Abstract
The use of plant biomass for biofuel production will require efficient utilization of the sugars in lignocellulose, primarily glucose and xylose. However, strains of Saccharomyces cerevisiae presently used in bioethanol production ferment glucose but not xylose. Yeasts engineered to ferment xylose do so slowly, and cannot utilize xylose until glucose is completely consumed. To overcome these bottlenecks, we engineered yeasts to coferment mixtures of xylose and cellobiose. In these yeast strains, hydrolysis of cellobiose takes place inside yeast cells through the action of an intracellular β-glucosidase following import by a high-affinity cellodextrin transporter. Intracellular hydrolysis of cellobiose minimizes glucose repression of xylose fermentation allowing coconsumption of cellobiose and xylose. The resulting yeast strains, cofermented cellobiose and xylose simultaneously and exhibited improved ethanol yield when compared to fermentation with either cellobiose or xylose as sole carbon sources. We also observed improved yields and productivities from cofermentation experiments performed with simulated cellulosic hydrolyzates, suggesting this is a promising cofermentation strategy for cellulosic biofuel production. The successful integration of cellobiose and xylose fermentation pathways in yeast is a critical step towards enabling economic biofuel production.
Keywords: biofuels, cellodextrin transporter, cofermentation, intracellular β-glucosidase
There is a long history of microbial fermentation of glucose derived from cornstarch or sugarcane to fuels and chemicals. Modifying this process to ferment sugars produced from the depolymerization of plant biomass is a central paradigm for cellulosic biofuel production (1). While hydrolyzates from cornstarch and sugarcane contain only hexoses, hydrolyzates from cellulosic biomass contain various hexoses and pentoses. Hexoses are produced following the hydrolysis of cellulose by fungal cellulases, which primarily generate cellobiose that can be further hydrolyzed to glucose by β-glucosidases. Pentoses are produced by acid treatment of hemicellulose, which releases xylose and arabinose. While the composition is variable, plant biomass hydrolyzates consists of ∼70% cellodextrins and glucose, and ∼30% of xylose (2). Successful conversion of cellulosic biomass into biofuel will therefore require organisms capable of efficient utilization of xylose as well as cellodextrins and glucose (3, 4).
The traditional glucose fermenting yeast, Saccharomyces cerevisiae, cannot ferment xylose because it does not possess a functional xylose assimilation pathway. However, as wild-type S. cerevisiae can metabolize xylulose using the pentose phosphate pathway, the introduction of genes (XYL1 and XYL2) encoding xylose reductase (XR) and xylitol dehydrogenase (XDH) from Pichia stipitis (taxonomic classification has been changed to Scheffersomyces stipitis) facilitated xylose assimilation in this yeast (5, 6). In addition, either overexpression of XKS1 coding for endogenous xylulokinase (7, 8), or introduction of XYL3 coding for P. stipitis xylulokinase (9), significantly improved the rate and yield of xylose fermentation in S. cerevisiae. Despite these endeavors, reported ethanol yields and productivities from xylose fermentation by engineered strains were much inferior to those of xylose fermentation by P. stipitis, as well as those of glucose fermentation by S. cerevisiae (10, 11). Moreover, both natural and engineered microorganisms showed reduced ethanol tolerance during xylose fermentation as compared to glucose fermentation (12). Combined with the lower fermentation rate, the reduced ethanol tolerance during xylose fermentation poses a significant problem in the fermentation of sugar mixtures containing high concentrations of glucose (∼70–100 g/L) and xylose (∼40–60 g/L) present in cellulosic hydrolyzates. Because microorganisms utilize glucose preferentially, at the time of glucose depletion (when cells begin to use xylose) the ethanol concentration is already high enough (∼35–45 g/L of ethanol) to further reduce the xylose fermentation rate. As a result, sequential utilization of xylose after glucose depletion because of so called “glucose repression” is a significant barrier to successful utilization of mixed sugars in cellulosic hydrolyzates.
