Summary
Activation of the eukaryotic replicative DNA helicase, the Mcm2-7 complex, requires phosphorylation by Cdc7/Dbf4 (Dbf4-dependent kinase or DDK), which, in turn, depends on prior phosphorylation of Mcm2-7 by an unknown kinase(s). We identified DDK phosphorylation sites on Mcm4 and Mcm6 and found that phosphorylation of either subunit suffices for cell proliferation. Importantly, prior phosphorylation of either S/T-P or S/T-Q motifs on these subunits is required for DDK phosphorylation of Mcm2-7 and for normal S phase passage. Phosphomimetic mutations of DDK target sites bypass both DDK function and mutation of the priming phosphorylation sites. Mrc1 facilitates Mec1 phosphorylation of the S/T-Q motifs of chromatin-bound Mcm2-7 during S phase to activate replication. Genetic interactions between priming site mutations and MRC1 or TOF1 deletion support a role for these modifications in replication fork stability. These findings identify new mechanisms to modulate origin firing and replication fork assembly during cell cycle progression.
Introduction
The replication process in eukaryotic cells temporally separates helicase loading and helicase activation to ensure that no origin of replication can initiate more than once per cell cycle. Helicase loading (also referred to as pre-replicative complex [pre-RC] formation or origin licensing) is restricted to the G1 phase of the cell cycle. This process requires the origin recognition complex (ORC), Cdc6 and Cdt1 to assemble a dimer of the eukaryotic helicase, the Mcm2-7 complex, around dsDNA (Remus et al., 2009). As cells enter S phase, the resulting head-to-head Mcm2-7 dimer is activated by initiation factor binding to the Mcm2-7 complex. Most notably, Cdc45 and the GINS complex activate Mcm2-7 ATPase and helicase activities (Ilves et al., 2010).
Two kinases trigger eukaryotic helicase activation: the Cdc7 kinase and S phase cyclin-dependent kinases (S-CDKs). Cdc7 directly phosphorylates the Mcm2-7 complex and is activated by the cell-cycle-regulated accessory subunit Dbf4 or the related Drf1 (Sclafani and Holzen, 2007). Thus, activated Cdc7 is known as Dbf4/Drf1-dependent kinase, or DDK. DDK phosphorylation of Mcm2-7 is required for the association of the helicase activators Cdc45 and GINS (Labib, 2010). The role of S-CDKs is best understood in S. cerevisiae cells, where they phosphorylate Sld2 and Sld3 leading to the recruitment of Sld2 and Dpb11 to the origin (Tanaka et al., 2007; Zegerman and Diffley, 2007). These three proteins also stimulate Cdc45 and GINS origin association (Labib, 2010), suggesting that S-CDKs and DDK function in parallel to trigger helicase activation.
Two additional kinases, Mec1 and Rad53, also coordinate the events of DNA replication in S. cerevisiae cells. The essential function of both kinases is to trigger the activation of ribonucleotide reductase (RNR) and the induction of deoxyribonucleotide synthesis as cells enter S phase (Zhao et al., 2001). This function can be bypassed by artificially increasing dNTP levels by a variety of means including the deletion of the RNR inhibitor SML1 (Zegerman and Diffley, 2009). The activation of Mec1 and Rad53 S phase entry is poorly understood but may involve the detection of ongoing DNA synthesis. Mec1 and Rad53 also mediate the checkpoint response to DNA damage and stalled DNA replication forks (Zegerman and Diffley, 2009). In each of these functions, Mec1 acts at the top of a signal transduction pathway that requires the Mrc1 or Rad9 proteins to activate Rad53 kinase.
Consistent with its role in activating loaded Mcm2-7 complexes, DDK associates with replication origins (Dowell et al., 1994) and preferentially binds and phosphorylates loaded Mcm2-7 complexes (Francis et al., 2009; Masai et al., 2006; Sheu and Stillman, 2006). Early in vitro studies using purified Mcm2-7 identified Mcm2 as a primary DDK target, whereas in vivo phosphorylation studies suggested that Mcm4 was the primary DDK target (Francis et al., 2009; Masai et al., 2000; Masai et al., 2006; Sheu and Stillman, 2006). DDK phosphorylation of Mcm4 and Mcm6 is stimulated in the context of the pre-RC (Francis et al., 2009), explaining the distinct in vivo and in vitro phosphorylation profiles. Intriguingly, DDK can not bind Mcm2-7 or phosphorylate Mcm4 or Mcm6 without prior Mcm2-7 phosphorylation by an unidentified kinase (Francis et al., 2009).
