Abstract
Many tissues in higher animals undergo dynamic homeostatic growth, wherein damaged or aged cells are replaced by the progeny of resident stem cells. To maintain homeostasis, stem cells must respond to tissue needs. Here we show that in response to damage or stress in the intestinal (midgut) epithelium of adult Drosophila, multiple EGFR ligands and rhomboids (intramembrane proteases that activate some EGFR ligands) are induced, leading to the activation of EGFR signaling in intestinal stem cells (ISCs). Activation of EGFR signaling promotes ISC division and midgut epithelium regeneration, thus maintaining tissue homeostasis. ISCs defective in EGFR signaling cannot grow or divide, are poorly maintained, and cannot support midgut epithelium regeneration following enteric infection by the bacterium, Pseudomonas entomophila. Furthermore, ISC proliferation induced by Jak/Stat signaling is dependent upon EGFR signaling. Thus the EGFR/Ras/MAPK signaling pathway plays central, essential roles in ISC maintenance and the feedback system that mediates intestinal homeostasis.
INTRODUCTION
Homeostasis and regeneration in adult tissue has long fascinated biologists and clinicians alike. The discovery of resident somatic stem cells identified the source of the remarkable regenerating ability in some of adult human tissues, such as blood, skin, hair and the digestive tract (Fuchs, 2009). However, how stem cells respond to tissue needs remains poorly understood (Pellettieri and Sanchez Alvarado, 2007). In particular, how stem cells are activated (for growth, proliferation and differentiation) to regenerate new tissues following tissue injury, stress, or normal wear and tear is still unclear in most cases.
Homeostasis in the human small intestine and colon is mediated by intestinal stem cells (ISCs) that reside in the crypts of Lieberkühn (Barker et al., 2007; Radtke and Clevers, 2005). ISCs proliferate and differentiate to give rise to new functional epithelial cells in order to replenish cell loss from the villi. This dynamic process is intimately linked to the development of colorectal carcinoma (CRC), the second leading cause of cancer mortality in the western world (Radtke and Clevers, 2005). Oncological studies have established a genetic model for CRC development involving multiple steps: mutations in the Adenomatous polyposis coli (Apc) gene result in the activation of WNT signaling, which promotes the formation of small adenomas in the form of polyps. Subsequent mutations in KRAS, BRAF, p53, MLH1 or TGF-β signaling promote the formation of carcinomas, and finally additional mutations drive tumor metastasis (Vogelstein et al., 1988; Walther et al., 2009). Activation of receptor tyrosine kinases, particularly the epidermal growth factor receptor (EGFR), is believed to be an early event in the development of colon adenomas. Ectopic activation of EGFR signaling can cause intestinal and colonic hyperplasia, a likely precursor to ademona formation (Calcagno et al., 2008; Sandgren et al., 1990). Consistently, genetic studies have shown that ectopic activation of the EGFR pathway can accelerate tumor progression in the ApcMin/+ genetic background (Bilger et al., 2008; Haigis et al., 2008; Phelps et al., 2009). Activating mutations in KRAS (codon 12, 13 or 61, which permanently lock it into the GTP-bound state) and BRAF (BRAFV600E) are among the most common mutations found in colon cancer samples (Andreyev et al., 1998; Fransen et al., 2004; Roth et al., 2010). Furthermore, partial loss of function of EGFR (Egfrwa2) severely impaired adenoma formation in ApcMin/+ mice (Roberts et al., 2002). Monoclonal antibodies against EGFR (panitumumab or cetuximab) are effective in treating CRC, provided that activating mutations in downstream KRAS or BRAF are not present, further emphasizing the critical role for EGFR signaling during CRC development (Amado et al., 2008; Di Nicolantonio et al., 2008). Developmentally, neonatal mice lacking EGFR function develop disorganized crypts in the gastrointestinal (Threadgill et al., 1995). Despite these many indications of its importance, the precise functions of EGFR signaling in normal gut homeostasis in mammals are poorly understood, making studies in model systems like Drosophila potentially informative.
As in the human intestine, the Drosophila adult midgut epithelium also undergoes rapid turnover, a dynamic process mediated by thousands of intestinal stem cells (ISC) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). In the fly midgut epithelium, basally localized intestinal stem cells (ISC) divide, renew themselves and give rise to progenitors called enteroblasts (EB). In contrast to transit amplifying cells in mammalian intestinal crypts, Drosophila EBs appear not to proliferate, but directly differentiate into two conserved cell types, the absorptive enterocytes (EC) and the secretory enteroendocrine cells (EE). Genetic studies show that the Drosophila Notch and WNT pathways play conserved roles in the self-renewal and proliferation of ISCs (Bardin et al., 2010; Lee et al., 2009; Lin et al., 2008; Ohlstein and Spradling, 2007). Using this simple model, we and others previously demonstrated a feedback regulatory mechanism for maintaining adult tissue homeostasis. In this case, cell loss, damage, or stress in the midgut epithelium triggers the expression of Unpaired (Upd) cytokines by differentiated enterocytes, and these signals activate Jak/Stat signaling in intestinal stem cells (ISCs) to promote their proliferation and differentiation (Amcheslavsky et al., 2009; Apidianakis et al., 2009; Biteau et al., 2008; Buchon et al., 2009a; Cronin et al., 2009; Jiang et al., 2009). This feedback provides a truly homeostatic mechanism for tissue maintenance in the Drosophila midgut, and may explain in general how stem cells respond to tissue needs in other organs and organisms.
In the present study we demonstrate that, in response to gut epithelial damage or stress in Drosophila, multiple EGFR ligands and several rhomboids are induced, and these activate the EGFR/RAS/MAPK pathway in ISCs. In parallel with Upd/Jak/Stat signaling, the activation of EGFR signaling promotes the proliferation of ISCs and their subsequent differentiation into mature midgut enterocytes (ECs), thus promoting gut self-renewal.