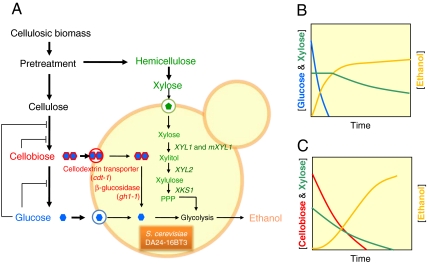
To bypass the problems caused by glucose repression in a mixed sugar fermentation, we developed a unique strategy to coferment cellobiose, a dimer of glucose, with xylose simultaneously (Fig. 1). Wild-type S. cerevisiae cannot assimilate cellobiose because it lacks both a cellobiose transporter and a β-glucosidase capable of hydrolyzing cellobiose into glucose. Therefore, we introduced a newly discovered cellodextrin transporter and intracellular β-glucosidase from the cellulolytic fungi, Neurospora crassa (13), into S. cerevisiae strains engineered to ferment xylose. A different approach enabling cofermentation of cellobiose and xylose through a display of β-glucosidase on the surface of xylose-fermenting S. cerevisiae has been reported (14). Although both approaches target cofermentation of cellobiose and xylose, the two differ in the location of cellobiose hydrolysis and its effect on xylose transport. The surface display of β-glucosidase generates glucose extracellularly. In contrast, in the present design, cellobiose is hydrolyzed intracellularly following transport. Because xylose uptake by S. cerevisiae is facilitated by hexose transporters which are severely inhibited by glucose (15–17), the extracellular glucose generated by surface display of β-glucosidase can inhibit xylose uptake (14) whereas the intracellular glucose from intracellular hydrolysis of cellobiose will not. Perhaps due to this fundamental distinction, our strategy achieves faster xylose and cellobiose coconsumption rates, and may prove optimal for biofuel production from cellulosic feedstocks.
Fig. 1.
Strategy for simultaneous cofermentation of cellobiose and xylose without glucose repression. (A) A strain improvement strategy to engineer yeast capable of fermenting two nonmetabolizable sugars (cellobiose and xylose). The cellodextrin assimilation pathway consists of a cellodextrin transporter (cdt-1) and an intracellular β-glucosidase (gh1-1) from the filamentous fungus N. crassa. The modified xylose metabolic pathway utilizes xylose reductase isoenzymes (wild-type XR and a mutant XRR276H), xylitol dehydrogenase (XYL2), and xylulokinase (XKS1) from the xylose-fermenting yeast P. stipitis. (B) Schematic fermentation profile of a sugar mixture containing glucose and xylose by the engineered S. cerevisiae. Glucose fermentation represses xylose fermentation completely so that xylose fermentation begins only after glucose depletion (analogous fermentation result shown in Fig. 5A). (C) Schematic fermentation profile of a sugar mixture containing cellobiose and xylose by the engineered S. cerevisiae. Cellobiose and xylose are simultaneously utilized, as neither carbon source represses consumption of the other (analogous fermentation result shown in Fig. 5B).
Results
Construction of Cellobiose-Fermenting S. cerevisiae Through Introduction of Cellodextrin Transporter and β-glucosidase from N. crassa.
Three N. crassa cellodextrin transporters (cdt-1, cdt-2, and NCU00809) were introduced into a S. cerevisiae strain expressing an intracellular β-glucosidase (gh1-1). All three transformants were able to grow and produce ethanol with cellobiose as the sole carbon source (Fig. S1), but the three transformants exhibited different cellobiose fermentation rates (cdt-1>cdt-2> NCU00809). The fastest cellulose-fermenting transformant (D801-130), expressing both cdt-1 and gh1-1, consumed 40 g/L of cellobiose within 24 h, producing 16.8 g/L of ethanol. The volumetric productivity of cellobiose fermentation (PEthanol/Cellobiose = 0.7 g/L·h) was lower than that of glucose fermentation (PEthanol/Glucose = 1.2 g/L·h), and ethanol yield from cellobiose (YEthanol/Cellobiose = 0.42 g/g or 0.26 mol/mol C) was slightly lower than ethanol yield from glucose (YEthanol/Glucose = 0.43 g/g or 0.28 mol/mol C) under the same culture conditions. However, the observed cellobiose consumption rate and ethanol yield by the D801-130 were an improvement over S. cerevisiae strains engineered to ferment cellobiose through surface display of β-glucosidase (14, 18). These results suggest that simultaneous expression of cdt-1 and gh1-1 in S. cerevisiae can result in an efficient cellobiose-fermenting yeast.
Construction of an Engineered S. cerevisiae Capable of Fermenting Xylose both Rapidly and Efficiently Through Rational and Combinatorial Strategies.
Attempts to engineer xylose fermentation into S. cerevisiae have suffered from low yields due to xylitol accumulation, and low productivities due to slow xylose fermentation rates (10, 11). In order to improve yield and productivity of xylose fermentation, we undertook both rational and combinatorial approaches. First, we drastically reduced xylitol accumulation through reconstituting an efficient xylose metabolic pathway. XR prefers NADPH to NADH while XDH solely uses NAD+ as cofactor (19, 20), resulting in a redox imbalance that is thought to result in xylitol accumulation. While this imbalance has been corrected by replacing wild-type XR with various mutant XRs exhibiting higher preferences to NADH (21, 22), these efforts resulted in much slower xylose assimilation rates despite reducing the amounts of xylitol produced, as the mutant XRs had reduced specific activities as compared to wild-type XR.