Two classes of DDK target sequences have been identified previously. “Intrinsic” sites (S/T-D/E) are characterized by an acidic residue at the +1 position (Charych et al., 2008; Cho et al., 2006; Montagnoli et al., 2006). In a second class of sites, a negative charge at the +1 position is provided by a phosphoserine or phosphothreonine. Thus, the recognition of these “phosphorylation-generated” (PG) sites by DDK requires prior phosphorylation by a second kinase. For example, DDK can phosphorylate the initial serine in an S-S-P motif when the second serine is previously phosphorylated by CDK, as with DDK phosphorylation of S. cerevisiae Mer2 (Sasanuma et al., 2008; Wan et al., 2008). Phosphorylation of similar sites by DDK on human Mcm2 and Mcm4 has been observed (Masai et al., 2006; Montagnoli et al., 2006), however, the biological relevance of these modifications remains unknown. S-CDK is unlikely to act as an essential priming kinase for DDK targeting of Mcm2-7 because Sld2 and Sld3 are the only essential S-CDK targets for DNA replication in budding yeast (Tanaka et al., 2007; Zegerman and Diffley, 2007). Despite our knowledge of potential DDK target sites, the essential Mcm2-7 sites targeted by DDK and how these modifications are stimulated by prior phosphorylation remain unknown.
In this study, we identify DDK phosphorylation sites on Mcm4 and Mcm6 as well as “priming” phosphorylation sites required for DDK phosphorylation of the same subunits. Phosphomimetic mutations of the identified DDK target sites bypass both DDK function and mutation of the priming sites. The majority of the priming sites are either S/T-P or S/T-Q motifs preceded by one or more S/T residues. Mutational analysis demonstrated overlapping function between the S/T-P and S/T-Q sites as well as between Mcm4 and Mcm6 phosphorylation. Mec1 phosphorylates the S/T-Q sites on chromatin-bound Mcm2-7 during S phase, whereas the S/T-P sites appear to be constitutively phosphorylated. Synthetic lethality between priming site mutations and MRC1 deletion suggests that incomplete priming site phosphorylation results in defective replication fork assembly. Our findings demonstrate that phosphorylation of Mcm4 and Mcm6 by Mec1 and a proline-directed kinase activates these proteins for phosphorylation by DDK and proper replication fork assembly.
Results
Identification of phosphorylation sites on Mcm2-7
We used mass spectrometry to map phosphorylation sites on the Mcm2-7 complex. Our previous studies indicated that DDK phosphorylation of Mcm2-7 is strongly influenced by pre-RC formation and by prior phosphorylation of Mcm2-7 by one or more kinases (Francis et al., 2009). For this reason, we used in vitro assembled pre-RCs as a DDK substrate to identify phosphorylated residues. Because Mcm2-7 is phosphorylated during an in vitro helicase loading reaction lacking DDK, we identified residues that were phosphorylated during pre-RC assembly with and without DDK treatment.
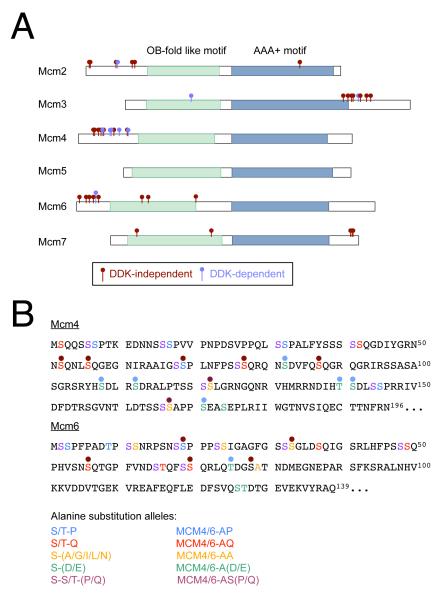
The majority of phosphorylation sites localized to four clusters outside the conserved OB-fold-like and AAA+ motifs of the Mcm subunits: the amino-termini of Mcm2, 4, and 6 and the carboxyl-terminus of Mcm3 (Fig. 1A; only unambiguously assigned phosphates are shown). The cluster of phosphosites in the Mcm3 C-terminus was mostly DDK-independent (10 of 12 sites) and corresponds to a CDK-regulated nuclear transport signal (Liku et al., 2005). The four DDK-independent sites found in the N terminus of Mcm2 are casein kinase 2 (CK2) consensus sites (S/T-D/E-X-D/E, Pinna, 2002) and depleting CK2 from extracts prior to pre-RC assembly eliminated Mcm2 phosphorylation (data not shown). In contrast to a recent study (Bruck and Kaplan, 2009), we found that these sites are not critical for the essential functions of Mcm2 (Fig. S1). Because Mcm4 is a critical DDK target in vivo (Sheu and Stillman, 2006, 2010) and because Mcm4 and Mcm6 are the two phosphorylation- and pre-RC-dependent targets of budding yeast DDK (Francis et al., 2009), we focused our analysis on the phosphosites in the Mcm4 and Mcm6 amino-termini.
