NK cell populations unstimulated or stimulated with anti-NKp46 (BAB281) and anti-NKp30 (AZ20) mAbs. Oligo(dT)-primed cDNA was prepared by standard technique using Transcriptor (Roche). For end-point PCR analysis, the following primers were used: β-actin, upstream, ACTCCATCATGAAGTGTGACG; β-actin, downstream, CATACTCCTGCTTGCTGATCC; IFN-γ, upstream, 5′ GCAGGTCATTCAGATGTAGCG; and IFN-γ, downstream, 5′ GGCAGGACAACCATTACTGG. Amplifications were carried out for 30 cycles (30 s at 95 °C, 30 s at 58 °C, 1 min at 68 °C) using Platinum TAQ (Invitrogen). Only for analysis of IFN-γ mRNA in sorted CD56dimCD16+ or CD56brightCD16+/− NK cells, 35 amplification cycles were performed. PCR products were run on a 0.8% agarose gel and visualized by ethidium bromide staining.
Relative quantification of IFN-γ expression was analyzed in real-time PCR using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). To internally standardize the levels of gene expression, we used the GAPDH housekeeping gene. Amplifications were performed with Mastercycler ep Realplex (Eppendorf) in a 20-μL final volume, using primers at 300 nM for 40 cycles (15 s at 95 °C, 30 s at 60 °C). A dissociation curve was performed at the end of 40 cycles to confirm specificity of amplification. The following primers were used: GAPDH, forward, 5′ GAAGGTGAAGGTCGGAGT; GAPDH, reverse, 5′ CATGGGTGGAATCATATTGGAA; IFN-γ, forward, 5′ AAAAATAATGCAGAGCCAAATTG; and IFN-γ, reverse, 5′ TAGCTGCTGGCGACAGTTCA. Each reaction was performed in triplicate and each sample was analyzed in two or three independent experiments. Relative expression of IFN-γ transcript was obtained using the ΔΔCT method.
Statistical analysis was performed by using the JMP 5.1.2 program (SAS)
Supplementary Material
Acknowledgments
We thank Emanuela Marcenaro and Alessia Agazzi for help in NK cell sorting, Guido Ferlazzo and Alessandro Moretta for advice and discussion, and Ms. Cinzia Miriello (University of Genova) for secretarial help. This work was supported by grants awarded by Istituto Superiore di Sanità (to I.S.S.); Programma nazionale di ricerca sull'AIDS, Accordi di collaborazione scientifica n. 40G.41 (to L.M. and A.D.M.) and 45G.11 (to A.D.M.); Italian Concerted Action for AIDS vaccine; Accordo di collaborazione scientifica n. 40D61 (to A.D.M.); Associazione Italiana per la Ricerca sul Cancro projects (L.M.); Ministero dell'Istruzione, dell'Università e della Ricerca MIUR-FIRB 2003 project RBLA039LSF-001 (to L.M.); Special Project 5x1000 from the Associazione Italiana per la Ricerca sul Cancro and Ministero della Salute RF2006–Ricerca Oncologica–Project of Integrated Program 2006-08, agreements n. RO strategici 3/07 (to L.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012356108/-/DCSupplemental.