In contrast to previous studies that relied on either wild-type XR or a mutant XR, we expressed both wild-type XR and a mutant XR (R276H) in S. cerevisiae, along with XDH and XK to construct a functional xylose metabolic pathway in S. cerevisiae. The XR mutant (R276H) had been reported to exhibit much higher preference for NADH whereas wild-type XR showed twofold higher preference for NADPH (22). The resulting xylose-fermenting S. cerevisiae strain (DA24) expressing both wild-type XR and mutant XR (R276H) exhibited comparable XR activities with both NADPH and NADH in an in vitro XR activity assay. Unlike other engineered strains showing higher XR activities with preferred cofactors, strain DA24 showed similar XR activities regardless of cofactors (Table 1).
Table 1.
Comparison of xylose reductase activity of DA24 with those of other engineered S. cerevisiae strains expressing wild-type or mutant XYL1
| Strains | Specific xylose reductase activity (U/mg) | Ratio of XR activity with NADH and NADPH | Reference | |
| NADH |
NADPH |
|||
| S. cerevisiae with wild-type XYL1 | 0.07 ± 0.03 | 0.09 ± 0.02 | 0.8 | Amore et al. (23) |
| 0.10 ± 0.02 | 0.12 ± 0.03 | 0.8 | Takuma et al. (24) | |
| - | 2.7 ± 2.5 | - | Walfridsson et al. (25) | |
| 2.3 ± 0.3 | 3.4 ± 0.5 | 0.68 | Tantirungkij et al. (5) | |
| - | 0.411 | - | Ho et al. (8) | |
| - | 0.43 ± 0.03 | - | Eliasson et al. (26) | |
| - | 1.1 ± 0.08 | - | Wahlbom et al. (27) | |
| S. cerevisiae with mutant XYL1 | 9.3 ± 0.1 | 0.4 ± 0.0 | 23.1 | Watanabe et al. (22) |
| DA24 | 0.32 ± 0.15 | 0.29 ± 0.10 | 1.1 | This study |
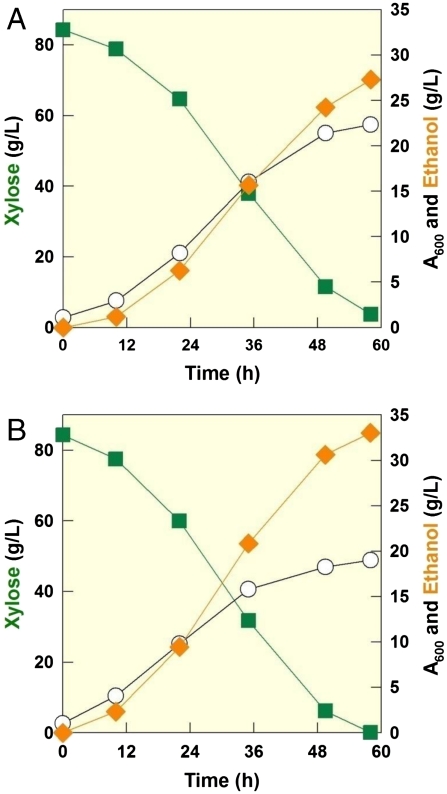
Second, we further improved the xylose fermentation rate of DA24 using an evolutionary engineering approach (28). The DA24 strain rapidly consumed xylose and produced ethanol with consistent ethanol yields (YEthanol/Xylose = 0.31–0.32 g/g or 0.21–0.22 mol/mol C) in both shaker-flask and bioreactor fermentation experiments (Fig. S2). The DA24 strain also produced negligible amounts of xylitol. Still, the DA24 strain consumed xylose slower than the naturally existing xylose-fermenting yeast, P. stipitis. Using evolutionary engineering, one strain (DA24-16) isolated after repeated subcultures of the DA24 on xylose containing medium showed much faster xylose fermentation rates when compared to the parental strain under various culture conditions (Table S1). The specific xylose uptake rates, ethanol yields, and ethanol production rates of the DA24 and DA24-16 strains were much higher than engineered S. cerevisiae strains reported previously (Table S2). Interestingly, the DA24-16 strain consumed xylose almost as fast as P. stipitis, the fastest xylose-fermenting yeast known. However, the ethanol yield from DA24-16 was slightly lower than that from P. stipitis (Fig. 2).