Figure 1. Identification of priming and target sites for DDK on Mcm2-7.
(A) Cartoon showing locations of phosphorylation sites on Mcm2-7 either before (red) or after (blue) phosphorylation by recombinant DDK. Pre-RCs were assembled on origin DNA-bound magnetic beads using whole cell extract. Origin-bound pre-RC proteins were separated by SDS/PAGE and the bands representing Mcm2-7 were analyzed for phosphorylation using LC/MS/MS.
(B) Phosphorylation sites on the Mcm4 and Mcm6 N-terminal tails. The colored residues correspond to sites mutated in this study, and are coded according to the residue at the +1 position. Blue, Pro; Red, Gln; Yellow, A/G/I/L/N; Green, Asp/Glu; Purple, Ser/Thr. Each illustration shows only unambiguously assigned sites of phosphorylation. See also Fig. S1.
We found 13 phosphorylation sites in the Mcm4 N-terminus and 6 sites in the Mcm6 N-terminus (Fig. 1B and Table S1). Seven of the Mcm4 and five of the Mcm6 phosphorylation sites were DDK-independent (Fig. 1B, red dots, and Table S1). We categorized the DDK-independent sites by the identity of the +1 residue: two S/T-P sites, six S/T-Q sites, and four sites characterized by an aliphatic residue (A, G, or L) at the +1 position. Intriguingly, many of these phosphoserines (7 of 12) were preceded by one or more serines or threonines, suggesting that these sites could be PG DDK target sites.
Six Mcm4 sites and one Mcm6 site were only observed in the DDK-treated reaction (Fig. 1B, blue dots, and Table S1). Each of these phosphosites is followed by an acidic residue at the +1 position as expected for intrinsic DDK sites. We also found evidence that DDK targeted PG sites in Mcm4 and Mcm6. In these cases, DDK targeted serines preceding a phospho-SP or phospho-SQ motif, although in all cases the abundance of serines in the analyzed peptides precluded unambiguous assignment of the phosphorylated residues. For example, mass spectrometry identified the tri-phosphorylated Mcm4 peptide AAIGSSPLNFPSSSQR (residues 64-79; Table S1). Because the SP and SQ sites in this peptide are both phosphorylated in a DDK-independent manner (Fig. 1B; Table S1; and see below), DDK must phosphorylate at least one of the serines at the −1 or −2 position relative to the DDK-independent sites. We conclude that DDK phosphorylates both intrinsic and PG sites in Mcm4 and Mcm6.
DDK phosphorylation of Mcm4 and Mcm6 has overlapping functions
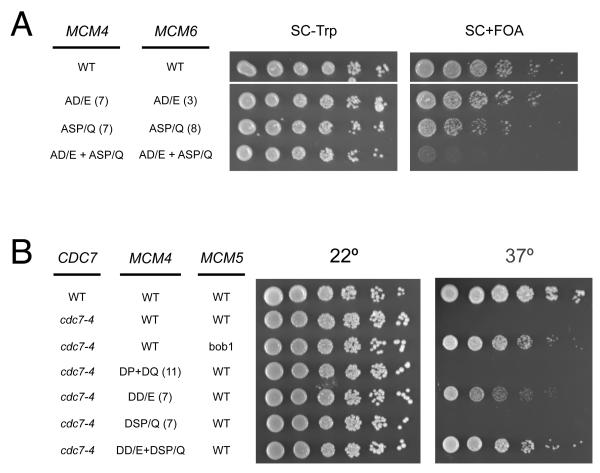
To test their in vivo relevance, we eliminated the potential DDK phosphorylation sites in Mcm4 and Mcm6. We hypothesized that there are two classes of DDK target sites: S/T-D/E sites (intrinsic sites) and the first serine in S-S-P/Q sites (PG sites). We created MCM4 and MCM6 mutations that eliminated each class of sites either individually or together. Mutation of intrinsic or PG sites alone or together in Mcm4 or Mcm6 resulted in no significant proliferation defect (Fig. S1). Similarly, no proliferation defect was observed in a strain when the intrinsic sites were eliminated (A-D/E) in Mcm4 and Mcm6 simultaneously (Fig. 2A). Elimination of the PG sites (A-S-P/Q) from both Mcm4 and Mcm6 resulted in a small proliferation defect. We observed a severe proliferation defect, however, when the intrinsic and PG sites in both MCM4 and MCM6 were mutated. This observation suggests that DDK phosphorylation of MCM4 and MCM6 can compensate for one another. Furthermore, phosphorylation of either the intrinsic or the PG sites is sufficient for cell proliferation.
