Abstract
Ribosomal proteins L2, L3 and L4, together with the 23S RNA, are the main candidates for catalyzing peptide bond formation on the 50S subunit. That L2 is evolutionarily highly conserved led us to perform a thorough functional analysis with reconstituted 50S particles either lacking L2 or harboring a mutated L2. L2 does not play a dominant role in the assembly of the 50S subunit or in the fixation of the 3′-ends of the tRNAs at the peptidyl-transferase center. However, it is absolutely required for the association of 30S and 50S subunits and is strongly involved in tRNA binding to both A and P sites, possibly at the elbow region of the tRNAs. Furthermore, while the conserved histidyl residue 229 is extremely important for peptidyl-transferase activity, it is apparently not involved in other measured functions. None of the other mutagenized amino acids (H14, D83, S177, D228, H231) showed this strong and exclusive participation in peptide bond formation. These results are used to examine critically the proposed direct involvement of His229 in catalysis of peptide synthesis.
Keywords: peptidyl transfer/ribosomal protein L2/ribosome/translation/tRNA
Introduction
Peptide bond formation is the central enzymatic activity of the ribosome. Despite intensive experimental investigations the question of which ribosomal components form the corresponding peptidyl-transferase center (PTC) remains unanswered. In the late 1960s, Monro and co-workers showed that the 50S subunit alone is able to catalyze peptide bond formation (Monro, 1967). The 50S subunit from the Escherichia coli ribosome consists of 35 different molecules: 33 proteins and two rRNAs. Many biochemical methods have been applied to limit the peptidyl-transferase candidates to a few molecules. Components that are in close proximity to the PTC have been identified by photocrosslinking studies employing photoreactive groups attached to the CCA-end of A-, P- or E-site-bound tRNAs. Since the peptidyl-transfer reaction takes place at the ends of A- and P-site-bound tRNAs, the crosslinked molecules must be a part of or in close proximity to the catalytic center. Ribosomal components identified by this approach are the proteins L2, L15, L16, L27 and L33, and 23S rRNA (for review see Wower et al., 1993). Photoaffinity labeling with antibiotics that inhibit the peptidyl-transfer reaction provided evidence for the presence of the proteins L2, L15, L16, L18, L22, L23 and L27 as well as the central loop of domain V of 23S rRNA at or near the PTC (Cooperman et al., 1990), and photolabile oligonucleotides complementary to domain V central loop sequences place proteins L2 and L3 in the PTC vicinity (Vladimirov et al., 2000).
Some of these components were excluded from catalyzing peptidyl transfer by single-omission tests: 50S subunits lacking one protein were reconstituted and tested in the puromycin reaction for peptidyl-transferase activity. It was shown that the proteins L2, L3 and L4 and 23S rRNA are essential, whereas the other proteins, as well as 5S rRNA, are dispensable (Schulze and Nierhaus, 1982; Franceschi and Nierhaus, 1990; Khaitovich and Mankin, 2000).
The discovery that not only proteins but also RNA molecules can have enzymatic activity places 23S rRNA in the center of interest in the search for the peptidyl transferase. The peptidyl-transferase activity of ribosomes from Thermus aquaticus withstands treatment with proteases, SDS and phenol (Noller et al., 1992). These components destroy the native conformation of isolated proteins. However, RNA–protein interactions in ribosomes are very stable and it was not possible to remove all proteins from 23S rRNA. Eight proteins (L2, L3, L13, L15, L17, L18, L21, L22) remained stoichiometrically bound to 23S rRNA (Khaitovich et al., 1999a), and L1 and L4 were later also found (Khaitovich and Mankin, 2000). The prime candidates for the peptidyl-transferase activity are still L2, L3, L4 and 23S rRNA. Evolutionary arguments favor L2 out of the proteins, since it is one of the most conserved proteins of those universally present within the large ribosomal subunit (Müller and Wittmann-Liebold, 1997). Moreover, in studies using reconstituted subunits, mutation of the highly conserved His229 in E.coli L2 to glutamine leads to a 50S particle devoid of peptidyl-transferase activity (Cooperman et al., 1995). Related in vivo studies by Ühlein et al. (1998) also indicate the importance of this His residue in the translational activity of ribosomes.
Recently, peptide bond formation was described to be catalyzed by naked mature or in vitro transcribed 23S rRNA (Nitta et al., 1998a,b), but this observation could not be reproduced by the authors and others (Khaitovich et al., 1999b; Nitta et al., 1999). Therefore, the question of whether or not 23S rRNA of the large ribosomal subunit can form peptide bonds remains unanswered. Of particular interest in this context are the results of Zhang and Cech (1998), who demonstrated that a 175 nucleotide ribozyme, selected from a random RNA library, not only can catalyze peptide bond formation but also shares secondary structure motifs with the PTC of 23S rRNA.
Alternatively, the catalytic core of serine proteases has been proposed as a molecular model for the PTC, since both active centers have evolved to catalyze hydrolysis and formation of peptide bonds, respectively. The active center of serine proteases consists of the catalytic triad serine, histidine and aspartic acid, and has been developed at least twice independently by convergent evolution, as seen in the eukaryal trypsin family and the bacterial subtilisin family (Nierhaus et al., 1980; Rychlik and Cerna, 1980). There is circumstantial evidence that a histidyl residue is involved in ribosomal peptide bond formation (Nierhaus et al., 1980; Cooperman et al., 1995 and references therein). Moreover, the ribosomal protein L2 contains universally conserved seryl, histidyl and aspartyl residues. Here we report the functional effects of mutating these amino acids in E.coli L2 to test this proposal as well as the possible involvement of L2 in other ribosome functions.
Results
In vivo analysis
In mutagenizing protein L2 we constructed two new groups of variants. Group 1, consisting of the six variants shown in Figure 1, focused on conserved histidines, serines and aspartic or glutamic acids. Group 2, consisting of the variants H14Q and H231Q, were made as controls for the previously studied H229Q. Mutations E144Q and D167N had no phenotype and are not considered further. In addition, as H229C had the same phenotype as H229A, we elected to pursue further studies only with H229A. For the Group 1 variants D83N, S177A, D228N and H229A, the gene for the ribosomal protein L2 and the mutagenized genes were cloned into the plasmid pQE-60, which plasmid adds eight additional codons for the amino acids Arg, Ser and His6 to the 3′-end of the cloned gene. Escherichia coli strain XL-1 was transformed with the plasmids. Variants H14Q, H229Q and H231Q (Group 2) were prepared from transformed E.coli cells using a conditional T7 RNA polymerase expression system as described earlier (Romero et al., 1990). DNA sequencing confirmed that the plasmid-encoded L2 genes only contain the desired mutations.
Fig. 1. Regions of L2 where mutations were introduced. Conserved amino acids are shaded. The mutations analyzed here are marked by an arrow. Organisms compared are E.coli (bacteria), Haloarcula marismortui (archea), Saccharomyces cerevisiae (low eukarya) and human.
Growth characteristics. Growth curves were monitored for Group 1 mutants in liquid LB medium in the absence and presence of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Without IPTG all mutants tested had similar generation times, between 30 and 35 min. In the presence of 0.1 mM IPTG the doubling time of the strain expressing L2-6×H (wild-type L2 with a C-terminal His tag) was 35 min. Mutation S177A has only a minor effect on the doubling time (40 min). Mutations D83N, D228N and H229A severely reduce the growth rate, prolonging the generation time at least 8-fold (Table I). These three mutants have a dominant deleterious (lethal) phenotype.