Fig. 2.
Comparisons of xylose consumption and ethanol production between DA24-16 (A) and P. stipitis (B). Symbols: xylose (green square), ethanol (orange diamond), and cell growth (open circle).
Cofermentation of Cellobiose and Xylose Lead to Increased Ethanol Yield and Productivity.
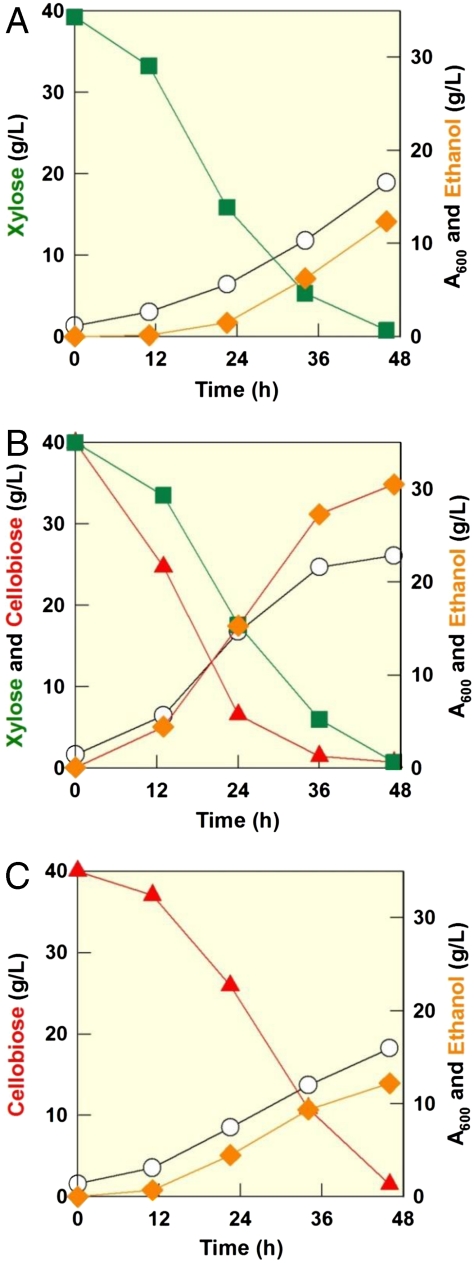
To further improve the newly engineered xylose-fermenting strains, we introduced genes coding for a cellodextrin transporter and a β-glucosidase (cdt-1 and gh1-1) into the efficient xylose-fermenting strain, DA24-16, to construct a strain capable of consuming cellobiose and xylose simultaneously. We hypothesized that glucose repression of xylose utilization may be alleviated in this strain, due to the intracellular hydrolysis of cellobiose. One resulting strain with cdt-1 integrated into the genome of DA24-16 and expressing gh1-1 from a multicopy plasmid, strain DA24-16-BT3, coconsumed cellobiose and xylose and produced ethanol with yields of 0.38–0.39 g/g in all conditions (Table S3). We also investigated potential synergistic effects of cofermentation by culturing DA2416-BT3 under three different conditions: 40 g/L of cellobiose, 40 g/L of xylose, and 40 g/L of both sugars (total 80 g/L of sugars). Interestingly, DA24-16BT3 was able to coconsume 80 g/L of a cellobiose/xylose mixture within the same period that was required to consume 40 g/L of cellobiose or 40 g/L xylose separately (Fig. 3). Moreover, DA24-16BT3 produced ethanol with a higher yield (0.39 g/g or 0.24 mol/mol C) from a mixture of cellobiose and xylose as compared to ethanol yields (0.31–0.33 g/g or 0.21–0.22 mol/mol C) from single sugar fermentations (cellobiose or xylose) (SI Text). Ethanol productivity also dramatically increased from 0.27 g/L·h to 0.65 g/L·h during cofermentation. These results suggest that cofermentation of cellobiose and xylose can enhance overall ethanol yield and productivity.
Fig. 3.
Synergistic effects from cofermentation of cellobiose and xylose by strain DA24-16BT3. Symbols: cellobiose (red triangle up), xylose (green square), ethanol (orange diamond), and cell growth (open circle). (A) 40 g/L of cellobiose, (B) a mixture of 40 g/L of cellobiose and 40 g/L of xylose, and (C) 40 g/L of xylose.