Figure 2. Phenotype of putative DDK target site mutants.
(A) Effect of alanine substitutions at putative DDK target sites. Strains yJCWR185, yJCWR176, yJCWR180, and yJCWR184 (see Table S2) carrying the indicated alleles of MCM4 and MCM6 in addition to a URA+ ARS/CEN MCM4+ plasmid were grown on ura+ media for 3-4 generations to allow loss of the URA+ MCM4+ plasmid. A five-fold serial dilution of cells was then spotted on plates without (left) or with (right) FOA selection against the URA+ MCM4+ plasmid. The numbers in parentheses adjacent to the allele name indicate the number of sites mutated in each allele (see also Fig. S1).
(B) CDC7 bypass by phosphomimetic MCM4 alleles. The indicated MCM4 alleles were introduced into strain OAy711 (cdc7-4) by direct replacement at the chromosomal MCM4 locus. Cells were spotted on YPD at 22° and 37°.
To confirm that the intrinsic and PG sites are targeted by DDK in vivo, we mutated these residues in MCM4 to the phosphomimetic amino acid aspartate and asked whether these mutations could bypass the temperature-sensitive proliferation defect of a cdc7-4 allele. We compared the proliferation of this strain to that of a previously-isolated CDC7 bypass allele, MCM5-bob1 (Hardy et al., 1997). Mutating the seven intrinsic (S/T-D/E) sites in MCM4 to D-D/E allowed proliferation of a cdc7-4 strain at the nonpermissive temperature (Fig. 2B). Phosphomimetic mutations in the 11 SP and SQ sites in MCM4 did not allow growth at elevated temperature, demonstrating that the DDK bypass conferred by the D-D/E mutations was not due to a general increase in the negative charge of the Mcm4 N-terminus. Phosphomimetic mutations in the PG class of MCM4 DDK target sites (S-S-P/Q to D-S-P/Q) did not bypass cdc7-4 function. Instead, combining the phosphomimetic mutations in the intrinsic and PG sites enhanced CDC7 bypass relative to the D-D/E mutations alone (Fig. 2B). Similar mutations in MCM6 did not bypass the cdc7-4 proliferation defect independently but did enhance proliferation in the MCM4 phosphomimetic background (data not shown). We conclude that both the S/T-D/E and the S-S-P/Q sites in Mcm4 and Mcm6 are targeted by DDK in vivo.
S/T-P and S/T-Q sites are priming sites for DDK phosphorylation
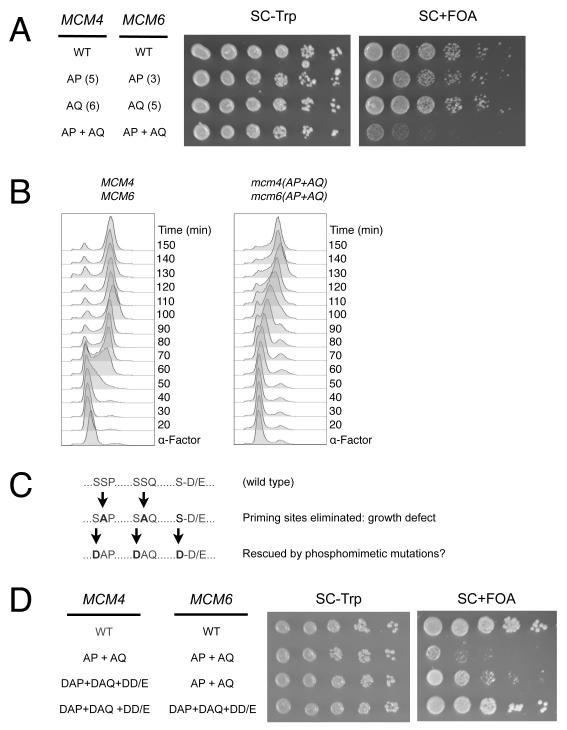
Having established the importance of the predicted DDK sites in vivo, we examined the importance of the proposed S/T-P and S/T-Q “priming” sites. We mutated each class of site either alone or in combination and assayed the effects of these mutations on cell viability. As with the DDK sites, mutating one or both classes of site in MCM4 or MCM6 alone had no effect on cell proliferation (Fig. S1). Similarly, mutating all of the MCM4 or MCM6 S/T-P sites to AP sites, or all of the MCM4 and MCM6 S/T-Q sites to AQ sites, revealed no phenotype (Fig. 3A). Consistent with the compensatory functions of Mcm4 and Mcm6 DDK phosphorylation, a severe proliferation defect was observed only when the AP and AQ mutations were simultaneously introduced into both Mcm4 and Mcm6 [mcm4(AP+AQ) mcm6(AP+AQ); Fig. 3A].