RESULTS
Damage or infection of the midgut induces EGFR signaling
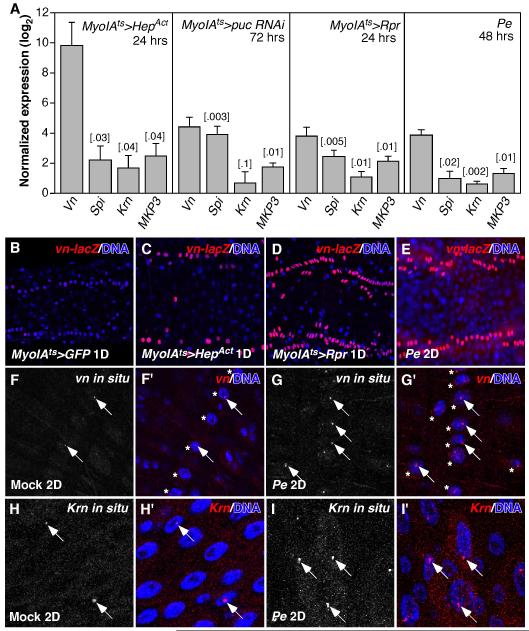
To test whether EGFR signaling is induced in the regenerating Drosophila adult midgut, we assayed the expression of EGFR ligands in whole midguts using RT-qPCR. We induced midgut epithelium regeneration by expressing the cell death gene reaper (Rpr), or activated JNKK (Drosophila HepAct), or RNAi against puckered (puc; a feedback inhibitor of JNK signaling) in the enterocytes (ECs) using the EC-specific inducible Gal4 driver, MyoIAts. Alternatively, we fed flies a pathogenic bacteria, Pseudomonas entomophila (Pe). As we showed previously, EC apoptosis, JNK activation and enteric Pe infection all induce compensatory ISC proliferation and midgut epithelial regeneration (Jiang et al., 2009). We found that three Drosophila EGFR ligands, vein (vn), spitz (spi) and Keren (Krn), were induced in these regenerating midguts (Figure 1A). Regenerating midguts also induced the expression of MAP Kinase Phosphatase 3 (MKP3), a downstream target of Drosophila EGFR signaling (Figure 1A). We examined the expression pattern of vn using the vn-lacZ reporter. Weak expression was observed exclusively in the visceral muscle cells (VM) of control midguts, similar to its expression in the larval midgut (Figure 1B) (Jiang and Edgar, 2009). vn-lacZ expression was highly induced in the VM of the regenerating midgut (Figure 1C-E). The induction of vn expression in response to Pe infection was further confirmed by vn fluorescent in situ hybridization (Figure 1F, G). The strongest signals were found in the nuclei of circular and longitudinal visceral muscle cells, appearing as intense foci, likely the loci of vn transcription (Figure 1F, G). Similarly, the activation of apoptosis and JNK signaling in the ECs also induced vn expression in the VM (data not shown). However, in the case of ectopic JNK activation (MyoIAts>HepAct), strong vn induction was also observed in the ECs (Figure S1A, B), where strong signals were found in the cytosol. Induction of vn in the ECs by HepAct is consistent with the much higher vn induction in these midguts detected by RT-qPCR (Figure 1A). Fluorescent in situ hybridization further revealed that Krn was induced in the ECs in response to Pe infection (Figure 1H, I). The strongest signal appeared as intense foci in EC nuclei. In contrast, a reporter for spi (spi-Gal4NP0261) was mainly expressed in small progenitor cells, with low levels of expression also observed in some ECs (Figure S1C, C’).
Figure 1. Drosophila EGFR ligands are induced in the regenerating adult midgut.
A. RT-qPCR quantification of Drosophila EGFR ligands (vn, spi and Krn) and MKP3 (MAP kinase phosphatase-3) mRNA expression in the regenerating midgut. The midgut was induced to regenerate by activating the JNK pathway in the ECs (MyoIAts>HepAct, 24hrs or puc RNAi, 72hrs), or inducing EC apoptosis (MyoIAts>Rpr, 24hrs), or Pe infection (48hrs). STDEV and P-value (t-test) were shown. B-E. Expression of vn-lacZ reporter in control (B) or regenerating posterior midguts (C-E). 2 of the 4 rows of circular visceral muscle cells (VM) were shown. F, G. vn fluorescent in situ hybridization. The strongest vn signals were in the nucleus (arrows) of VMs (asterisks), most likely the loci of Vn transcription. H, I. Krn fluorescent in situ hybridization. The strongest Krn signals were in the nucleus of ECs (arrows). In mock-infected control midguts, vn and Krn were expressed at low levels in the VM and ECs respectively (F, H).
Drosophila rhomboids encode intramembrane proteases that cleave and activate some EGFR ligands, including Spi and Krn (Urban et al., 2002). We quantified the expression of all 7 rhomboid-like genes in the midgut by RT-qPCR and observed modest upregulation of rho, rho2, 4 and 6 in regenerating midguts (Figure S2A). We also examined the expression of rho using the rhoX81-lacZ reporter. rho-lacZ was weakly expressed in the VM (data not shown), but not in the epithelial cells of controls (Figure S2B). Although rho-lacZ expression in the VM did not change following infection (data not shown), its expression was induced in the ECs (Figure S2C-E). The induction of rho in the ECs in response to Pe infection was confirmed by in situ hybridization (Figure S2F, G).
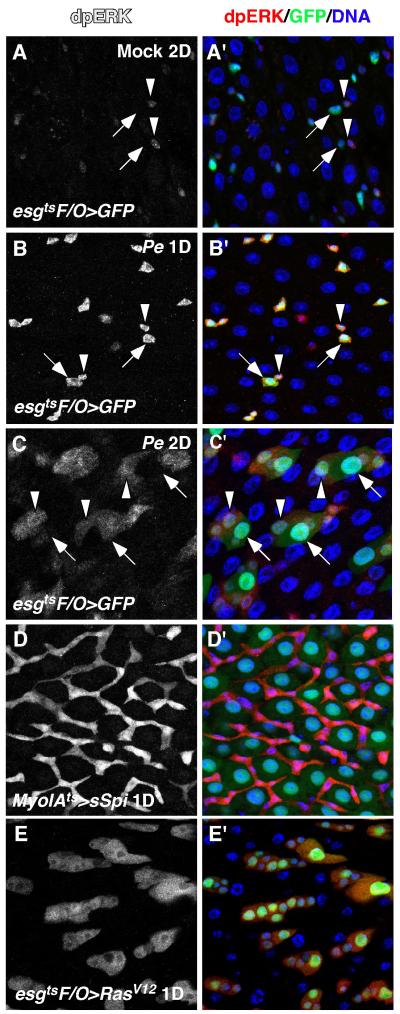
The induction of multiple EGFR ligands and rhos in the midgut was also detected when flies were infected with another pathogenic bacteria, ECC15 (Buchon et al., 2009b). We reasoned that the induction of these factors likely activates EGFR signaling. To test this, we examined the activity of mitogen activated protein kinase (MAPK), a downstream effector of EGFR, using antibodies against the di-phosphorylated, active form of MAPK, termed dpERK (Gabay et al., 1997). Staining for dpERK in control midguts revealed that MAPK was mainly active in ISCs, but was weak or absent in the EBs (Figures 2A, and S3A-A”). Brief Pe infection (1 Day) led to increased dpERK in both ISCs and EBs (Figure 2B, B’), suggesting that Pe infection induced the activation of MAPK in midgut progenitor cells. Interestingly, MAPK activity in the progenitor cells decreased after 2 days of Pe infection, and ectopic MAPK activity was observed in newly formed pre-ECs (Figure 2C, C’). This downregulation in progenitors is likely the result of increased expression of MKP3, a negative regulator of MAPK (Rintelen et al., 2003)(Figure 1A). Consistent with the activation of MAPK in midgut progenitors, ectopic induction of strong EGFR ligands (MyoIAts>sSpi) activated MAPK only in the progenitor cells, but not in the mature ECs (Figure 2D, D’). However, activated Ras (esgtsF/O>RasV12) led to strong cell-autonomous activation of MAPK in both progenitors and large polyploid ECs (Figure 2E, E’). This suggests that differentiated ECs lack a critical component of the EGFR pathway upstream of Ras, and are therefore unable to respond to EGFR ligands. One possibility is that ECs downregulate EGFR as they differentiate.