Table I. Generation times of L2 mutants.
| L2 mutant | Generation time (min) |
|---|---|
| 50S[L2] | 35 |
| 50S[D83N] | 280 |
| 50S[S177A] | 40 |
| 50S[D228N] | 320 |
| 50S[H229A] | 290 |
Cells were grown in LB/ampicillin medium at 37°C. At 0.4 A560/ml, IPTG was added to a final concentration of 0.1 mM. At 0.8 A560/ml, cells were diluted 100-fold in fresh LB/ampicillin medium supplemented with 0.1 mM IPTG and cell growth was monitored. 50S[L2], wild type.
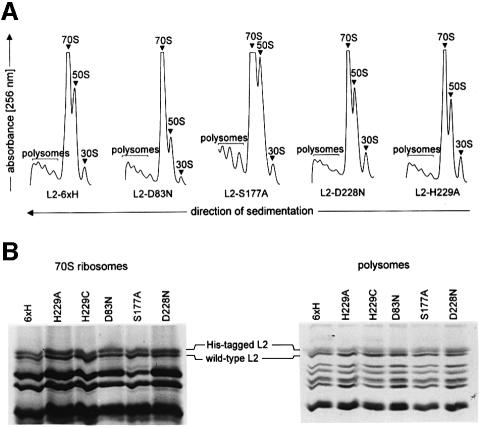
Incorporation of Group 1 L2 mutants into 70S ribosomes and polysomes. Lysates of L2-overproducing cells were separated on a sucrose gradient (Figure 2A). The UV profiles of all mutants look similar. They show symmetrical peaks for 50S subunits, 70S ribosomes and polysomes in comparable ratios. No precursors of the 50S assembly were detected between the 30S and 50S peaks, indicating a normal assembly of the 50S subunit. 70S ribosomes and polysomes were isolated and their proteins were separated by SDS–PAGE. Overproduced, His-tagged L2 can be distinguished from wild-type L2 by its increased molecular weight. All L2 mutants are incorporated into 70S ribosomes and polysomes in similar amounts to His-tagged L2 without mutation (Figure 2B).
Fig. 2. Incorporation of mutagenized L2 into 70S ribosomes and polysomes. (A) Lysates from cells overproducing L2 were separated on a sucrose gradient. L2-6×H, wild-type L2 with a C-terminal His tag. (B) L2 content of 70S ribosomes and polysomes isolated from L2-overproducing cells. 70S ribosomes and polysomes shown in (A) were loaded on SDS–PAGE gels. The region of the gels between 20 and 30 kDa is shown.
In vitro analysis of reconstituted 50S particles
Mutagenized L2 was overproduced and purified using either the high-affinity interaction between the C-terminal stretch of six histidine residues and a Ni-NTA matrix (Group 1) or using a procedure combining streptomycin sulfate precipitation and RP-HPLC (Group 2), as described (Cooperman et al., 1995). In order to test the effect of the mutagenized L2 proteins in an in vitro translation system we constructed 50S subunits in which the wild-type L2 is replaced by the mutagenized variant. Proteins from the 50S subunit (TP50) were isolated and L2 was removed by either ion-exchange chromatography and gel filtration (Group 1) or RP-HPLC and gel filtration (Group 2) as described (Cooperman et al., 1995). The mutagenized L2, L2-depleted TP50 (TP50-L2) and rRNA were reconstituted into 50S particles that were purified from non-reconstituted material by sucrose-density centrifugation. Before testing the functional activities of the reconstituted subunits, we analyzed whether the amino acid exchange in L2 or the absence of L2 affected the 50S reconstitution or 70S formation in vitro.
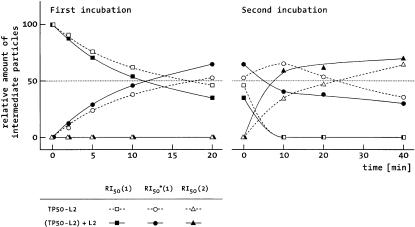
In vitro reconstitution of 50S subunits lacking L2. Two-dimensional gel electrophoresis revealed that the preparation TP50-L2 contained all ribosomal proteins in stoichiometric amounts, except L2, which was quantitatively absent (data not shown). TP50-L2 and total rRNAs derived from 70S ribosomes were incubated under reconstitution conditions with and without L2. At various time points of the first and the second incubation step, the formation of the three reconstitution intermediates RI50(1), RI50*(1) and RI50(2) was monitored by analyzing an aliquot of the reconstitution mixture on sucrose gradients as described previously (Dohme and Nierhaus, 1976). Figure 3 demonstrates that in the absence of L2 the formations of the intermediate particles RI50*(1) and RI50(2) were retarded, but that the final yield of 50S particles was the same as that formed in the presence of L2. The final 50S particles formed in the absence and presence of L2 had identical S values (not shown) and, except for L2, the same protein content as determined by two-dimensional (2D) gel electrophoresis. Only the amount of L16 was somewhat reduced; since the intensity of the L16 spot in the 2D gel can vary significantly even if derived from native 50S subunits, its amount cannot be estimated precisely, but is certainly >50%.
Fig. 3. Kinetics of the formation of the reconstitution intermediates. Particles were reconstituted from rRNA and TP50 with or without protein L2 (filled and open symbols, respectively). Aliquots were withdrawn from the reconstitution mixture during the first (left) or second (right) incubation step and subjected to a sucrose gradient centrifugation. The relative areas of the peaks of the reconstitution intermediates were determined from the A260 profile.
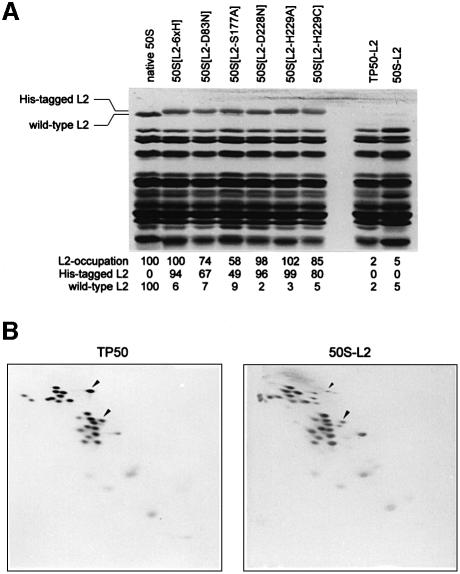
L2 occupation in purified reconstituted 50S particles. For the Group 1 variants, purified reconstituted 50S particles were applied to SDS–PAGE in order to determine the amount of L2 incorporated (Figure 4A). Small amounts of wild-type L2 are still present in L2-depleted 50S particles (50S-L2; Figure 4B) and in all 50S containing mutagenized L2; the residual wild-type L2 stems from the 23S rRNA preparation that still contained <10% of L2. The subunits containing His-tagged L2 and the mutants D228N and H229A show L2 bands of similar intensities to those of native 50S subunits, indicating that the L2 incorporation is not hampered by the amino acid exchange or the His tag. The intensities of the mutagenized L2 bands in 50S[D83N] and 50S[S177A] are reduced to 67 and 49%, respectively. This reduced L2 occupation affects the interpretation of the in vitro assay results (see below).
Fig. 4. Protein content of reconstituted 50S subunits. (A) SDS–PAGE analysis of 50S subunits reconstituted with mutagenized L2 and purified by sucrose-density centrifugation. The protein equivalent of 0.3 A260 units of reconstituted particles was applied onto the gel. The intensities of the bands corresponding to mutagenized and wild-type L2 band were normalized to the intensities of the bands of L1 and the double band L3/L4 below the L2 bands, and the content of L2 is given as a percentage of the amount of L2 found in native 50S. (B) Two-dimensional gel electrophoresis of the proteins derived from native 50S subunits (left) and purified 50S particles reconstituted in the absence of L2 (right). The arrowheads indicate the spots of L2 (above) and L16 (below). In the 50S-L2 particle a residual amount of L2 of <10% is seen.