Figure 3. Phenotype of putative priming site mutants.
(A) Effect of alanine substitutions at putative priming sites. mcm4/mcm6 double-mutant strains yJCWR185, yJCWR167, yJCWR171, and yJCWR175 were analyzed as in Fig. 2A. The numbers in parentheses adjacent to the allele name indicate the number of sites mutated in each allele (see also Fig. S1).
(B) S-phase progression defect of mcm4(AP+AQ) mcm6(AP+AQ) strain. DNA content was measured by FACS analysis after release from alpha factor at 25°.
(C) Schematic of bypass strategy used in (D).
(D) Rescue of priming site mutations by MCM4 and/or MCM6 phosphomimetic mutations at DDK target sites. Analysis of strains yJCWR185, yJCWR175, yJCWR236, and yJCWR237 was performed as in Fig. 2A.
The priming site deficient strain [mcm4(AP+AQ) mcm6(AP+AQ)] was defective for passage through S phase (Fig. 3B). By monitoring DNA content of synchronous cell populations we found that relative to wild-type cells, the priming site deficient strain showed a 10 to 20 minute delay in the onset of DNA replication and a doubling of S phase length (60 vs 30 minutes). Thus, eliminating the S/T-P and S/T-Q priming phosphorylation sites altered both the onset of bulk DNA replication and the time required to complete DNA synthesis.
If phosphorylation of the S/T-Q and S/T-P sites facilitates DDK targeting, then mutations that bypass DDK activity should also bypass the defects of the AP and AQ mutations. Indeed, the proliferation defect of the priming site deficient strain was partially rescued by incorporating phosphomimetic mutations in the predicted DDK sites of MCM4 (D-A-P/Q and D-D/E mutations, Figs. 3C and 3D). This rescue was strongly enhanced when DDK phosphomimetic mutations were also incorporated into MCM6. These studies strongly suggest that the primary role of the S/T-P and S/T-Q sites is to activate Mcm2-7 for targeting by DDK during replication initiation.
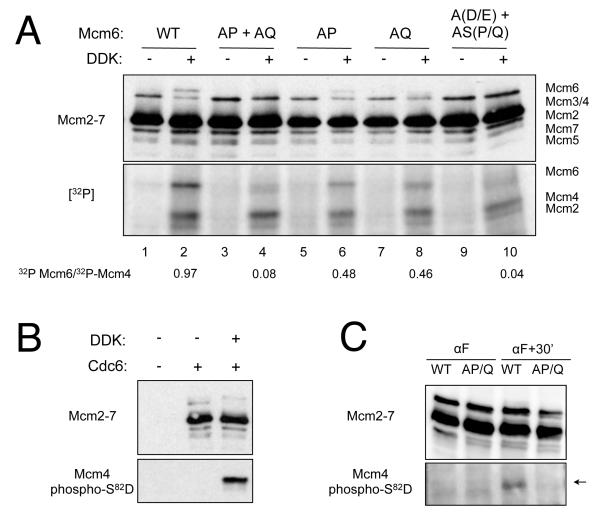
To test the hypothesis that phosphorylation of the S/T-P or S/T-Q sites influences DDK targeting, we assembled pre-RCs in extracts derived from strains in which the Mcm6 priming sites were mutated. The resulting pre-RCs were treated with DDK in the presence of radiolabeled ATP. We also performed this experiment using a mcm6 mutant lacking the putative DDK targets to test if these mutations eliminated DDK phosphorylation. None of the mutations affected Mcm2-7 loading (Fig. 4A).