Figure 2. MAPK is activated in the regenerating midgut.
The activity of Drosophila MAPK was assayed by anti-dpERK staining. A, A’. MAPK activity in the mock-infected control midgut. B, B’. MAPK activity after infecting with Pe for 1 day. ISCs and EBs were marked by esgGal4-driven GFP expression and indicated by arrowheads and arrows respectively (A, B). C, C’. MAPK activity after infecting with Pe for 2 days. Differentiating ECs (pre-ECs, medium nucleus) and newly formed mature ECs (large nucleus) were indicated by arrowheads and arrows respectively. D, D’. MAPK activation induced by ectopic expression of sSpi (MyoIAts>sSpi). E, E’. Cell autonomous MAPK activation induced by activated Ras (esgtsF/O>RasV12).
EGFR activates ISCs through RAS/RAF/MAPK signaling
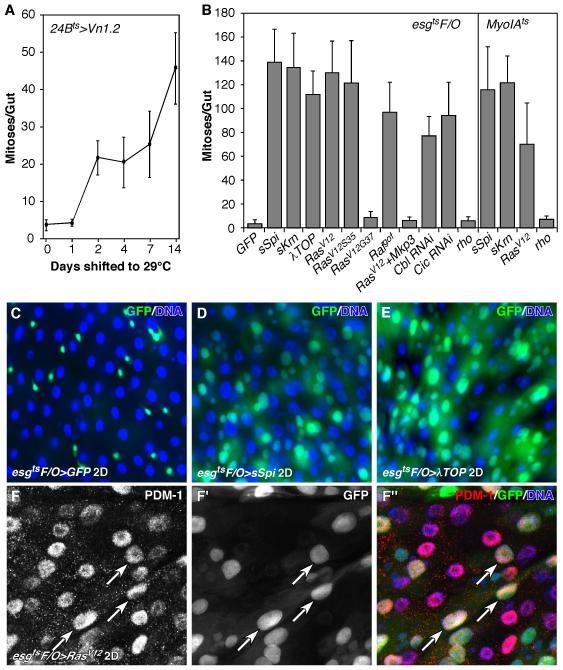
We previously reported that EGFR signaling drives the proliferation of adult midgut progenitors (AMPs) in the larval gut, and showed that VM-derived Vn is required for AMP proliferation during early larval development (Jiang and Edgar, 2009). Using an inducible visceral muscle driver, 24Bts, we over-expressed Vn specifically in adult VM and observed a mild increase of mitotic ISCs (Figure 3A). Thus VM-derived Vn is sufficient to induce ISC proliferation. The mild effect on ISC proliferation is likely because Vn is a weak EGFR ligand (Schnepp et al., 1998). Next, we ectopically activated EGFR signaling in the ISCs by expressing the strong EGFR ligands, sSpi or sKrn (Reich and Shilo, 2002; Schweitzer et al., 1995), activated Egfr (λTOP) (Queenan et al., 1997), or activated Ras (RasV12) (Karim and Rubin, 1998) using a lineage induction system, esgtsF/O. In the esgtsF/O system, progenitor cells and all of their newborn progeny express Gal4 and UAS-linked Gal4 targets, including the UAS-GFP marker (Jiang et al., 2009). We then examined their effects on ISC proliferation. Activation of EGFR signaling induced increased ISC division (Figure 3B), resulting in the generation of many new midgut cells, including EC-like GFP+ cells (Figure 3D-F). Most of these large GFP+ cells were positive for PDM-1, a marker for fully differentiated ECs (Figure 3F-F”). Therefore, EGFR/Ras signaling does not suppress EC differentiation. In addition, we found that knocking down Cbl, a negative regulator of EGFR signaling (Hime et al., 1997; Meisner et al., 1997), by Cbl RNAi (esgtsF/O>Cbl RNAi), also induced ISC proliferation (Figures 3B & S4B). Prolonged activation of EGFR signaling resulted in severely hyperplasic midguts (Figure S8D).
Figure 3. EGFR signaling promotes ISC proliferation and midgut growth.
A. Ectopic ISC proliferation induced by Vn. Vn was induced in the midgut using the inducible VM-specific driver, 24Bts. B. ISC proliferation induced by activated EGFR signaling. Transgenes were induced in the midgut for 2 days using the esgtsF/O or MyoIAts system. Midguts were scored for PH3+ mitotic figures in both A and B. C-E. Adult midgut growth measured using the esgtsF/O system. Both sSpi (D) and λTOP (E) promoted significant new midgut cell formation. F-F”. RasV12 also promoted the formation of new mature midgut cells. Most of the newly formed large polyploid midgut cells (GFP+, arrows) were positive for mature EC marker, PDM-1.
We also induced EGFR ligands in mature ECs (MyoIAts>sSpi or sKrn). This treatment similarly promoted ISC proliferation, demonstrating that paracrine EGF signaling is able to activate ISC division (Figure 3B). In fact, the source of ectopic EGFR ligands did not seem to be important. No matter where Vn, sSpi or sKrn were induced (VMs, ECs or progenitors), they were always capable of inducing dramatic ISC proliferation (data not shown).
To ask which downstream effectors of EGFR are responsible for inducing ISC proliferation, we ectopically expressed pathway-specific Ras variants (RasV12S35 or RasV12G37) in midgut progenitor cells (Karim and Rubin, 1998). RasV12S35, which specifically activates the MAPK pathway, was able to promote ISC proliferation, whereas induction of RasV12G37, which preferentially activates the PI3K/AKT pathway, had no effect on ISC proliferation (Figure 3B). Activated Raf (Rafgof) also promoted ISC proliferation (Figure 3B), and co-expressing MKP3 largely inhibited ectopic ISC proliferation induced by RasV12 (Figure 3B). Furthermore, depleting Capicua (Cic) (esgtsF/O>Cic RNAi), a transcriptional repressor downstream of MAPK pathway (Astigarraga et al., 2007), also induced ISC proliferation (Figures 3B & S4C). We conclude that EGFR signaling induces ISC proliferation specifically through Ras, Raf, and MAPK, rather than via PI3K or another effector pathway.