For the Group 2 variants, the TP50-L2 pool used for reconstitution was virtually devoid of L2 (<2%). The amount of the L2 variant H229Q incorporated into reconstituted 50S subunits was 98% of that seen with wild-type L2, as estimated by densitometric scanning of a 2D PAGE analysis of extracted protein (Geyl et al., 1981).
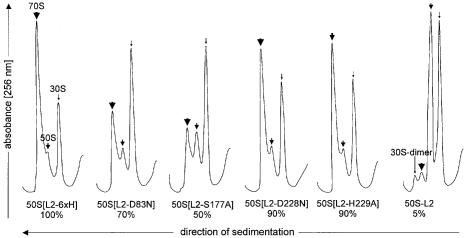
70S formation of the purified 50S mutants. The association capability of the 50S Group 1 mutants was examined by incubating them with a 2-molar excess of 30S subunits under conditions that strongly favor the association of 30S and 50S subunits. 70S formation was analyzed by sucrose-density centrifugation (Figure 5). 50S subunits containing His-tagged L2 associate almost quantitatively to 70S ribosomes. In contrast, 50S subunits lacking L2 are not able to form 70S ribosomes at all. We conclude that L2 is absolutely required for the association of the 30S and 50S subunits to form the 70S ribosome. Earlier we showed that the H229Q mutation has little effect on 50S association with 30S subunits (Cooperman et al., 1995). The D228N and H229A mutations slightly impair the association (90% active), the D83N and S177A mutations are less active (70 and 50%, respectively). The reduced activities of the latter mutations can be explained by the decreased incorporation of mutagenized L2 into the 50S particles.
Fig. 5. Sucrose gradients of reassociated 70S ribosomes. Reconstituted 50S particle (1 A260) was incubated with a 2-molar excess of 30S subunits and loaded onto a sucrose gradient. Approximately 90% of the 50S subunits containing His-tagged L2 are associated to 70S particles. The percentages below the mutants are the area of the 70S peak compared with the 70S area of the 70S[L2-6×H] particle. Thick arrow, 70S ribosomes; intermediate arrow, 50S subunits; thin arrow, 30S subunits.
Functional assays: the puromycin reaction and dipeptide formation with 50S subunits, and poly(Phe) synthesis with 70S ribosomes. Both Group 1 and Group 2 50S subunits were assayed in the puromycin reaction, whereas only Group 1 subunits were assayed for poly(Phe) synthetic activity and dipeptide formation. The His tag at the C-terminus of the recombinant L2 does not affect activities in these assays, since 50S subunits containing wild-type L2 or His-tagged L2 have the same activities (data not shown). Poly(Phe) synthesis requires 70S ribosomes and tests all reactions of the elongation cycle (A-site occupation, peptidyl transfer, translocation). The puromycin reaction and dipeptide formation are simpler systems, working with 50S subunits alone, and testing the binding of substrates to A and P sites as well as peptidyl transfer. In the puromycin reaction, fMet-tRNA (or N-AcPhe-tRNAPhe) and puromycin are substrates for P and A sites, respectively. In the case of dipeptide formation, fMet-tRNA is the P-site substrate and Phe-tRNAPhe is the A-site substrate.
The results of these assays are presented in Table II. We exploited the inability of 50S-L2 to form 70S ribosomes for removal of traces of 50S particles containing L2. 50S-L2 particles were incubated under association conditions with an excess of 30S subunits, and 50S-L2 was separated from 70S and 30S by sucrose-density centrifugation. No L2 was detectable in the purified 50S-L2 by SDS–PAGE analysis and such subunits had no activity in assays of the puromycin reaction, poly(Phe) synthetic activity or dipeptide formation (Table II).
Table II. Activities (in %) of 50S mutants in functional assays relative to the activity of 50S subunits reconstituted with wild-type L2.
| |
Group 1 variants |
Group 2 variants |
||||||
|---|---|---|---|---|---|---|---|---|
| D83N | S177A | D228N | H229A | 50S-L2 | H229Q | H14Q | H231Q | |
| L2 content | 74 | 58 | 98 | 102 | 0a | 105b | ||
| 70S association | 95 | 86 | 90 | 88 | 0a | 100b | ||
| Poly(Phe) synthesis | 53 | 35 | 64 | 62 | 0a | |||
| Dipeptide formation | 40 | 25 | 51 | 33 | 0a | |||
| Puromycin reaction with complete tRNAs | 40 | 33 | 57 | 8 | 0a | 0b | 105 | 106 |
| Binding to the | ||||||||
| P site: | ||||||||
| AcPhe-tRNA | 72 | 75 | 88 | 78 | 2 | |||
| CACCA-AcPhe | 65 | 72 | ||||||
| A site: | ||||||||
| Phe-tRNA | 95 | 88 | 95 | 100 | 19 | |||
| CACCA-Phe | 84 | 72 | ||||||
| CACCA-Phe | 90c | 20c | ||||||
| Puromycin reaction with CACCA-AcPhe | 14 | 4 | ||||||
For Group 1 variants, experimentally determined activities were corrected for the content of mutant L2 in the reconstituted 50S particles (see Figure 4), and for the presence of residual amounts of wild-type L2 (<10%) in the 23S rRNA preparation used for the reconstitution of the particles containing variant L2. For 100% values see Materials and methods. In all functional assays, native 50S subunits were included; they showed a 1.7-fold higher activity on average than the control particle reconstituted with wild-type L2.
aDetermined after removal of the minor fraction of 50S ribosomes still containing L2. For details see text.
bEarlier results (Cooperman et al., 1995).
cThe stimulation of the binding of CACCA-Phe upon addition of deacylated tRNAPhe was measured. Addition of deacylated tRNAPhe leads to a 3.1-fold increase (= 100%) in CACCA-Phe binding to reconstituted 50S subunits containing L2-6×His, consistent with earlier results (Ulbrich et al., 1978).
Variants D83N, S177A and D228N give consistent results in all three assay systems: 40–53% activity for D83N, 25–35% for S177A and 51–64% for D228N. The inhibition seen for 50S[S177A] is likely to be due to a structural distortion of the reconstituted 50S subunit rather than a functional defect, since the corresponding mutant has a normal growth rate. Differentiated activities are obtained for the H229A mutation, with activity in the puromycin reaction (8%) being substantially lower than in dipeptide formation (33%) or poly(Phe) synthesis (62%). Furthermore, the H229Q variant is totally devoid of puromycin activity (Cooperman et al., 1995), whereas the corresponding mutation in the non-conserved His14 and less well-conserved His231 residues is without effect on the puromycin reaction.
Functional assays: binding of tRNAs and tRNA fragments to various reconstituted 50S particles. To test the possibility that His229 might be involved in the binding of adenosine 76 of the A-site-bound tRNA, we investigated the relative binding affinities of tRNAs and tRNA fragments to the various reconstituted 50S particles under conditions similar to those of the puromycin reaction. The results (Table II) show that the presence of L2 is important for the binding of both P- and A-site tRNAs, with the 50S-L2 subunit having relative binding activities of 2 and 19%, respectively. In contrast, L2 is relatively unimportant for the binding of the 3′-terminal fragments CACCA-AcPhe and CACCA-Phe, which bind virtually exclusively to the P and A sites, respectively (Ulbrich et al., 1978), with the 50S-L2 subunit having relative binding activities of 72% to both sites. These contrasting effects suggest that the strong binding of tRNAs in the presence of L2 results from interactions of 50S to regions other than the 3′-ends of the tRNAs, i.e. outside the immediate PTC. However, the 3′-fragments of tRNA bind well to the 50S-L2 particles. Therefore, a puromycin reaction was performed in the presence of the fragment CACCA-AcPhe; the 50S-L2 particle has basically no and the H229A very little PTF activity (Table II).