Fermentation of Simulated Cellulosic Hydrolyzates by the Engineered S. cerevisiae Capable of Cofermenting Cellobiose and Xylose.
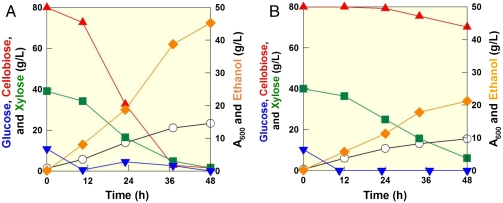
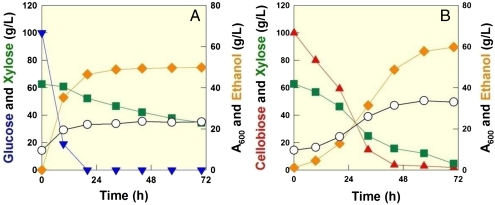
We also performed fermentation experiments comparing our engineered S. cerevisiae strain (DA24-16BT3) to P. stipitis, a yeast capable of cofermenting cellobiose and xylose efficiently. Using a simulated hydrolyzate (10 g/L of glucose, 80 g/L of cellobiose, and 40 g/L of xylose) based on the composition of energycane (SI Text), the DA24-16BT3 consumed glucose first and coconsumed cellobiose, and xylose rapidly. A total of 130 g/L of sugars were consumed within 60 h even though small inoculums were used (OD600 = 1.3, or ∼0.4 g dried cell/L). In contrast, P. stipitis could not finish fermenting the sugar mixture within the same period under identical culture conditions (Fig. 4). DA24-16BT3 produced 48 g/L of ethanol within 60 h (YEthanol/Sugars = 0.37 g/g or 0.23 mol/mol C, and PEthanol/Sugars = 0.79 g/L·h). Fermentation experiments using a bioreactor with the same or higher inoculums yielded consistent results (Table S4). We also performed fermentation experiments of sugar mixtures containing extreme concentrations of glucose and xylose (100 g/L of glucose and 60 g/L of xylose) and cellobiose and xylose (100 g/L of cellobiose and 60 g/L xylose). As illustrated in Fig. 1B, xylose fermentation is severely delayed after glucose consumption when the mixture of glucose and xylose was used. However, cofermentation of cellobiose and xylose showed a slower ethanol production initially, but resulted in higher ethanol production at the end (Fig. 5).
Fig. 4.
Cofermentation of glucose, cellobiose, and xylose by strain DA24-16BT3 and P. stipitis. (A) DA24-16BT3 fermentation profile of a sugar mixture containing 80 g/L of cellobiose, 40 g/L of xylose, and 10 g/L of glucose. (B) P. stipitis fermentation of a sugar mixture containing 80 g/L of cellobiose, 40 g/L of xylose, and 10 g/L of glucose. Symbols: cellobiose (red triangle up), xylose (green square), ethanol (orange diamond), OD (open circle), and glucose (blue triangle down).
Fig. 5.
Fermentation profiles by DA24-16BT3 during the fermentation of sugar mixtures containing extreme concentrations of glucose and xylose, (A) 100 g/L of glucose and 60 g/L of xylose and cellobiose and xylose, (B) 100 g/L of cellobiose and 60 g/L xylose. Actual fermentation profiles are similar to the patterns of sugar utilization and ethanol production illustrated in Fig. 1. Symbols: cellobiose (red triangle up), xylose (green square), ethanol (orange diamond), OD (open circle), and glucose (blue triangle down).
Discussion
Although we obtained promising results from our cofermentation experiments, further improvements to these yeast strains can likely be made. For example, we observed transient accumulation of cellodextrins in the medium during cellobiose fermentation (Figs. S3 and S4). It is likely that the accumulated cellodextrins were generated by the transglycosylation activity (29) of β-glucosidase (gh1-1), and secreted by the cellodextrin transporter (cdt-1), which likely facilitates the transport of cellodextrins in both directions (intracellular↔extracellular). This transient cellodextrin accumulation may not reduce product yields because the accumulated cellodextrins are consumed by our engineered yeast after cellobiose depletion. However, the accumulated cellodextrins might decrease productivity because the transport rates of cellotriose and cellotetraose might be slower than that of cellobiose. We also observed that small amounts of glucose were constantly detected in the medium during cofermentation. Because even low amounts of glucose accumulation can repress xylose fermentation, glucose accumulation must be controlled at the lowest possible levels. We hypothesize that the relative expression level of the cellodextrin transporter and β-glucosidase is likely to affect glucose accumulation, because more glucose is accumulated in the medium when cdt-1 is introduced on a multicopy plasmid than when cdt-1 is integrated into the genome. For example, the strain (DA24-16-BT) with both cdt-1 and gh1-1 on a multicopy plasmid showed relatively slower xylose utilization than DA24-16-BT3, potentially because of glucose repression (Fig. S5). Future optimization of cellodextrin transporter and β-glucosidase expression levels, or the identification of β-glucosidases with reduced transglycosylastion activities, may eliminate the accumulation of glucose and cellodextrins during cofermentation.