References
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moretta A, Marcenaro E, Parolini S, Ferlazzo G, Moretta L. NK cells at the interface between innate and adaptive immunity. Cell Death Differ. 2008;15:226–233. doi: 10.1038/sj.cdd.4402170. [DOI] [PubMed] [Google Scholar]
- 3.Moretta L, et al. Surface NK receptors and their ligands on tumor cells. Semin Immunol. 2006;18:151–158. doi: 10.1016/j.smim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Bottino C, Castriconi R, Moretta L, Moretta A. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26:221–226. doi: 10.1016/j.it.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Moretta L, Moretta A. Killer immunoglobulin-like receptors. Curr Opin Immunol. 2004;16:626–633. doi: 10.1016/j.coi.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Moretta L, Moretta A. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 2004;23:255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper MA, Fehniger TA, Fuchs A, Colonna M, Caligiuri MA. NK cell and DC interactions. Trends Immunol. 2004;25:47–52. doi: 10.1016/j.it.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Ferlazzo G, et al. Distinct roles of IL-12 and IL-15 in human natural killer cell activation by dendritic cells from secondary lymphoid organs. Proc Natl Acad Sci USA. 2004;101:16606–16611. doi: 10.1073/pnas.0407522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moretta A, et al. Early liaisons between cells of the innate immune system in inflamed peripheral tissues. Trends Immunol. 2005;26:668–675. doi: 10.1016/j.it.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Vacca P, et al. Regulatory role of NKp44, NKp46, DNAM-1 and NKG2D receptors in the interaction between NK cells and trophoblast cells. Evidence for divergent functional profiles of decidual versus peripheral NK cells. Int Immunol. 2008;20:1395–1405. doi: 10.1093/intimm/dxn105. [DOI] [PubMed] [Google Scholar]
- 11.Vacca P, et al. Crosstalk between decidual NK and CD14+ myelomonocytic cells results in induction of Tregs and immunosuppression. Proc Natl Acad Sci USA. 2010;107:11918–11923. doi: 10.1073/pnas.1001749107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehniger TA, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood. 2003;101:3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 13.Romagnani C, et al. CD56brightCD16- killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. 2007;178:4947–4955. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
- 14.Moretta L, et al. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev. 2006;214:219–228. doi: 10.1111/j.1600-065X.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- 15.Fauriat C, Long EO, Ljunggren H-G, Bryceson YT. Regulation of human NK cell cytokine and chemokine productionby target cell recognition. Blood. 2010;115:2167–2176. doi: 10.1182/blood-2009-08-238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferlazzo G, et al. The interaction between NK cells and dendritic cells in bacterial infections results in rapid induction of NK cell activation and in the lysis of uninfected dendritic cells. Eur J Immunol. 2003;33:306–313. doi: 10.1002/immu.200310004. [DOI] [PubMed] [Google Scholar]
- 17.Ferlazzo G, et al. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J Exp Med. 2002;195:343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedlund G, Dohlsten M, Ericsson PO, Sjögren HO. Rapid response to Con A by CD4+CD45R- rat memory lymphocytes as compared to CD4+CD45R+ lymphocytes. Cell Immunol. 1989;119:317–326. doi: 10.1016/0008-8749(89)90247-5. [DOI] [PubMed] [Google Scholar]
- 19.Valitutti S, Dessing M, Aktories K, Gallati H, Lanzavecchia A. Sustained signaling leading to T cell activation results from prolonged T cell receptor occupancy. Role of T cell actin cytoskeleton. J Exp Med. 1995;181:577–584. doi: 10.1084/jem.181.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velardi A, Ruggeri L, Alessandro , Moretta L, Moretta L. NK cells: A lesson from mismatched hematopoietic transplantation. Trends Immunol. 2002;23:438–444. doi: 10.1016/s1471-4906(02)02284-6. [DOI] [PubMed] [Google Scholar]
- 21.Moretta A, Locatelli F, Moretta L. Human NK cells: from HLA class I-specific killer Ig-like receptors to the therapy of acute leukemias. Immunol Rev. 2008;224:58–69. doi: 10.1111/j.1600-065X.2008.00651.x. [DOI] [PubMed] [Google Scholar]
- 22.De Maria A, et al. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur J Immunol. 2003;33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 23.Bozzano F, et al. Functionally relevant decreases in activatory receptor expression on NK cells are associated with pulmonary tuberculosis in vivo and persist after successful treatment. Int Immunol. 2009;21:779–791. doi: 10.1093/intimm/dxp046. [DOI] [PubMed] [Google Scholar]
- 24.De Maria A, et al. Increased natural cytotoxicity receptor expression and relevant IL-10 production in NK cells from chronically infected viremic HCV patients. Eur J Immunol. 2007;37:445–455. doi: 10.1002/eji.200635989. [DOI] [PubMed] [Google Scholar]
- 25.Fogli M, et al. Significant NK cell activation associated with decreased cytolytic function in peripheral blood of HIV-1-infected patients. Eur J Immunol. 2004;34:2313–2321. doi: 10.1002/eji.200425251. [DOI] [PubMed] [Google Scholar]
- 26.Sivori S, et al. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human NK cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur J Immunol. 1999;29:1656–1666. doi: 10.1002/(SICI)1521-4141(199905)29:05<1656::AID-IMMU1656>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.