The effects of the various L2 mutations are more pronounced on P-site binding, but are in no case dramatic. Thus, binding of AcPhe-tRNA to the P site of 50S subunits containing mutated L2 is impaired by ∼25%, whereas the binding of Phe-tRNA to the A site is hardly reduced. Similarly, the H229A variant has a stronger effect (35% reduction) on CACCA-AcPhe binding than on CACCA-Phe binding (16% reduction).
Cooperative effects between the binding of A- and P-site substrates of the PTC have been described. For example, addition of the P-site substrate deacylated tRNA increases the binding of the A-site substrate CACCA-Phe several-fold under fragment assay conditions (Ulbrich et al., 1978). In the experiments reported in the second last line of Table II the stimulation was 3.1-fold. Addition of deacylated tRNA increases binding of the A-site CACCA-Phe in the presence of the H229A mutation almost as strongly (2.9-fold), corresponding to a 90% stimulatory effect as compared with the wild-type control L2-6×H (Table II). This observation indicates that H229 is not involved in the fixation of the terminal A of the A-site-bound tRNA. 50S-L2 subunits show a weak stimulation of 1.4-fold in the presence of deacylated tRNA (20%; Table II). This is probably due to a poor deacylated tRNA binding in the absence of L2. Note that deacylated tRNA binds specifically to the P site under fragment assay conditions as does AcPhe-tRNA (Ulbrich et al., 1978), and, as mentioned above, P-site binding of whole tRNAs is abolished in 50S-L2 particles (see AcPhe-tRNA binding in Table II).
Discussion
Structural and functional analysis of 50S subunits lacking L2
Our current studies demonstrate that L2 is not essential for in vitro 50S assembly. All the reconstitution intermediates are formed, albeit more slowly than in the presence of L2 (Figure 3), and the final particle has a sedimentation constant of 50S and a full set of ribosomal L-proteins except L2, although the incorporation of L16 is somewhat reduced (Figure 4). The dependence of L16 incorporation on the presence of L2 was also observed in vivo and seems to be even stronger than during the in vitro reconstitution: 50S subunits with a deletion mutant of L2 (Thr222–Asp228) completely lack L16 and partly lack the proteins L28, L33 and L34 (Romero et al., 1990). Earlier (Cooperman et al., 1995), some of us, using a reconstitution procedure analogous to that used in this paper, had demonstrated an apparent requirement for L2 in 50S reconstitution. We have not as yet identified the specific reasons for this difference.
In contrast to 50S assembly, association of 30S and 50S subunits to 70S ribosomes is completely dependent on L2 (Figure 5). The involvement of L2 in subunit association was previously suggested on the basis of L2 crosslinks to several proteins of the 30S subunit (Lambert and Traut, 1981). The coupling is probably mediated by an interaction between L2 and S20: (i) S20 was previously assigned as L26, since it can be found on either subunit depending on the isolation conditions; and (ii) proteins L2 and S20 form a very stable complex even in the presence of 6 M urea. In cation-exchange chromatography of TP50, L2 is always contaminated with S20, although S20 alone elutes at a much lower salt concentration (Diedrich et al., 1997).
The 50S-L2 subunit is completely inactive in poly(Phe) synthesis, puromycin reaction and dipeptide formation (Table II). The inactivity in poly(Phe) synthesis is explained by the inability to form 70S ribosomes, which are required for programmed protein synthesis. However, the puromycin reaction and dipeptide formation assay work with 50S subunits alone in the presence of alcohol. 50S-L2 is inactive in both systems, indicating that at least one of the following reactions is dependent on L2: (i) binding of the P-site substrate (fMet-tRNA); (ii) binding of the A-site substrate (puromycin or aminoacyl-tRNA); or (iii) peptidyl transfer. The results of further functional assays suggest that L2 is involved in all three activities. The binding of AcPhe-tRNA to the P site is abolished and that of Phe-tRNA to the A site is strongly reduced in the absence of L2. However, the binding of 3′-fragments of acylated tRNAs is hardly impaired when L2 is absent (Table II). Therefore, the puromycin reaction was tested in the presence of the CACCA-AcPhe fragment, and practically no activity was observed (4%).
The presence of L2 directly at the PTC near to the acyl residues of both aminoacyl-tRNA and peptidyl-tRNA is documented by a wealth of data. (i) L2 is crosslinked to the peptidyl residue and to A73 of P-site-bound peptidyl-tRNA (Wower et al., 1993). (ii) The main crosslink site of bromamphenicol, a derivative of the antibiotic chloramphenicol, is L2 (Sonenberg et al., 1973). Chlor amphenicol inhibits the binding of the CCA-aminoacyl terminus of the aminoacyl-tRNA in the A site but not the binding of substrates of the P site (Pestka, 1969; Celma et al., 1971; Contreras and Vazquez, 1977; Ulbrich et al., 1978). Recently, however, effects of chloramphenicol at the tRNA 3′-ends at both A and P sites have been observed (S.V.Kirillov, B.Porse and R.A.Garrett, unpublished). (iii) Photolysis of 50S complexes with photolabile oligonucleotides complementary to 23S rRNA sequences 2448–2458 and 2604–2612, both of which fall within the central loop of domain V, affords target site-specific photoincorporation into protein L2 (Vladimirov et al., 2000).
These indications of the topographical neighborhood of L2 and of the 3′-ends of the tRNAs in A and P sites contrast with our finding that L2 is required not for the binding of the peptidyl-transferase substrates CACCA-AcPhe and CACCA-Phe (Table II) but rather for tRNA binding to A and P sites. Therefore, L2 might be important for binding the tRNAs, e.g. at the elbow region, rather then being involved in the fixation of the 3′-ends of the tRNAs at the PTC. The involvement of L2 in tRNA binding to both A and P sites as well as in 70S association makes it likely that this protein is an important component of the most massive bridge connecting the two subunits within the 70S ribosome, viz. bridge 2 in the terminology of Frank and co-workers (Frank et al., 1995; Lata et al., 1996). Indeed, the localization of L2 within the 50S subunit as well as the 70S ribosome by means of neutron scattering indicates that one end of the elongated L2 is located at the interface side of the 50S subunit and might even protrude into the inter-subunit space (R.Willumeit, S.Forthmann, J.Beckmann, G.Diedrich, H.B.Stuhrmann and K.H.Nierhaus, manuscript submitted). Bridge 2, which can split into various branches, connects the two functional hot spots of the ribosome, the decoding center on the small ribosomal subunit and the PTC on the large subunit. In the frame of the α–ε model, this bridge is the prime candidate for the α–ε domain of the 70S ribosome that binds two tRNAs tightly. We have proposed that this domain carries the two tRNAs during the translocation reaction from the A and P sites to the P and E sites, respectively (Nierhaus et al., 1995; Dabrowski et al., 1998). Our results suggest that L2 is part of bridge 2, thus explaining the essential involvement of this protein in association of the ribosomal subunits. The fact that L2 was positioned relatively far from the interface of the 50S subunit in a recent low-resolution X-ray structure of the 50S subunit (Ban et al., 1999) is not necessarily in conflict with the neutron scattering results, since only an L2 fragment of half the size of L2 was fitted into the 50S structure.