EGFR signaling is required for ISC proliferation and midgut regeneration
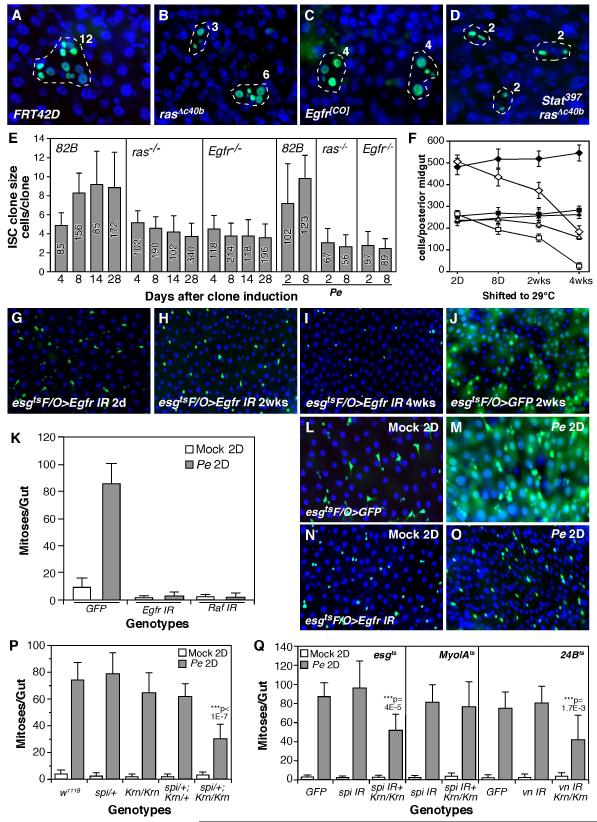
To further explore the role of EGFR signaling in the midgut, we generated mosaic ISC clones homozygous for rasΔc40b, a null allele (Schnorr and Berg, 1996), or Egfr (Egfr null, Egfr[CO]) (Clifford and Schupbach, 1989), or both ras and stat function (ras and Stat92E double null mutants, rasΔc40b, stat397) (Silver and Montell, 2001) using the MARCM system (Lee and Luo, 2001). We then quantified the size of marked ISC clones at intervals following clone induction. Although the initial growth of ras and Egfr mutant ISC clones was normal, their long-term proliferation was severely compromised (Figure 4A-E). For ras and stat double mutant, the clones were not only small, but also lacked ECs (Figure 4D), a phenotype consistent with Jak/Stat’s critical role for ISC differentiation (Beebe et al., 2010; Jiang et al., 2009). Consistent with the EGFR pathway’s essential role in ISC proliferation, midgut renewal following Pe infection was completely inhibited when EGFR signaling was suppressed in the progenitor cells by Egfr RNAi (Figure 4G-J). Furthermore, prolonged EGFR suppression in healthy animals (4 weeks) lead to almost complete loss of enteroblasts (esg+, Su(H)+) and ~33% reduction of intestinal stem cells (esg+, Su(H)−) (Figure 4F, I). In the short term however, EGFR suppression did not significantly alter the number of ISCs, but likely only prevented their growth and division. Interestingly, old ECs generated before the induction of lineage marking were still present in these aged midguts (~1 month, Figure 4I), suggesting that EC loss were also partially inhibited.
Figure 4. Drosophila EGFR signaling is required for midgut homeostasis and regeneration.
A-D. MARCM analysis of ISC clones. Wildtype (A) and mutant ISC clones (B-D) were induced using the MARCM system, and examined 8 days later. The number of cells in each clone were indicated. E. Quantification of ISC clone sizes. The number of clones counted for each genotype were indicated inside each bar. F. Quantification of progenitor cells in the posterior midguts of GFP and EGFR knockdown. Progenitor cells (esg+) were indicated by diamonds, EBs (esg+, Su(H)+) were indicated by squares and presumed ISCs (esg+, Su(H)−) were indicated by triangles. Filled symbols, esgts>GFP; Open symbols, esgts>EGFR RNAi. G-J. Midgut epithelium turnover assay. EGFR suppression inhibited midgut turnover (H, esgtsF/O>Egfr RNAi). Furthermore, GFP+ progenitor cells were depleted after long-term EGFR knockdown (I). In control midgut, GFP were present in both progenitors and large polyploid cells (likely ECs) after 2 weeks (J, esgtsF/O>GFP). K. Quantification of compensatory ISC proliferation induced by Pe infection. EGFR signaling was suppressed in the progenitor cells by esgtsF/O-driven Egfr or Raf RNAi. L-O. Midgut turnover in mock (L, M) or Pe-infected (N, O) animals. Midgut turnover was assayed using the esgtsF/O system. P, Q. Quantification of compensatory ISC proliferation in spi, vn and Krn mutants. We used viable Krn null mutant (Krn27-7-B), lethal spi null mutant (spiA14, in a heterozygous background), spi RNAi knockdown in progenitors (esgts>spi IR) or ECs (MyoIAts>spi IR), or vn RNAi knockdown in VMs (24Bts>vn IR). IR, inverted repeats.
Next we tested whether EGFR signaling is required for compensatory ISC proliferation and midgut epithelium regeneration induced by Pe infection. We first examined the growth of control ISC clones in Pe-infected midgut and observed large ISC clones (~7 cells/clone) 2 days after clone induction (Figure 4E). However, the ISC clones lacking ras or Egfr function were much smaller (~3 cells/clone). Like the long-term ras or Egfr mutant ISC clones in non-infected midguts, these clones did not grow even after the flies had recovered from Pe infection for about a week (Figure 4E). Quantification of midgut mitotic indices revealed that Pe-induced compensatory ISC proliferation was completely inhibited when Egfr or Raf was knocked down (esgtsF/O>Egfr RNAi or Raf RNAi, Figure 4K). Furthermore, while Pe infection almost completely eliminated old ECs and induced midgut epithelial regeneration in controls (Figure 4L, M), suppression of EGFR signaling largely inhibited midgut epithelium regeneration (Figures 4N, O & S5). In both cases, however, large numbers of progenitor cells expressing these RNAi’s survived for the duration of the experiment. In summary, EGFR signaling is required for ISC proliferation during both normal midgut homeostasis and regeneration, such as that induced by Pe infection.
Multiple EGFR ligands function redundantly to activate ISC proliferation
To examine the function of EGFR ligands and rhomboid during Drosophila midgut homeostasis and regeneration, we knocked down spi, vn and rho individually in the midgut using RNAi and several midgut-specific drivers, including esgts, MyoIAts and 24Bts. Inducing spi RNAi in midgut progenitors (esgts>spi RNAi), vn RNAi in visceral muscle cells (24Bts>vn RNAi) or rho RNAi in the ECs (MyoIAts>rho RNAi) all significantly knocked down target gene expression (Figure S6A). In each case, however, these RNAi-depleted midguts appeared to be normal, even after long periods of gene knockdown (data not shown). We then orally infected the flies with Pe and quantified ISC proliferation. Pe infection-induced ISC proliferation also appeared normal in these RNAi-depleted midguts (Figures 4Q, S6B). Finally we examined the regenerative response in the midguts of Krn (krn27-7-B, viable null), rho (rhoA0544, viable partial loss-of-function) and Star (Sd01624, viable partial loss-of-function) mutants (Corl et al., 2009; McDonald et al., 2006). In these cases ISC proliferation induced by Pe infection was also normal (Figures 4P, S6B).