Here, we demonstrated a unique strategy to allow the cofermenation of hexose and pentose sugars by S. cerevisiae. By combining an efficient xylose utilization pathway with a cellobiose transport system, we bypassed problems caused by glucose repression. As a result, the engineered yeast cofermented two nonmetabolizable sugars in cellulosic hydrolyzates, synergistically, into ethanol. The unique cofermentation method advances lignocellulosic technologies on both the saccharification and fermentation fronts. Most traditional fungal cellulase cocktails require extra β-glucosidase enzyme to be added to fully convert cellobiose into glucose for yeast fermentation. The cellobiose/xylose cofermentation yeast makes it possible to use cellulase cocktails with limited β-glucosidase activities, lowering the enzyme usage and cost associated with the cellulose saccharification process. On the other hand, the synergy between the cellobiose and xylose cofermentation significantly increases ethanol productivity, improving the fermentation economics. Furthermore, the presence of small amounts of glucose from the pretreatment and hydrolysis of lignocellulosic materials should not affect the capacity of the engineered yeast to convert the hexose and pentose sugar mixtures into ethanol.
In this study, we measured the capacity of our engineered strain to ferment various mixtures of sugars meant to mimic hydrolyzates from plant biomass. However, we predict that the capacity of our strain to coferment cellodextrins and xylose will make it particularly useful during the Simultaneous Saccharification and CoFermentation (SSCF) of pretreated plant biomass (30). During SSCF, hemicellulose would first be hydrolyzed by acid pretreatment, resulting in xylose and cellulose. Then, fungal cellulases and the yeast strain developed here would be added, allowing the coconversion of xylose and cellobiose released by the cellulases, into ethanol. Because little extracelluar glucose is produced in this scheme, repression of xylose utilization will be minimized, and cofermentation will proceed rapidly and synergistically.
Although the S. cerevisiae strain used in this study was a laboratory strain, the fermentation performance of the engineered strain was very impressive as compared to published results (31). We envision that key fermentation parameters (yield and productivity) can be improved further if we employ industrial strains as a platform. Finally, applications of our cofermentation strategy would not be limited to ethanol production. Because it is a foundational technology, the strategy presented here can be combined with any other product diversification technologies to produce commodity chemicals and advanced biofuels.
Materials and Methods
Strains and Plasmid Constructions.
S. cerevisiae D452-2 (MATalpha, leu2, his3,ura3, and can1) was used for engineering of xylose and cellobiose metabolism in yeast. Escherichia coli DH5 (F–recA1 endA1 hsdR17 [rK– mK+] supE44 thi-1 gyrA relA1) (Invitrogen) was used for gene cloning and manipulation. P. stipitis CBS 6054 (32) was obtained from Thomas Jeffries at University of Wisconsin-Madison. Strains and plasmids used in this work are described in Table S5. The primers used for confirming the transformation of expression cassettes containing cdt-1, cdt-2, and gh1-1 are listed in Table S6.
Medium and Culture Conditions.
E. coli was grown in Luria-Bertani medium; 50 μg/mL of ampicillin was added to the medium when required. Yeast strains were routinely cultivated at 30 °C in YP medium (10 g/L yeast extract, 20 g/L Bacto peptone) with 20 g/L glucose. To select transformants using an amino acid auxotrophic marker, yeast synthetic complete (YSC) medium was used, which contained 6.7 g/L yeast nitrogen base plus 20 g/L glucose, 20 g/L agar, and CSM-Leu-Trp-Ura (Bio 101) which supplied appropriate nucleotides and amino acids.
Fermentation Experiments.