Since the 50S-L2 particles bind 3′-fragments of acylated tRNAs well, a distortion of the CCA binding region at the PTC seems to be unlikely. The non-perturbed binding of the substrates to the PTC on one hand and the complete block of peptide bond formation (Table II) on the other suggest either a direct involvement of L2 in peptide bond formation or a subtle distortion of the active center in the absence of L2.
Are conserved His, Ser and Asp residues of L2 involved in peptide bond formation?
A strong indication for an active role of L2 in peptide bond formation would be a mutation of a single amino acid that specifically destroyed the peptidyl-transferase activity. To explore this possibility, we constructed several mutants of L2 and tested their effects on the ribosomal functions. The selection of the mutagenized amino acids was based on a hypothetical mechanism for the peptidyl-transfer reaction (Nierhaus et al., 1980; Rychlik and Cerna, 1980) according to which peptidyl transfer occurs via a charge-relay system similar to serine protease cleavage of peptide bonds. The catalytic core of a serine protease consists of the active triad of the amino acids serine, histidine and aspartic acid. In the frame of the hypothesis, the acyl serine intermediate in serine protease catalysis is replaced by the P-site-bound peptidyl-tRNA and, in addition, the PTC active site includes a histidine and an aspartic or glutamic acid.
Although the involvement of an aspartic or glutamic acid residue in the peptidyl-transfer reaction has never been demonstrated, there is evidence supporting histidine side-chain involvement. (i) The pH dependence of peptide bond formation suggests the need for a neutral pH of an imidazole residue (Maden et al., 1968; Fahnestock et al., 1970). (ii) Histidine-modifying agents inhibit the peptidyl-transferase activity of the 50S subunit (Rychlik and Cerna, 1980 and references therein). (iii) The first-order rate of inactivation of the peptidyl transferase indicates that modification of one histidine residue is sufficient for the loss of activity (Wan et al., 1975).
To test the catalytic triad hypothesis we examined the effects of mutation of the conserved serine (S177A), histidine (H299A) and aspartic acid residues (D83N, D228N) of protein L2 (Figure 1) on both cell growth and ribosomal function. Two additional conserved glutamic and aspartic acids at positions 144 and 167 were also replaced by glutamine and asparagine, respectively, and two less well-conserved histidine residues (H14 and H231) were replaced by glutamine, but these mutants did not show significant effects in functional assays. Expression of three of these L2 variants, D83N, D228N and H229A, but not of S177A, retards cell growth >8-fold (Table I). As none of these three mutations impairs the assembly of L2 into 50S subunits, and 50S subunits containing these mutations flow easily into 70S ribosomes and also show up in polysomes (Figure 2) so that neither the association of ribosomal subunits nor the initiation is affected, we conclude that the mutations cause a dramatic defect of the elongation and/or termination phase.
50S particles containing the mutagenized L2 were reconstituted and analyzed with in vitro assays to define further the defects of the mutations (Table II). In contrast to the in vivo assembly, mutations D83N and S177A impair the incorporation of L2 into 50S subunits in vitro. The reduced L2 content of these 50S mutants must be taken into consideration for a comparison of the functional activities of the mutants, since 50S subunits lacking L2 are inactive in most in vitro systems. The synopsis of the results obtained in the various functional tests reveals that all mutants are fully active in Phe-tRNA binding to the A site (88–100%) and show an only slightly reduced AcPhe-tRNA binding to the P site by (∼30%). D83N is active at a 40–55% level in all other activities, except for retaining full capability in 70S formation, and therefore plays no specific role in one distinct function. A similar pattern is seen with S177A and D228N, which display activity ranges of 25–35 and 50–65%, respectively. The reduced activity of S177A is probably an in vitro artifact, since no retardation of the growth rate is observed with this mutant. A different functional behavior is seen with H229A. Dipeptide formation is sharply reduced (residual activity 33%) and the activity of the puromycin reaction is reduced even further, to ∼10% (and to 0% with the H229Q variant). Surprisingly, poly(Phe) synthesis is only marginally reduced (60% activity), although peptide bond formation is one of the reactions of the elongation cycle tested with this system.
A similar effect, drastic reduction of peptide bond formation and much less impairment of poly(Phe) synthesis, has been observed with a series of 23S rRNA mutations, in particular G2581A. G2581 is thought to interact with C75 of a tRNA at the P site (Spahn et al., 1996a,b). At least two hypotheses may be offered to explain this apparently paradoxical behavior. (i) His229 might be involved in the binding of adenosine 76 of the A-site-bound tRNA. Mutation of this residue would strongly reduce the affinity of puromycin, whereas the affinity of the whole aminoacyl-tRNA would be only moderately reduced, because it is bound via multiple contacts to the 50S subunit (M.A.Schäfer and K.H.Nierhaus, manuscript in preparation). Accordingly, the inhibitory effect of the mutation would be stronger for puromycin than for the whole aminoacyl-tRNA. However, our results do not support this hypothesis: if H229A mutation impaired either puromycin binding or that of the 3′-end of aminoacyl-tRNAs, a stronger inhibition of CACCA-Phe binding would be expected than the slight reduction observed (Table II). We also note the recent demonstration that the base adjacent to the terminal adenosine, namely C75 of an A-site-bound tRNA, probably forms a Watson–Crick base pair with the universally conserved G2553 of 23S rRNA (Kim and Green, 1999). (ii) The poly(Phe) synthesis and puromycin reaction assays have different rate-limiting steps. The slowest reaction of the elongation cycle is the A-site occupation, which is much slower than peptidyl transfer (Bilgin et al., 1988; Schilling-Bartetzko et al., 1992). For the puromycin reaction, however, peptide bond formation is probably the rate-limiting step, rather than the expected fast binding of puromycin to the A-site region of the PTC. As a result, a severe reduction of the rate of peptidyl-transferase activity would more strongly decrease the puromycin reaction rate than the rate of poly(Phe) synthesis. This point is probably also related to the fact that the drastic defects of cell growth are reflected by the severe effects seen in the assays for peptidyl-transferase activity in contrast to poly(Phe) synthesis. Poly(Phe) synthesis was performed at 6 mM Mg2+ with an incorporation rate of 0.5 Phe residues per ribosome per second; the in vivo incorporation rate is >10 times faster. Under conditions of fast protein synthesis a severe retardation of peptide bond formation will have serious consequences for cell growth.
In support of the importance of H229 for ribosomal function are the results of Ühlein et al. (1998), who demonstrated that L2 from the archaeon Haloarcula marismortui, when expressed in E.coli, could be incorporated into active ribosomes in vivo, whereas the H229G mutant of H.marismortui yielded inactive ribosomes. On the other hand, the result that the mutant H229E in yeast mitochondria behaved normally at 30°C, even though it was conditionally lethal and caused a strong respiratory-deficient phenotype, led Pan and Mason (1997) to conclude that H229 is not essential for peptide bond formation.
These seemingly contradictory results can be reconciled taking into account the present results showing that a mutation of H229 drastically impairs peptide bond formation but does not completely abolish the activity, and assuming that protein synthesis in yeast mitochondria at 30°C is very slow and rather insensitive to the rate of peptide bond formation, i.e. even severe damage of the PTC could be masked by a slow translational apparatus. As discussed for our relatively slow poly(Phe) synthesis, likewise a slow in vivo rate might be insensitive to even pronounced decreases in the rate of peptide bond formation.