Figure 4. S/T-P and S/T-Q sites reduces DDK targeting of Mcm2-7.
(A) Elimination of S/T-P and S/T-Q sites in Mcm6 inhibits DDK phosphorylation. Pre-RCs were assembled using G1-arrested extracts from strains yJCWR128, yRH163, yJCWR129, yJCWR130, and yJCWR135 carrying the indicated sets of alanine substitutions in MCM6. All strains are wild-type for MCM4. The resulting pre-RCs were then phosphorylated by DDK using radiolabelled ATP and the labeled proteins were analyzed by immunoblotting (top panel) or autoradiography (bottom panel).
(B) Mcm4-S82D83 is targeted by DDK. Pre-RCs assembly reactions were performed using G1-arrested extracts with or without Cdc6 and DDK. The resulting DNA associated proteins were analyzed by immunoblotting with anti-Mcm2-7 (UM-174, top) or phosph-S82-D83 antibodies. See also Fig. S2.
(C) S/T-P and S/T-Q sites are required for DDK targeting of Mcm4 in vivo. MCM4 MCM6 or mcm4(AP+AQ) mcm6(AP+AQ) cultures (yJCWR205 or yJCWR208) were arrested in G1 phase with alpha factor. Cells were washed and released into medium lacking alpha factor for 30 minutes at 30°. Total cell protein was collected by TCA precipitation and analyzed by immunoblotting using anti-Mcm2-7 (top) and a phosphospecific antibody targeting Mcm4-S82-D83 (bottom).
DDK treatment of the loaded, mutant Mcm2-7 complexes demonstrated that the S/T-P and S/T-Q sites control DDK phosphorylation of both classes of DDK target sites. As expected, simultaneous mutation of the DDK target residues in the intrinsic and PG (S/T-S-P/Q) sites resulted in a ~20-fold reduction in Mcm6 phosphorylation (Fig. 4A, compare lanes 2 and 10). Importantly, elimination of both S/T-P and S/T-Q sites on Mcm6 resulted a similar reduction in DDK phosphorylation of Mcm6 (Fig. 4A, compare lanes 2, 4 and 10). In both cases, the Mcm6 mutations did not affect Mcm4 phosphorylation, indicating that these mutations acted in cis. Interestingly, pre-RCs containing either Mcm6-AP or Mcm6-AQ mutations reduced DDK phosphorylation equivalently (~50%, lanes 6 and 8). Thus, in contrast to their apparently redundant function in the proliferation studies, direct phosphorylation assays showed that S/T-P and S/T-Q sites both contribute to full DDK phosphorylation of Mcm6.
We addressed the role of priming phosphorylation in DDK targeting in vivo using a phosphospecific antibody that recognized the phosphorylation of Mcm4-S82-D83, an intrinsic DDK target site. Studies using in vitro assembled pre-RCs showed that recognition of this site by the phosphospecific antibody was dependent on treatment of the pre-RCs with DDK (Fig. 4B). In vivo, phosphorylation of this intrinsic DDK site was detected in wild-type S phase cells but was eliminated in the priming deficient strain (Fig. 4C). Thus, as observed in the in vitro studies, phosphorylation of the S/T-P and S/T-Q sites was critical for DDK targeting in vivo, even for an intrinsic DDK target site.