Yeast cultures were grown in YP medium containing 20 g/L of glucose or 20 g/L of cellobiose to prepare inoculums for xylose or cellobiose fermentation experiments, respectively. Cells at midexponential phase from YP media containing 20 g/L of glucose or cellobiose were harvested and inoculated after washing twice by sterilized water. Flask fermentation experiments were performed using 50 mL of YP medium containing appropriate amounts of sugars in 250 mL flask at 30 °C with initial OD600 of ∼1 or 10 under oxygen limited conditions. All of the flask fermentation experiments were repeated independently. The variations between independent fermentations were less than 5%. Fermentation profiles shown in figures are from one representative fermentation. Bioreactor fermentations were performed in 400 mL of YP medium containing appropriate amounts of sugars using Sixfors Bioreactors (Appropriate Technical Resources, Inc) at 30 °C with an agitation speed of 200 rpm under oxygen limited conditions. Initial cell densities were adjusted to OD600 = ∼ 1 or 10.
Yeast Transformation.
Transformation of expression cassettes for constructing xylose and cellobiose metabolic pathways was performed using the yeast EZ-Transformation kit (BIO 101). Transformants were selected on YSC medium containing 20 g/L glucose. Amino acids and nucleotides were added as necessary. For the construction of cellobiose consuming recombinant S. cerevisiae, transformations of cdt-1 and gh1-1 were selected on YSC medium containing 20 g/L cellobiose. Introduction of expression cassettes into yeast was confirmed by colony PCR with specific primers (Table S6).
Analytical Methods.
Cell growth was monitored by optical density (OD) at 600 nm using UV-visible Spectrophotometer (Biomate 5). Glucose, xylose, xylitol, glycerol, acetate, and ethanol concentrations were determined by high performance liquid chromatography (HPLC, Agilent Technologies 1200 Series) equipped with a refractive index detector using a Rezex ROA-Organic Acid H+ (8%) column (Phenomenex Inc.). The column was eluted with 0.005 N of H2SO4 at a flow rate of 0.6 mL/ min at 50 °C. The analysis of cellodextrin in fermentation samples was performed using high performance anion exchange chromatography (HPAEC) analysis. HPAEC analysis was performed with an analytical column for carbohydrate detection (CarboPac PA100, Dionex Co.) and an electrochemical detector (ED40, Dionex Co.). Filtered samples were eluted with a linear gradient from 100% buffer A (100 mM NaOH in water) to 60% buffer B (500 mM of sodium acetate in buffer A) over 70 min. The flow rate of the mobile phase was maintained at 1.0 mL/ min.
Supplementary Material
Acknowledgments.
The authors thank Dr. Huimin Zhao for sharing his preliminary results, Dr. Isaac Cann for HPAEC analysis, and William Beeson for helpful discussions. We also thank Dr. Thomas Jeffries for generously providing P. stipitis CBS 6054. This work was supported by funding from Energy Biosciences Institute to J.H.D.C. and Y.-S.J. Strain DA26-16 will be deposited in the American Type Culture Collection.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010456108/-/DCSupplemental.
References
- 1.Wyman CE. What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol. 2007;25:153–157. doi: 10.1016/j.tibtech.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol. 2009;60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 3.Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315:801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 4.Ragauskas AJ, et al. The path forward for biofuels and biomaterials. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 5.Tantirungkij M, Nakashima N, Seki T, Yoshida T. Construction of xylose-assimilating Saccharomyces cerevisiae. J Ferment Bioeng. 1993;75:83–88. [Google Scholar]
- 6.Kotter P, Ciriacy M. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1993;38:776–783. [Google Scholar]
- 7.Toivari MH, Aristidou A, Ruohonen L, Penttila M. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: importance of xylulokinase (XKS1) and oxygen availability. Metab Eng. 2001;3:236–249. doi: 10.1006/mben.2000.0191. [DOI] [PubMed] [Google Scholar]
- 8.Ho NWY, Chen Z, Brainard AP. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol. 1998;64:1852–1859. doi: 10.1128/aem.64.5.1852-1859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin YS, Ni H, Laplaza JM, Jeffries TW. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl Environ Microbiol. 2003;69:495–503. doi: 10.1128/AEM.69.1.495-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeffries TW, Jin YS. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol. 2004;63:495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- 11.Hahn-Hagerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol. 2007;108:147–177. doi: 10.1007/10_2007_062. [DOI] [PubMed] [Google Scholar]