Does H229 belong to a charge-relay system similar to the His–Asp system in the active center of serine proteases? We did not find strong evidence for the existence of such a system within the L2 protein, since only mutations at H229, and not at several conserved Asp and Glu residues, efficiently inhibit peptide bond formation. The participation of an acidic residue elsewhere in the ribosome cannot, however, be excluded. Possible candidates would include acidic residues from another protein, e.g. L4, which like L2 is universally present in the ribosomes of various organisms (Planta and Mager, 1998), or a well placed phosphate group of 23S rRNA. Alternatively, it may be that the H229 mutation has an allosteric, rather than direct, effect on peptidyl transferase. Such an effect would be consistent with a forthcoming high resolution structure of the 50S subunit showing that proteins are still some 25 Å away from the PTC. Nevertheless, such an interpretation must be considered with caution. Isolated 50S subunits are totally inactive for peptide bond formation in the absence of 30% alcohol, and achieving an active conformation could require major structural rearrangement within the PTC region.
The results presented here demonstrate that L2 is essential for the association of the ribosomal subunits and tRNA binding and is probably an important element of the α–ε domain proposed to translocate the tRNAs. They further suggest that, within L2, H229 plays an important role specifically in peptide bond formation, since the H229A mutant has a dominant lethal phenotype but no assembly defects. This mutant strongly blocks the peptidyl-transferase reaction while having little effect either on the binding of the substrates for the PTC, viz. CACCA-AcPhe and CACCA-Phe, or on the cooperativity between the P- and A-site regions of the PTC.
Materials and methods
Mutagenesis, cloning and sequence analysis
For Group 1 variants, the gene for the ribosomal protein L2 was amplified by PCR using total E.coli genome as template and GAGTAATAC CATGGCAG and AGCTTAGATCTTTTGCTACGGA as primers. The gene was cloned into plasmid pQE-60 (Qiagen) using the NcoI and BglII restriction sites. Mutagenesis of the L2 gene was performed by PCR. The following primers and their reverse complements were used: CGGTTC GGATTGTACTCAA for the D83N mutation, CGTCTGCGTGCT GGTGAAAT for the S177 mutation, AACCCGGTAAACCACCCA CAT for the D228N mutation and AACCCGGTAGACGCCCCACAT GGTGGT for the H229A mutation. The mutagenized DNA was ligated into plasmid pQE-60 and transformed into E.coli strain XL1 blue (Stratagene). The presence of the desired mutation was confirmed by sequencing.
Group 2 variants were prepared as described (Cooperman et al., 1995; Romero et al., 1990).
Overproduction and purification of L2 variants
For Group 1, the transformed cells were grown in 400 ml of LB medium at 37°C. At a cell density of 0.5 A560, overproduction was induced by adding IPTG to a final concentration of 1 mM. The cells were harvested 10 h after induction by centrifugation, resuspended in 10 ml of buffer A (10 mM Tris–HCl, 100 mM NaH2PO4, 6 M guanidine hydrochloride pH 8.0) and incubated at 4°C for 2 h. The supernatant was collected by centrifugation (20 min at 15 000 r.p.m., SA600 rotor) and added to the Ni-NTA column (3 ml matrix volume) equilibrated to buffer A. Weakly bound proteins were eluted with 20 ml of buffer B (10 mM Tris–HCl, 100 mM NaH2PO4, 6 M urea pH 5.9). His-tagged L2 was eluted with buffer C (10 mM Tris–HCl, 100 mM NaH2PO4, 6 M urea pH 4.5). The L2 containing fractions were unified, dialyzed against 500 ml of water (4× 1 h) and lyophilized. The proteins were resuspended in 2 ml of buffer rec4-6U [rec4 buffer containing 20 mM HEPES pH 7.6 (4°C), 4 mM MgOAc2, 400 mM NH4Cl, 0.2 mM EDTA, 4 mM 2-mercaptoethanol in the presence of 6 M urea], dialyzed against 500 ml of the same buffer for at least 6 h and finally dialyzed three times against 500 ml of rec4 buffer for 45 min.
Group 2 variant L2s were purified as described (Cooperman et al., 1995).
Isolation of polysomes and 70S ribosomes
The isolation of polysomes and 70S ribosomes is described in Remme et al. (1989) or Cooperman et al. (1990).
Preparation of TP50-L2 and rRNA
The procedure for the preparation of TP50-L2 is described in Diedrich et al. (1997) and Cooperman et al. (1990). Ribosomal RNA was isolated by phenol extraction of crude 70S ribosomes (according to Nierhaus, 1990) or of 50S subunits (Cooperman et al., 1990).
Reconstitution of 50S subunits and the poly(U)-dependent poly(Phe)-synthesis system
Both assay systems are described in Nierhaus (1990). Eighteen picomoles of 50S were used for the poly(Phe) synthesis system.
Isolation of reconstituted 50S particles and characterization of their protein composition
Seventy-five A260 RNA, 4.6 A230 TP50-L2 and the optimized amount of L2 were reconstituted. 50S subunits were isolated by sucrose-density centrifugation {10–40% sucrose in 38 ml of rec20 [20 mM HEPES–KOH pH 7.6 (4°C), 20 mM MgOAc2, 400 mM NH4Cl, 0.2 mM EDTA, 4 mM 2-mercaptoethanol]; 16 h at 22 000 r.p.m., SW27 rotor}. The 50S peak was pooled and centrifuged (20 h at 35 000 r.p.m., 60Ti rotor). 50S particles were resuspended in 100 µl of buffer D [20 mM HEPES pH 7.6 (4°C), 6 mM MgOAc2, 30 mM NH4Cl, 4 mM 2-mercaptoethanol]. The incorporation of mutagenized L2 was analyzed by applying 0.7 A260 of 50S subunits on a 15% polyacrylamide gel. Two-dimensional gel electrophoresis was performed according to Geyl et al. (1981).
Association of ribosomal subunits to 70S ribosomes
One A260 of each subunit was incubated in 100 µl of buffer E (20 mM HEPES pH 7.4, 20 mM MgOAc2, 30 mM KCl, 4 mM β-mercaptoethanol) for 30 min at 4°C. The mixture was separated by sucrose-density centrifugation (SW40 rotor, 10–40% sucrose in buffer E, 22 000 r.p.m. for 16 h at 4°C). The absorbance of the gradient was monitored at 256 nm.
Puromycin reaction with complete tRNAs
For Group 1, 5 pmol of 50S and 20 pmol of [35S]fMet-tRNA in 200 µl of buffer rec20 were mixed with 80 µl of methanol containing 1 mg/ml puromycin and incubated for 15 min on ice. The ionic conditions are those of buffer rec20. One hundred percent values (50S subunits containing wild-type L2 with a His tag) were 6820 c.p.m.; the background value (minus ribosomes; 1274) has been subtracted. For further processing see Schulze and Nierhaus (1982).
For Group 2, the reaction was carried out as described earlier (Nowotny et al., 1988; Cooperman et al., 1990).
Dipeptide formation with 50S particles
This system is described in Spahn et al. (1996b) except that 15 pmol of 50S (instead of 70S) and 20 pmol of fMet-tRNA (instead of pA-fMet) were used. One hundred percent values (50S subunits containing wild-type L2 with a His tag) were 4289 c.p.m.; the background value (minus ribosomes; 424) has been subtracted.
Poly(Phe) synthesis
Poly(Phe) synthesis followed Bommer et al. (1996). One hundred percent values (50S subunits containing wild-type L2 with a His tag and native 30S subunits) were 3944 c.p.m.; the background value (minus ribosomes; 127) has been subtracted.
Preparation of tRNA fragments
Ac[14C]Phe-tRNA and [14C]Phe-tRNA were incubated with T1 RNase, phenolized and applied on an HPLC column (C4). The fragments were eluted with a gradient (buffer A: 50 mM potassium acetate pH 4.5; buffer B: 50 mM potassium acetate pH 4.5 and 30% acetonitrile), and the fractions containing the labeled fragments used directly without further treatment in the binding assays.