In further tests we quantified Pe-induced ISC proliferation in spi and Krn double mutants. In this case we found that heterozygosity for spi in a Krn homozygous mutant background (spiA14/+; Krn27-7-B/Krn27-7-B) significantly reduced Pe-induced ISC proliferation (Figure 4P). Our previous analysis indicated that this double mutant does not affect the development of the adult midgut progenitor (AMPs) in larvae (Jiang and Edgar, 2009), and quantification of esg+ cells indicated that these midguts had normal numbers of progenitor cells (data not shown). Hence, the suppression of ISC mitotic response suggests that spi and Krn function redundantly during midgut epithelium regeneration. To test which cell types are the source of spi expression, we knocked down spi expression using RNAi, driven either by the esgts driver (progenitor-specific) or the MyoIAts driver (EC-specific) in a Krn mutant background. Knocking down spi in progenitor cells (esgts>spi IR, Krn27-7-B/Krn27-7-B), but not ECs (MyoIAts>spi IR; Krn27-7-B/Krn27-7-B), significantly reduced midgut mitoses induced by Pe ingestion (Figure 4Q). We surmise that autocrine spi (from progenitor cells) and paracrine Krn (from ECs) function redundantly to promote ISC proliferation during midgut epithelium regeneration.
We next tested vein function, using RNAi to deplete vn in the visceral muscle of Krn mutant animals, using the 24Bts driver. Simultaneous loss of Krn and vn (24Bts>vn IR, Krn27-7-B/Krn27-7-B) significantly reduced the ISC proliferation (Figure 4Q), suggesting that vn and Krn also have overlapping function during midgut epithelium regeneration.
EGFR signaling is required for ISC proliferation induced by Jak/Stat signaling
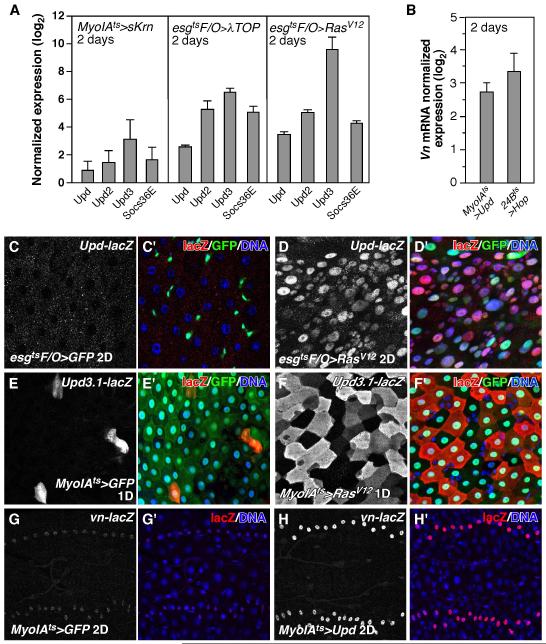
Since both EGFR and Jak/Stat signaling are sufficient and required for midgut epithelium regeneration and both pathways are induced in the regenerating midgut (Figures 1-4) (Buchon et al., 2009a; Cronin et al., 2009; Gabay et al., 1997; Jiang et al., 2009), we examined their epistatic relationship. We first ectopically activated EGFR signaling and examined the expression of the Upd cytokines by RT-qPCR. When activated EGFR ligand (MyoIAts>sKrn), activated Egfr (esgtsF/O>λTOP) or activated Ras (esgtsF/O>RasV12) were expressed in the midgut, all three Upd cytokines were induced, along with downstream target gene, Socs36E (Figure 5A). Consistently, the upd-lacZ reporter was induced in the midgut epithelial cells by RasV12 (Figure 5C, D). Similarly, when we ectopically activated EGFR signaling (MyoIAts>RasV12), the upd3 reporter, upd3.1-lacZ, was induced in the enterocytes (ECs) (Figure 5E, F). Accordingly, RasV12 expression in the ECs was capable of inducing ISC proliferation (Figure 3B). The induction of cytokines and subsequent activation of Jak/Stat signaling likely depends on the levels of EGFR activation since the inductions by sKrn were much lower than that by activated EGFR (λTOP), or RasV12 (Figure 5A). Moreover ectopic expression of Vn (24Bts>Vn), a weak EGFR ligand, did not induce cytokine expression (data not shown), though it did promote mild ISC proliferation (Figure 3A).
Figure 5. Induction of EGFR and Jak/Stat signaling in the midgut.
A. Activating EGFR signaling induced Jak/Stat signaling in the midgut. The expression levels of Drosophila cytokines (upds) and downstream target gene, Socs36E, in the midgut were analyzed by RT-qPCR. B. Induction of vn expression in the midgut by Jak/Stat signaling as quantified by RT-qPCR. Jak/Stat signaling was activated in the VM by ectopic expression of Upd in the ECs (MyoIAts>Upd) or Hop directly in the VM (24Bts>Hop). C, D. Induction of the upd-lacZ reporter in the midgut epithelium by activated Ras (esgtsF/O>RasV12, D). E, F. Induction of the Upd3.1-lacZ reporter in ECs by activated Ras (MyoIAts>RasV12, F). G, H. Induction of the vn-lacZ reporter in the VM by ectopic expression of Upd (MyoIAts>Upd, H).
We next asked what signals might induce Vn expression in the visceral muscle. We observed increased nuclear STAT92E staining in the VM of Pe-infected midguts (Figure S7A, B), suggesting that Jak/Stat signaling was activated in the VM. Consistent with this, expression of the Jak/Stat reporter 10XSTAT-DGFP increased dramatically in the VM after Pe infection (Figure S7C, D). Since the induction of vn coincided with enhanced cytokine signaling in the VM, we speculated that it might be the result of Upds (cytokine) released from the midgut epithelium. In testing this idea, we found that vn and the vn-lacZ reporter could be induced in the VM in response to EC-specific expression of Upd (MyoIAts>Upd) (Figure 5B, G, H). Activating Jak/Stat signaling directly in the VM via the expression of Drosophila Jak (24Bts>Hop) also induced comparable vn expression (Figure 5B). These experiments indicate that midgut epithelium-derived cytokines can activate Jak/Stat signaling and induce vn expression in the VM. However, we found that Pe infection could induce vn upregulation in the midguts of Jak mutants (hop25, partial loss-of-function) or when stat was depleted in the VM (24Bts>Stat RNAi, Figure S7E). These data indicate that, although activated Jak/Stat signaling can induce vn, Jak/Stat signaling is not required for vn induction in response to Pe infection.