Cell cycle regulation of Mcm2-7 phosphorylation
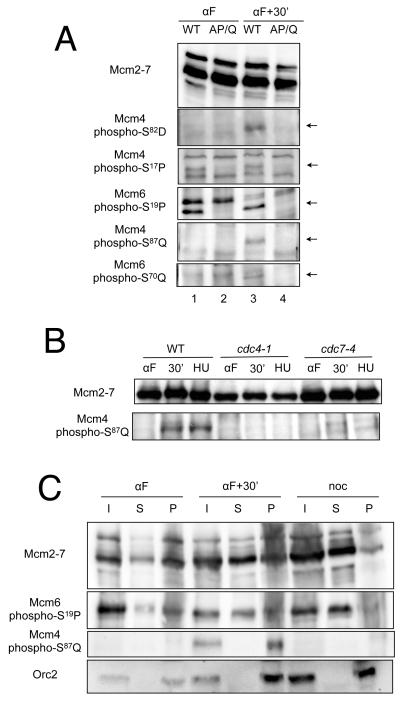
We next addressed the cell cycle regulation of Mcm2-7 phosphorylation. To this end we used both the Mcm4-phosphoS82-D83 antibody as well as additional phospho-specific antibodies that recognized specific SP and SQ sites in Mcm4 and Mcm6. Analysis of the phosphoSP and phosphoSQ antibodies showed that their recognition of Mcm2-7 was inhibited by phosphatase treatment (Fig. S2) and by mutations that eliminated their target sites (Fig. 5A). As expected for a DDK target, phosphorylation of Mcm4-S82-D83 was only seen in the S phase sample (Fig. 5A, lane 3). The S/T-P and S/T-Q sites had distinct patterns of phosphorylation during the cell cycle. Two SP sites (Mcm4-S17-P18 and Mcm6-S19-P20) were phosphorylated in G1, S and M phase cells (Fig. 5A, lanes 1 and 3 and Fig. 5C). In contrast, both SQ sites analyzed (Mcm4-S87-Q88 and Mcm6-S70-Q71) were phosphorylated in S phase cells but not in G1 or M phase cells. To further define the time of SQ phosphorylation, we arrested cells at the G1/S transition using cdc7-4 or cdc4-1 temperature-sensitive alleles. For both mutants, Mcm4-S87-Q88 was not phosphorylated in cells arrested at the nonpermissive temperature (Fig. 5B). Thus, unlike the phosphorylation of SP sites, SQ phosphorylation on Mcm4 and Mcm6 occurs after entry into S phase.
Figure 5. Mcm2-7 SQ sites are phosphorylated on chromatin in S phase.
(A) S/T-P and S/T-Q phosphorylation in G1 and S phase. Extracts of MCM4 MCM6 or mcm4(AP+AQ) mcm6(AP+AQ) cells were prepared as described in 4C and analyzed by immunoblotting using anti-Mcm2-7 and the indicated phosphospecific antisera. Arrows indicate the phosphospecific bands.
(B) SQ phosphorylation in cells arrested at the G1/S transition. Wild-type, cdc4-1 or cdc7-4 cells were grown to mid-log phase at 25°, arrested in G1 using α-factor, then released at 37°. TCA-precipitated protein was analyzed by immunoblotting using anti-Mcm2-7 or anti-Mcm4-phospho-S87-Q88 sera.
(C) S/T-Q phosphorylation is specific for chromatin-associated Mcm2-7. W303BLa cells were arrested in α-factor (αF) or nocodazole (noc) with or without 30 min release at 30°. Total cell protein was separated by a chromatin fractionation procedure into input (I), supernatant (S), and pelleted chromatin (P) fractions and analyzed by immunoblotting with sera detecting Mcm2-7, Mcm6-phospho-S19-P20, Mcm4-phospho-S87-Q88, or Orc2.
To ask if phosphorylation of S/T-P or S/T-Q sites on Mcm2-7 is dependent on helicase loading, we fractionated chromatin-bound and -unbound proteins from G1, S, and G2/M-phase cells and detected Mcm4 and Mcm6 phosphorylation using the phosphospecific antisera (Fig. 5C). Mcm6-S19-P20 was phosphorylated both on and off chromatin and the amount of phosphorylation correlated with total Mcm2-7 levels. In contrast, phosphorylation of Mcm4-S87-Q88 in S phase was exclusive to chromatin associated Mcm2-7 (Fig. 5C, lanes 4-6).