- 12.Jeffries TW, Jin YS. Ethanol and thermotolerance in the bioconversion of xylose by yeasts. Adv Appl Microbiol. 2000;47:221–268. doi: 10.1016/s0065-2164(00)47006-1. [DOI] [PubMed] [Google Scholar]
- 13.Galazka JM, et al. Cellodextrin transport in yeast for improved biofuel production. Science. 2010;330:84–86. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura N, et al. Effective xylose/cellobiose co-fermentation and ethanol production by xylose-assimilating S. cerevisiae via expression of β-glucosidase on its cell surface. Enzyme Microb Tech. 2008;43:233–236. [Google Scholar]
- 15.Lee WJ, Kim MD, Ryu YW, Bisson LF, Seo JH. Kinetic studies on glucose and xylose transport in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2002;60:186–191. doi: 10.1007/s00253-002-1085-6. [DOI] [PubMed] [Google Scholar]
- 16.Jojima T, Omumasaba CA, Inui M, Yukawa H. Sugar transporters in efficient utilization of mixed sugar substrates: current knowledge and outlook. Appl Microbiol Biotechnol. 2010;85:471–480. doi: 10.1007/s00253-009-2292-1. [DOI] [PubMed] [Google Scholar]
- 17.Runquist D, Hahn-Hèagerdal B, Rêadstrèom P. Comparison of heterologous xylose transporters in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2010;3:5. doi: 10.1186/1754-6834-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotaka A, et al. Direct ethanol production from barley β-glucan by sake yeast displaying Aspergillus oryzae β-glucosidase and endoglucanase. J Biosci Bioeng. 2008;105:622–627. doi: 10.1263/jbb.105.622. [DOI] [PubMed] [Google Scholar]
- 19.Verduyn C, et al. Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem J. 1985;226:669–677. doi: 10.1042/bj2260669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizzi M, Harwart K, Bui-Thanh N-A, Dellweg H. A kinetic study of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J Ferment Bioeng. 1989;67:25–30. [Google Scholar]
- 21.Bengtsson O, Hahn-Hagerdal B, Gorwa-Grauslund MF. Xylose reductase from Pichia stipitis with altered coenzyme preference improves ethanolic xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2009;2:9. doi: 10.1186/1754-6834-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe S, et al. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein-engineered NADH-preferring xylose reductase from Pichia stipitis. Microbiology. 2007;153:3044–3054. doi: 10.1099/mic.0.2007/007856-0. [DOI] [PubMed] [Google Scholar]
- 23.Amore R, Kötter P, Küster C, Ciriacy M, Hollenberg CP. Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (XYL1) from the xylose-assimilating yeast Pichia stipitis. Gene. 1991;109:89–97. doi: 10.1016/0378-1119(91)90592-y. [DOI] [PubMed] [Google Scholar]
- 24.Takuma S, et al. Isolation of xylose reductase gene of Pichia stipitis and its expression in Saccharomyces cerevisiae. Appl Biochem Biotechnol. 1991;28–29:327–340. doi: 10.1007/BF02922612. [DOI] [PubMed] [Google Scholar]
- 25.Walfridsson M, Anderlund M, Bao X, Hahn-Hägerdal B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilization. Appl Microbiol Biotechnol. 1997;48:218–224. doi: 10.1007/s002530051041. [DOI] [PubMed] [Google Scholar]
- 26.Eliasson A, Christensson C, Wahlbom CF, Hahn-Hägerdal B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol. 2000;66:3381–3386. doi: 10.1128/aem.66.8.3381-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahlbom CF, Van Zyl WH, Jonsson LJ, Hahn-Hägerdal B, Cordero Otero RR. Generation of the improved recombinant xylose-utilizing Saccharomyces cerevisiae TMB 3400 by random mutagenesis and physiological comparison with Pichia stipitis CBS 6054. FEMS Yeast Res. 2003;3:319–326. doi: 10.1016/S1567-1356(02)00206-4. [DOI] [PubMed] [Google Scholar]
- 28.Sauer U. Evolutionary engineering of industrially important microbial phenotypes. Adv Biochem Eng Biot. 2001;73:129–169. doi: 10.1007/3-540-45300-8_7. [DOI] [PubMed] [Google Scholar]
- 29.Christakopoulos P, Bhat MK, Kekos D, Macris BJ. Enzymatic synthesis of trisaccharides and alkyl beta-D-glucosides by the transglycosylation reaction of beta-glucosidase from Fusarium oxysporum. Int J Biol Macromol. 1994;16:331–334. doi: 10.1016/0141-8130(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 30.Doran-Peterson J, et al. Simultaneous saccharification and fermentation and partial saccharification and co-fermentation of lignocellulosic biomass for ethanol production. Springer Protocols. 2009;581:263–280. doi: 10.1007/978-1-60761-214-8_17. [DOI] [PubMed] [Google Scholar]
- 31.Hahn-Hagerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- 32.Jeffries TW, et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat Biotechnol. 2007;25:319–326. doi: 10.1038/nbt1290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.