Puromycin reaction with tRNA fragments
50S (22.5 pmol) was incubated with 2–4 pmol of 14C-labeled CACCA-AcPhe (1000 d.p.m./pmol) for 10 min at 37°C in 50 µl. The mixture was cooled on ice and the reaction was started by addition of 25 µl of 3 mM puromycin in EtOH. The assay (final ionic conditions HEPES–KOH pH 7.4, 135 mM NH4Cl, 250 mM KCl, 20 mM MgCl2, 33% EtOH) was incubated for 60 min on ice and stopped by the addition of 75 µl of 0.3 M NaOAc solution (saturated with MgSO4). The product was extracted with 1 ml of EtOAc and the radioactivity was determined by liquid scintillation counting (background, 80 d.p.m.; 100%, 850–1000 d.p.m.).
Binding of tRNA fragments and tRNAs to ribosomal particles
50S (30 pmol) in 25 µl of rec20 buffer was incubated for 10 min at 37°C. If indicated, tRNA (molar ratio tRNA:50S = 2:1) was added prior to the incubation. The sample was cooled on ice for 10 min and 2–4 pmol of 14C-labeled fragment (CACCA-Phe or CACCA-AcPhe, specific activity was ∼1000 d.p.m./pmol) were added. Fragment binding was initiated by the addition of 25 µl of ice-cold ethanol (50% vol/vol) and left for 30 min on ice. The 50S subunits with the bound fragment were centrifuged (14 000 r.p.m. for 60 min at 4°C). The supernatant was carefully removed and the pelleted subunits were resuspended in 200 µl of H2O by shaking at room temperature for 1 h. The radioactivity in the supernatant and in the resuspended pellet was measured. One hundred percent values correspond to 300–500 d.p.m. in various experiments. Background values (minus particles) were typically 40 d.p.m. and were subtracted. The error was <±10%.
Binding of AcPhe-tRNA and Phe-tRNA was performed under fragment assay conditions in the presence of 40% ethanol and in the absence of mRNA (25 µl with 1.5 pmol of 50S particles and 1.5 pmol of acylated tRNA in rec20 buffer plus 25 µl of 80% ethanol). Particles and bound tRNA were pelleted by centrifugation. The remaining radioactivity in the supernatant indicates how much tRNA did not bind to 50S subunits. When no 50S subunit was added, >90% of the tRNA input remained in the supernatant (1313 d.p.m. AcPhe-tRNA, 1080 d.p.m. Phe-tRNA). In the presence of the reconstituted control particle 50S[L2-6×H], 0.64 AcPhe-tRNA (278 d.p.m. in supernatant) and 0.58 Phe-tRNA (320 d.p.m.) were bound per 50S, respectively. A control with a puromycin reaction in the presence of 50S[L2-6×H] particles indicated site-specific binding of the charged tRNAs: 81–100% of the bound AcPhe-tRNA was in the P site in various assays, and at least 84% of the Phe-tRNA in the A site.
Acknowledgments
Acknowledgements
We thank Sean Connell for help and discussion and Detlev Kamp for technical assistance in protein purification. This work was supported by the Deutsche Forschungsgemeinschaft (grant Ni 174/8-2 to K.H.N.) and the National Institutes of Health, GM 53146 to B.S.C. and GM 17924 to R.R.T.
References
- Ban N., Nissen,P., Hansen,J., Capel,M., Moore,P.B. and Steitz,T.A. (1999) Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature, 400, 841–847. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Kirsebom,L.A., Ehrenberg,M. and Kurland,C.G. (1988) Mutations in ribosomal proteins L7/L12 perturb EF-G and EF-Tu functions. Biochimie, 70, 611–618. [DOI] [PubMed] [Google Scholar]
- Bommer U., Burkhardt,N., Jünemann,R., Spahn,C.M.T., Triana-Alonso,F.J. and Nierhaus,K.H. (1996) Ribosomes and polysomes. In Graham,J. and Rickwoods,D. (eds), Subcellular Fractionation. A Practical Approach. IRL Press at Oxford University Press, Oxford, UK, pp. 271–301. [Google Scholar]
- Celma M.L., Monro,R.E. and Vazquez,D. (1971) Substrate and antibiotic binding sites at the peptidyl transferase centre of E.coli ribosomes: binding of UACCA-leu to 50S subunits. FEBS Lett., 13, 247–251. [DOI] [PubMed] [Google Scholar]
- Contreras A. and Vazquez,D. (1977) Cooperative and antagonistic interactions of peptidyl-tRNA and antibiotics with bacterial ribosomes. Eur. J. Biochem., 74, 539–547. [DOI] [PubMed] [Google Scholar]
- Cooperman B.S., Weitzmann,C.J. and Fernandez,C.L. (1990) Antibiotic probes of E.coli ribosomal peptidyl transferase. In Hill,W.E., Dahlberg,A., Garrett,R.A., Moore,P.B., Schlessinger,D. and Warners,J.R. (eds), The Ribosome: Structure, Function and Evolution. American Society for Microbiology, Washington, DC, pp. 491–501. [Google Scholar]
- Cooperman B.S., Wooten,T., Romero,D.P. and Traut,R. (1995) Histidine 229 in the protein L2 is apparently essential for 50S peptidyl transferase activity. Biochem. Cell Biol., 73, 1087–1094. [DOI] [PubMed] [Google Scholar]
- Dabrowski M., Spahn,C.M.T., Schäfer,M.A., Patzke,S. and Nierhaus,K.H. (1998) Contact patterns of tRNAs do not change during ribosomal translocation. J. Biol. Chem., 273, 32793–32800. [DOI] [PubMed] [Google Scholar]
- Diedrich G., Burkhardt,N. and Nierhaus,K.H. (1997) Large-scale isolation of proteins of the large subunit from Escherichia coli ribosomes. Protein Expr. Purif., 10, 42–50. [DOI] [PubMed] [Google Scholar]
- Dohme F. and Nierhaus,K.H. (1976) Total reconstitution and assembly of 50S subunits from E.coli ribosomes in vitro. J. Mol. Biol., 107, 585–599. [DOI] [PubMed] [Google Scholar]
- Fahnestock S.R., Neumann,H., Shashua,V. and Rich,A. (1970) Ribosome-catalyzed ester formation. Biochemistry, 9, 2477–2483. [DOI] [PubMed] [Google Scholar]
- Franceschi F. and Nierhaus,K.H. (1990) Ribosomal proteins L15 and L16 are mere late assembly proteins of the large ribosomal subunit. J. Biol. Chem., 265, 16676–16682. [PubMed] [Google Scholar]
- Frank J. et al. (1995) A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem. Cell Biol., 73, 757–765. [DOI] [PubMed] [Google Scholar]
- Geyl D., Böck,A. and Isono,K. (1981) An improved method for 2-d gel electrophoresis: analysis of mutationally altered ribosomal proteins of E.coli. Mol. Gen. Genet., 181, 309–312. [DOI] [PubMed] [Google Scholar]
- Khaitovich P. and Mankin,A.S. (2000) Reconstitution of the 50S subunit with in vitro-transcribed 23S rRNA: a new tool for studying peptidlytransferase. In Garrett,R., Douthwaite,S., Liljas,A., Matheson,A., Moore,P. and Noller,H. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 229–243. [Google Scholar]