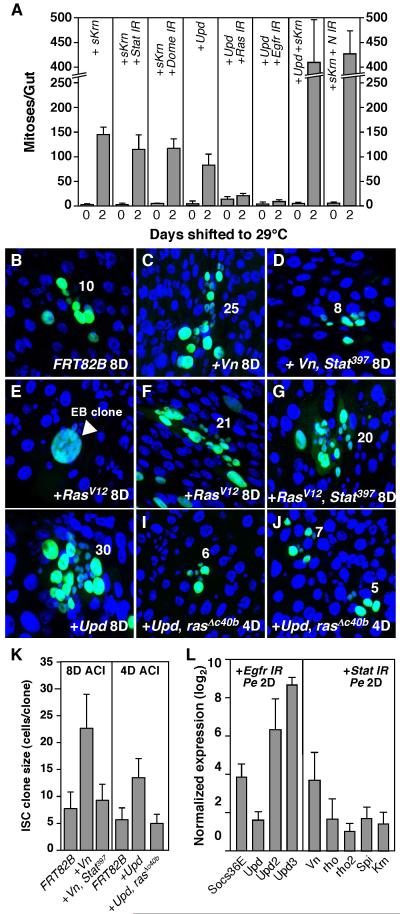
Further epistasis tests showed that when EGFR signaling was activated in the background of reduced Jak/Stat signaling (esgtsF/O>sKrn + Stat or Dome RNAi), its stimulatory effect on ISC proliferation was not diminished (Figures 6A, S8D-F). Similar results were obtained when activated Egfr (λTOP) or Ras (RasV12) was co-expressed with Stat or Dome RNAi (data not shown). Using the MARCM technique, we induced activated Ras in ISCs mutant for Stat (+RasV12, stat397) and analyzed their clonal growth. Loss of Jak/Stat signaling did not affect RasV12’s ability to drive the growth of large ISC clones (Figure 6F, G). However, in a similar experiment, clonal growth induced by the weak EGFR ligand, Vn, was largely inhibited by loss of Stat (Figure 6C, D and K). These data suggest that the requirement of Jak/Stat signaling for ISC proliferation likely depends on the levels of EGFR activation, such that high level EGFR activation is able to induce ISC proliferation independent of Jak/Stat signaling, whereas ISC proliferation induced by low level EGFR activation (such as that induced by Vn) is largely dependent on Jak/Stat signaling.
Figure 6. Jak/Stat-induced ISC proliferation requires EGFR signaling.
A. ISC proliferation induced by EGFR and Jak/Stat signaling. With the exception of co-expressing sKrn and Upd in the ECs (MyoIAts>Upd + sKrn), all the other ectopic expression experiments were performed using the esgtsF/O driver. Midgut mitotic indices (PH3+) were quantified after activating the transgenes for 2 days. B-J. ISC clonal assay. GFP-marked ISC clones were induced using the MARCM system and analyzed 4 or 8 days later. The sizes of the ISC clones were indicated. Vn-induced ISC proliferation is dependent on Jak/Stat signaling (B-D). Activated Ras (RasV12)-induced ISC proliferation is independent of Jak/Stat signaling (F-G). Some EB clones overexpressing RasV12 underwent extra round of endoreplication (E). Upd-induced ISC proliferation is dependent on EGFR signaling (H-J). K. Quantification of ISC clone sizes. The sizes of ISC clones were measured 4 or 8 days after clone induction (ACI) using the MARCM system. L. RT-qPCR analysis of the induction of Jak/Stat and EGFR signalings by Pe infection in the absence of either pathway (esgtsF/O>Stat or Egfr RNAi).
In further experiments we found that ISC proliferation induced by ectopic Upd was completely inhibited when EGFR signaling was downregulated in the ISCs (Figure 6A). Knocking down Egfr or Ras completely inhibited the midgut hyperplasia phenotype that results from ectopic Upd expression (esgtsF/O>Upd + Egfr or Ras RNAi, Figure S8G-I). Similar results were obtained in a clonal setting, using the rasΔc40b mutant allele (Figure 6I-K). Thus EGFR signaling is required for ISC proliferation induced by Jak/Stat signaling. However, activating Jak/Stat and EGFR signaling simultaneously induced a much higher ISC mitotic index than that induced by the activation of either pathway alone (MyoIAts>Upd + sSpi, Figure 6A), indicating that the two pathways can function synergistically to induce ISC proliferation. Like the Jak/Stat signaling (Beebe et al., 2010), EGFR signaling can also induce much higher rate of ISC proliferation when Notch signaling is inhibited (esgtsF/O>sKrn + N IR, Figure 6A). Since Notch suppression increases stem cell pools, this suggests that both pathways primarily regulate ISC division, rather than ISC numbers.
Finally, we examined whether the induction of Upd/Jak/Stat and EGFR signaling by Pe infection depended on each other. We inhibited Pe-induced midgut epithelium regeneration by knocking down Egfr (esgtsF/O>Egfr RNAi) or Stat (esgtsF/O>Stat RNAi) and examined the expression of upds and Socs36E or Egfr ligands and rhos by RT-qPCR. The induction of Jak/Stat and EGFR signaling by Pe was normal in both cases (Figure 6L), suggesting that these two signaling pathways can be induced independently of each other by midgut damage (Figure 7).
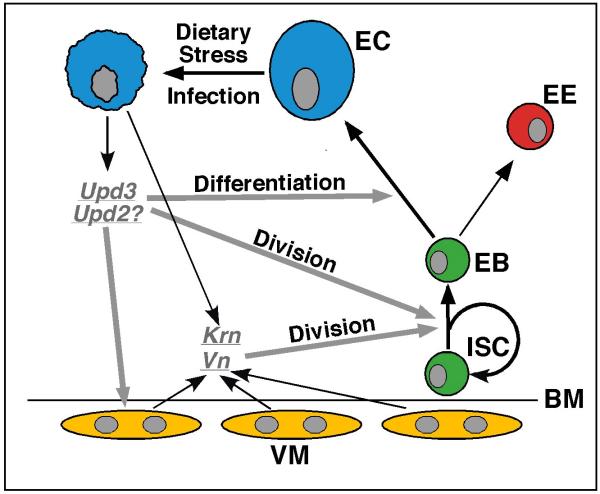
Figure 7. Updated model for midgut homeostasis and regeneration in Drosophila.
Stressed or dying ECs induce the expression of fly cytokines (such as Upd3 and Upd2) and EGFs (such as Krn and Vn) in the midgut, which activate the Jak/Stat and EGFR pathways in the midgut progenitor cells. While EGFR signaling functions mainly to promote ISC proliferation, Jak/Stat signaling functions to promote both ISC proliferation and EB differentiation.
DISCUSSION
EGFR signaling is essential for ISC growth and division
These studies show that Drosophila EGFR pathway provides an essential mitogenic signal for ISC proliferation during midgut homeostasis and regeneration (Figure 4). Furthermore, ISC proliferation induced by Jak/Stat signaling depends on functional EGFR signaling (Figures 6A, H-K & S8G-I). The critical role of EGFR signaling in the fly midgut is consistent with its role during mammalian gut homeostasis and colorectal cancer development. EGFR signaling is required for the development, maintenance and tumorigenesis of mucosal epithelium in the mouse GI tract (Roberts et al., 2002; Threadgill et al., 1995; Troyer et al., 2001). Antibodies targeting EGFR have been shown to be effective in treating colorectal cancer provided there are no activating mutations in downstream signaling components, such as KRAS or BRAF (Amado et al., 2008; Di Nicolantonio et al., 2008).
Our data also demonstrate that EGFR signaling is induced in response to damage in the Drosophila midgut, and functions to promote ISC proliferation during midgut epithelium regeneration (Figures 1-3). In this capacity it is a central and essential component of the feedback mechanism for adult tissue homeostasis that we described previously (Figure 7) (Jiang et al., 2009). Like EGFR ligands in Drosophila, two mammalian EGFR ligands, epiregulin and amphiregulin, have been reported to be upregulated in the gut epithelium following damage (Lee et al., 2004; Nishimura et al., 2008). Their expression is also increased in neoplastic lesions in the colon, suggesting a possible role in colon cancer development (Nishimura et al., 2008).
One of our more unexpected findings was that, whereas differentiating immature cells (preECs) were often positive for MAPK activity, fully differentiated midgut cells such as ECs were not (Figure 2C, C’). A potential explanation for this is that mature ECs lose EGFR or downstream effector and thereby become un-responsive to EGFR ligands. This is consistent with our data showing that MAPK could be activated only in progenitor cells (ICS and EBs) even when activated EGFR ligands (such as sSpi) were ectopically expressed at high levels (Figure 2D, D’). A similar mechanism may confine the activity of Jak/Stat signaling to the midgut progenitor cells (Beebe et al., 2010; Buchon et al., 2009a; Jiang et al., 2009). In this case Domeless, the receptor for the Upd cytokines, is expressed in the midgut progenitor cells but not in their progeny (Jiang et al., 2009). Switching off receptor expression for cytokines or growth factors may be one way to ensure that mature differentiated cells do not respond to these mitogenic cues. Despite this failsafe mechanism, the expression of RasV12 was able to induce the cell autonomous activation of MAPK (Figure 2E, E’) and the expression of Upd3 in the ECs (Figure 5E, F), leading to a non-cell autonomous stimulation of ISC proliferation (Figure 3B). This suggests that the downregulation of mitogen receptors upon differentiation may be important to throttle EGFR←→Jak/Stat positive feedback that might otherwise result in run-away signaling and ISC proliferation.
As with the Upd cytokines, we know little about how the Drosophila EGFR ligands are induced by stress or damage to the midgut epithelium. In the case of the Upds, potential activating stress signals span a very wide range, including induced apoptosis, autophagic cell death, JNK signaling, infection by pathogenic bacteria and colonization by non-pathogenic enteric bacteria, ingestion of detergents, oxidative stress inducers, DNA damaging agents, and even physical “pinching” of the epithelium (Amcheslavsky et al., 2009; Apidianakis et al., 2009; Biteau et al., 2008; Buchon et al., 2009a; Cronin et al., 2009; Jiang et al., 2009). The signals capable of activating the EGFR ligands are likely to be just as diverse. Further genetic studies in the fly should be able to determine whether these stress responses are cell autonomous or a property of the epithelium as a tissue, and to identify the genes and pathways involved. Given the critical roles of the mammalian Jak/Stat and EGFR pathways in regulating tissue homeostasis and cancer development, such studies should have some clinical relevance.
Is visceral muscle a niche for ISCs?
Expression of wingless (wg, a Drosophila Wnt) from the visceral muscle (VM) has been reported to regulate ISC proliferation and self-renewal, leading to the proposal that visceral muscle serves as a niche for ISCs (Lin et al., 2008). However, while Drosophila Wnt signaling appears to be required for ISC survival (Lin et al., 2008), its role in promoting ISC self-renewal was not confirmed in another independent study (Lee et al., 2009). In addition, ISC proliferation induced by ectopic Wnt signaling is much weaker than that induced by Jak/Stat or EGFR signaling (Jiang et al., 2009; Lee et al., 2009; Lin et al., 2008). Thus, although the role of VM-derived Wg in midgut homeostasis and regeneration has not been rigorously tested, the data suggest that other signaling systems play more critical roles.
Pertinent to the function of the visceral muscle, we discovered that the EGFR ligand vn was induced in the VM during gut regeneration (Figure 1), and that VM-derived Vn was capable of inducing ectopic ISC proliferation (Figure 3A). This suggested that the VM might serve as a part of the ISC niche by providing a mitogenic signal. However, Pe-induced compensatory ISC proliferation was not affected when we specifically downregulated vn in the VM (Figure 4Q), suggesting that VM-derived Vn is probably not by itself an essential EGFR ligand during midgut epithelium regeneration. In fact, we also observed the induction of two other EGFR ligands (spi and Krn) in midgut epithelial cells during regeneration (Figure 1). Although the concurrent expression of multiple EGFR ligands complicated our efforts to identify the exact role of each ligand, single and double mutant analysis suggested that all three ligands have overlapping function in activating EGFR signaling (Figure 4P, Q). Importantly, a significant fraction of the mitogenic EGFR signals probably come from the epithelium itself. Similarly, the Upd cytokines are induced primarily in midgut epithelial cells (Buchon et al., 2009a; Jiang et al., 2009). Moreover, the self-renewal and differentiation of Drosophila intestinal stem cells are regulated by Notch signaling, which occurs between the two daughter cells produced following ISC division, and is not known to directly involve the VM (Bardin et al., 2010; Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006, 2007).
Hence we propose that the most important component of the niche for fly intestinal stem cells may be the midgut epithelium itself. In this context it is interesting to note that an epithelial niche has also been proposed for mouse intestinal stem cells (Sato et al., 2009). The murine Lgr5+ ISCs reside at the bottom of the crypts, juxtaposed directly with Paneth cells (Barker et al., 2007). In vitro culture of individual Lgr5+ ISCs has demonstrated that they can form self-organizing organoids in the absence of mesenchymal cells. Lgr5+ ISCs are normally always in contact with Paneth cells, which have been proposed to be a niche for ISCs (Sato et al., 2009). Interestingly, EGF is one of the factors required in the media to support the growth of intestinal organoids (Sato et al., 2009). However, it is not yet clear which cells are the endogenous source for EGFR ligands in the mouse intestine or colon, nor which specific ligands are expressed or functionally important. It is tempting to speculate that Paneth cells, as a critical niche component, might be one of the sources of mitogenic signals, such as EGFs and cytokines, for mammalian intestinal stem cells.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Celeste Berg, Denise Montell, Gyeong-Hun Baeg, Erika Bach, Jocelyn McDonald, Matthew Freeman, and the VDRC (Austria), NIG (Japan), Bloomington (USA) Drosophila Stock Centers for fly stocks; the Moen’s lab for confocal imaging; Xiaohang Yang for anti-Pdm-1 antibody; David D. O’Keefe for advice on anti-dpERK staining and members of the Edgar lab for comments. This work was supported by NIH grant R01 GM51186 to B.A.E.
Appendix
MATERIALS AND METHODS
Fly genetics
See supplemental methods for fly stocks used in this study.
Upd3-lacZ reporters
To generate upd3-lacZ reporters, 4 genomic PCR fragments (upd3.1-4, see primer sequences in the supplemental materials) covering the original ~4kb upd3 promoter region (Agaisse et al., 2003) were digested with BamH I/Kpn I and cloned into the same restriction sites of pH-Pelican vector. Transgenic lines were established through standard P-element mediated transformation.