- Khaitovich P., Mankin,A.S., Green,R., Lancaster,L. and Noller,H.F. (1999a) Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc. Natl Acad. Sci. USA, 96, 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P., Tenson,T., Mankin,A.S. and Green,R. (1999b) Peptidyl transferase activity catalyzed by protein-free 23S ribosomal RNA remains elusive. RNA, 5, 605–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. and Green,R. (1999) Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell, 4, 859–864. [DOI] [PubMed] [Google Scholar]
- Lambert J.M. and Traut,R.R. (1981) The subunit interface of the E.coli ribosome. Identification of proteins at the interface between the 30S and 50S subunits by crosslinking with 2-iminothiolane. J. Mol. Biol., 149, 451–476. [DOI] [PubMed] [Google Scholar]
- Lata K.R., Agrawal,R.K., Penczek,P., Grassucci,R., Zhu,J. and Frank,J. (1996) Three-dimensional reconstruction of the Escherichia coli 30S ribosomal subunit in ice. J. Mol. Biol., 262, 43–52. [DOI] [PubMed] [Google Scholar]
- Maden B., Traut,R. and Monro,R. (1968) Ribosome-catalysed peptidyl transfer: the polyphenylalanine system. J. Mol. Biol., 35, 333–345. [DOI] [PubMed] [Google Scholar]
- Monro R. (1967) Catalysis of peptide bond formation by 50S ribosomal subunits from Escherichia coli. J. Mol. Biol., 26, 147–151. [DOI] [PubMed] [Google Scholar]
- Müller E.C. and Wittmann-Liebold,B. (1997) Phylogenetic relationship of organisms by ribosomal protein comparison. Cell. Mol. Life Sci., 53, 34–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus K. (1990) Reconstitution of ribosomes. In Spedding,G. (ed.), Ribosomes and Protein Synthesis. A Practical Approach. IRL Press at Oxford University Press, Oxford, UK, pp. 161–189. [Google Scholar]
- Nierhaus K.H., Schulze,H. and Cooperman,B.S. (1980) Molecular mechanisms of the ribosomal peptidyltransferase center. Biochem. Int., 1, 185–192. [Google Scholar]
- Nierhaus K.H. et al. (1995) The elongating ribosome: structural and functional aspects. Biochem. Cell Biol., 73, 1011–1021. [DOI] [PubMed] [Google Scholar]
- Nitta I., Kamada,Y., Noda,H., Ueada,T. and Watanabe,K. (1998a) Reconstitution of peptide bond formation with Escherichia coli 23S ribosomal RNA domains. Science, 281, 666–669. [DOI] [PubMed] [Google Scholar]
- Nitta I., Ueda,T. and Watanabe,K. (1998b) Possible involvement of Escherichia coli 23S ribosomal RNA in peptide bond formation. RNA, 4, 257–267. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nitta I., Kamada,Y., Noda,H., Ueada,T. and Watanabe,K. (1999) Peptide bond formation: retraction. Science, 283, 2019–2020. [DOI] [PubMed] [Google Scholar]
- Noller H.F., Hoffarth,V. and Zimniak,L. (1992) Unusual resistance of peptidyltransferase to protein extraction procedures. Science, 256, 1416–1419. [DOI] [PubMed] [Google Scholar]
- Nowotny P., Nowotny,V., Voss,H. and Nierhaus,K. (1988) Preparation and activity measurements of deuterated 50S subunits for neutron-scattering analysis. Methods Enzymol., 164, 131–147. [DOI] [PubMed] [Google Scholar]
- Pan C. and Mason,T.L. (1997) Functional analysis of ribosomal protein L2 in yeast mitochondria. J. Biol. Chem., 272, 8165–8171. [DOI] [PubMed] [Google Scholar]
- Pestka S. (1969) Studies on the formation of transfer ribonucleic acid–ribosome complexes. X. Phenylalanyl-oligonucleotide binding to ribosomes and the mechanism of chloramphenicol action. Biochem. Biophys. Res. Commun., 36, 589–595. [DOI] [PubMed] [Google Scholar]
- Planta R. and Mager,W. (1998) The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast, 14, 471–477. [DOI] [PubMed] [Google Scholar]
- Remme J., Margus,T., Villems,R. and Nierhaus,K.H. (1989) The third ribosomal tRNA-binding site, the E site, is occupied in native polysomes. Eur. J. Biochem., 183, 281–284. [DOI] [PubMed] [Google Scholar]
- Romero D.P., Arredondo,J.A. and Traut,R.R. (1990) Identification of a region of Escherichia coli ribosomal protein L2 required for the assembly of L16 into the 50S ribosomal subunit. J. Biol. Chem., 265, 18185–18191. [PubMed] [Google Scholar]
- Rychlik I. and Cerna,J. (1980) Peptidyl transferase—involvement of histidine in substrate binding and peptide bond formation. Biochem. Int., 1, 193–200. [Google Scholar]
- Schilling-Bartetzko S., Bartetzko,A. and Nierhaus,K.H. (1992) Kinetic and thermodynamic parameters for transfer RNA binding to the ribosome and for the translocation reaction. J. Biol. Chem., 267, 4703–4712. [PubMed] [Google Scholar]
- Schulze H. and Nierhaus,K.H. (1982) Minimal set of ribosomal components for the reconstitution of the peptidyltransferase activity. EMBO J., 1, 609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Wilchek,M. and Zamir,A. (1973) Mapping of Escherichia coli ribosomal components involved in peptidyl tranferase activity. Proc. Natl Acad. Sci. USA, 70, 1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M.T., Remme,J., Schäfer,M.A. and Nierhaus,K.H. (1996a) Mutational analysis of two highly conserved UGG sequences of 23S rRNA from Escherichia coli. J. Biol. Chem., 271, 32849–32856. [DOI] [PubMed] [Google Scholar]
- Spahn C.M.T., Schäfer,M.A., Krayevsky,A.A. and Nierhaus,K.H. (1996b) Conserved nucleotides of 23S rRNA located at the ribosomal peptidyltransferase center. J. Biol. Chem., 271, 32857–32862. [DOI] [PubMed] [Google Scholar]
- Ühlein M., Weglöhner,W., Urlaub,H. and Wittmann-Liebold,B. (1998) Functional implications of ribosomal protein L2 in protein biosynthesis as shown by in vivo replacement studies. Biochem. J., 331, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich B., Mertens,G. and Nierhaus,K.H. (1978) Cooperative binding of 3′ fragments of tRNA to the peptidyltransferase centre of E.coli ribosomes. Arch. Biochem. Biophys., 190, 149–154. [DOI] [PubMed] [Google Scholar]
- Vladimirov S.N., Druzina,Z., Wang,R. and Cooperman,B.S. (2000) Identification of 50S components neighboring 23S rRNA nucleotides A2448 and U2604 within the peptidyl transferase center of Escherichia coli ribosomes. Biochemistry, 39, 183–193. [DOI] [PubMed] [Google Scholar]
- Wan K.K., Zahid,N.D. and Baxter,R.M. (1975) The photochemical inactivation of peptidyl transferase activity. Eur. J. Biochem., 58, 397–402. [DOI] [PubMed] [Google Scholar]
- Wower J., Sylvers,L.A., Rosen,K.V., Hixon,S.S. and Zimmermann,R.A. (1993) A model of the tRNA binding sites on the E.coli ribosome. In Nierhaus,K.H., Franceschi,F., Subramanian,A.D., Erdmann,V.A. and Wittmann-Liebold,B. (eds), The Translational Apparatus. Plenum Press, New York, NY, pp. 455–464. [Google Scholar]
- Zhang B. and Cech,T.R. (1998) Peptidyl-transferase ribozymes: trans reactions, structural characterization and ribosomal RNA-like features. Chem. Biol., 5, 539–553. [DOI] [PubMed] [Google Scholar]