RNA in situ hybridization in the adult midgut
RNA fluorescent in situ hybridization (FISH) in the midgut was performed as described (Raj et al., 2008) with a few modifications. Briefly, 40-48 20-mer DNA oligos complementing the coding region of the target genes (vn, krn, and rho) were designed using the online software (http://www.singlemoleculefish.com/designer.html). The oligos were synthesized with 3′ amine modification (Biosearch Technologies), then manually pooled and coupled with Alexa-568, carboxylic acid, succinimidyl ester (Invitrogen A-20003). The labeled oligos were purified using HPLC (Reverse phase C-18 column) and vacuum dried and resuspended in 100 μl H2O. For RNA in situ hybridization, the midguts were first dissected and fixed in 8% Paraformalhyde overnight at 4°C, then washed with PBS and Triton X-100 (0.1%) for 3 times (15 minutes each). The samples were further permeablized in 70% ethanol over night at 4°C. The probes were used at dilution 1:2,000-10,000. The hybridization was then performed following the online protocol (http://www.singlemoleculefish.com/protocols.html).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
Supplemental Data include eight figures, supplemental materials and methods.
REFERENCES
- Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Developmental cell. 2003;5:441–450. doi: 10.1016/s1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell stem cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. Journal of the National Cancer Institute. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- Apidianakis Y, Pitsouli C, Perrimon N, Rahme L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proceedings of the National Academy of Sciences of the United States of America. 2009 doi: 10.1073/pnas.0911797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astigarraga S, Grossman R, Diaz-Delfin J, Caelles C, Paroush Z, Jimenez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. The EMBO journal. 2007;26:668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Perdigoto CN, Southall TD, Brand AH, Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development (Cambridge, England) 2010;137:705–714. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Beebe K, Lee WC, Micchelli CA. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Developmental biology. 2010;338:28–37. doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Bilger A, Sullivan R, Prunuske AJ, Clipson L, Drinkwater NR, Dove WF. Widespread hyperplasia induced by transgenic TGFalpha in ApcMin mice is associated with only regional effects on tumorigenesis. Carcinogenesis. 2008;29:1825–1830. doi: 10.1093/carcin/bgn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell stem cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes & development. 2009a;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell host & microbe. 2009b;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Calcagno SR, Li S, Colon M, Kreinest PA, Thompson EA, Fields AP, Murray NR. Oncogenic K-ras promotes early carcinogenesis in the mouse proximal colon. International journal of cancer. 2008;122:2462–2470. doi: 10.1002/ijc.23383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford RJ, Schupbach T. Coordinately and differentially mutable activities of torpedo, the Drosophila melanogaster homolog of the vertebrate EGF receptor gene. Genetics. 1989;123:771–787. doi: 10.1093/genetics/123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, Leibbrandt A, Simoes Rde M, Gruber S, Puc U, et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science (New York, NY. 2009;325:340–343. doi: 10.1126/science.1173164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- Fransen K, Klintenas M, Osterstrom A, Dimberg J, Monstein HJ, Soderkvist P. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527–533. doi: 10.1093/carcin/bgh049. [DOI] [PubMed] [Google Scholar]
- Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development (Cambridge, England) 1997;124:3535–3541. doi: 10.1242/dev.124.18.3535. [DOI] [PubMed] [Google Scholar]
- Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nature genetics. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime GR, Dhungat MP, Ng A, Bowtell DD. D-Cbl, the Drosophila homologue of the c-Cbl proto-oncogene, interacts with the Drosophila EGF receptor in vivo, despite lacking C-terminal adaptor binding sites. Oncogene. 1997;14:2709–2719. doi: 10.1038/sj.onc.1201223. [DOI] [PubMed] [Google Scholar]
- Jiang H, Edgar BA. EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development (Cambridge, England) 2009;136:483–493. doi: 10.1242/dev.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development (Cambridge, England) 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- Lee D, Pearsall RS, Das S, Dey SK, Godfrey VL, Threadgill DW. Epiregulin is not essential for development of intestinal tumors but is required for protection from intestinal damage. Molecular and cellular biology. 2004;24:8907–8916. doi: 10.1128/MCB.24.20.8907-8916.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in neurosciences. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lee WC, Beebe K, Sudmeier L, Micchelli CA. Adenomatous polyposis coli regulates Drosophila intestinal stem cell proliferation. Development (Cambridge, England) 2009;136:2255–2264. doi: 10.1242/dev.035196. [DOI] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Pinheiro EM, Kadlec L, Schupbach T, Montell DJ. Multiple EGFR ligands participate in guiding migrating border cells. Developmental biology. 2006;296:94–103. doi: 10.1016/j.ydbio.2006.04.438. [DOI] [PubMed] [Google Scholar]
- Meisner H, Daga A, Buxton J, Fernandez B, Chawla A, Banerjee U, Czech MP. Interactions of Drosophila Cbl with epidermal growth factor receptors and role of Cbl in R7 photoreceptor cell development. Molecular and cellular biology. 1997;17:2217–2225. doi: 10.1128/mcb.17.4.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Andoh A, Inatomi O, Shioya M, Yagi Y, Tsujikawa T, Fujiyama Y. Amphiregulin and epiregulin expression in neoplastic and inflammatory lesions in the colon. Oncology reports. 2008;19:105–110. [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science (New York, NY. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Sanchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annual review of genetics. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K, Broadbent T, Sarkar S, Burt RW, Jones DA. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schupbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development (Cambridge, England) 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science (New York, NY. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich A, Shilo BZ. Keren, a new ligand of the Drosophila epidermal growth factor receptor, undergoes two modes of cleavage. The EMBO journal. 2002;21:4287–4296. doi: 10.1093/emboj/cdf439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintelen F, Hafen E, Nairz K. The Drosophila dual-specificity ERK phosphatase DMKP3 cooperates with the ERK tyrosine phosphatase PTP-ER. Development (Cambridge, England) 2003;130:3479–3490. doi: 10.1242/dev.00568. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Min L, Washington MK, Olsen SJ, Settle SH, Coffey RJ, Threadgill DW. Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1521–1526. doi: 10.1073/pnas.032678499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- Sandgren EP, Luetteke NC, Palmiter RD, Brinster RL, Lee DC. Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schnepp B, Donaldson T, Grumbling G, Ostrowski S, Schweitzer R, Shilo BZ, Simcox A. EGF domain swap converts a drosophila EGF receptor activator into an inhibitor. Genes & development. 1998;12:908–913. doi: 10.1101/gad.12.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr JD, Berg CA. Differential activity of Ras1 during patterning of the Drosophila dorsoventral axis. Genetics. 1996;144:1545–1557. doi: 10.1093/genetics/144.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes & development. 1995;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science (New York, NY. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- Troyer KL, Luetteke NC, Saxon ML, Qiu TH, Xian CJ, Lee DC. Growth retardation, duodenal lesions, and aberrant ileum architecture in triple null mice lacking EGF, amphiregulin, and TGF-alpha. Gastroenterology. 2001;121:68–78. doi: 10.1053/gast.2001.25478. [DOI] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M. A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. The EMBO journal. 2002;21:4277–4286. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. The New England journal of medicine. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nature reviews. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.