Mrc1 facilitates Mec1 phosphorylation of Mcm4
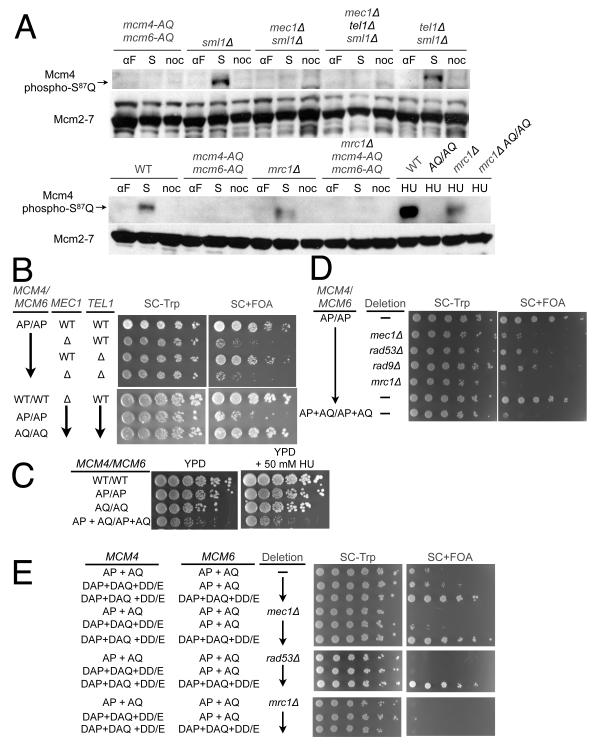
S/T-Q sites are frequently targets of PI3 kinases, which in S. cerevisiae include the Mec1 and Tel1 kinases. To assess whether Mec1 or Tel1 is responsible for the phosphorylation of S/T-Q sites on Mcm4 in vivo, we deleted these kinases and assessed phosphorylation of Mcm4-S87-Q88 at various cell cycle stages. SML1 was deleted in all strains to rescue the lethality of MEC1 deletion (Zhao et al., 2001). S phase phosphorylation of Mcm4-S87-Q88 was eliminated in the mec1Δ and mec1Δ tel1Δ but not the tel1Δ strain (Fig. 6A), strongly suggesting that Mec1 is the principal kinase responsible for phosphorylating this site in vivo. Given their association with the Mcm2-7 complex and known roles in Mec1 function (Mordes et al., 2008; Navadgi-Patil and Burgers, 2008; Naylor et al., 2009), we asked if Mrc1 or Dpb11 was required for Mcm4-SQ phosphorylation. Elimination of Mrc1 reduced but did not eliminate phosphorylation of Mcm4-S87-Q88 during normal S phase passage and upon HU treatment (Fig. 6A). In contrast, a temperature sensitive mutation in Dpb11 had no effect on the same phosphorylation event (Fig. S3).
Figure 6. Mec1 targets Mcm4 in vivo independent of checkpoint activation.
(A) Effect of deletion of MEC1, TEL1 and MRC1 on Mcm4 SQ phosphorylation. Extracts were prepared from cells after arrest in α-factor (αF), hydroxyurea (HU), nocodazol (noc) or 30 minutes after α-factor release (S-phase, S) as indicated and analyzed by immunoblotting using anti-Mcm2-7 and the indicated phosphospecific antisera. Upper panel: yCC29, yRH109, yRH110, yJCWR79, yJCWR78; lower panel: yJCWR200, yJCWR220, yCC98, yCC100.
(B) Deletion of MEC1 reduces viability of mcm4-AP mcm6-AP cells. Strains yJCWR214, yJCWR216, yJCWR217, yJCWR218 yJCWR214, yJCWR216, and yJCWR221 were analyzed as in Fig. 2A. The relevant genotype is indicated to the left of each strain.
(C) Eliminating Mec1 target sites on Mcm4 and Mcm6 does not render cells sensitive to hydroxyurea. Strains yJCWR205, yJCWR206, yJCWR207, and yJCWR208 carrying the indicated WT or mutant alleles of MCM4 and MCM6 as the only copy of those genes were plated on YPD in the absence or presence of 50 mM hydroxyurea (HU), as indicated.
(D) Deletion of checkpoint genes reduces viability of mcm4-AP mcm6-AP cells. Strains yJCWR214, yJCWR216, yJCWR219, yCC48, yCC55, yJCWR167, and yJCWR175 were analyzed as in Fig. 2A. All of these strains include a deletion of SML1. See also Fig. S3.
(E) Phosphomimetic mutations at DDK target sites rescue the deletion of MEC1 andRAD53 but not MRC1 in the mcm4(AP+AQ) mcm6(AP+AQ) strain background. Strains yJCWR175, yJCWR236, yJCWR237, yCC90, yCC94, yCC95, yCC114, yCC116, yCC117, yCC73, yCC75, and yCC77 were analyzed as in Fig. 2A.
If Mec1 is the SQ-directed Mcm4/6 priming kinase, deleting MEC1 should phenocopy the effect of mutating the S/T-Q sites in these subunits. Because the proliferation role of S/T-Q sites in MCM4 and MCM6 is overlapping with that of the S/T-P sites (Fig. 3A), we deleted MEC1 in a mcm4-AP mcm6-AP strain. Deletion of MEC1 in the context of the AP mutations caused the same severe proliferation defect as combining the AP and AQ mutations in MCM4 and MCM6 (compare Fig. 6B to Fig. 3A). Deletion of TEL1 in the same strain background caused no defect either in the presence or absence of MEC1 (Fig. 6B). Similarly, no proliferation defect was observed when MEC1 was deleted in a strain carrying the wild-type or AQ alleles of MCM4 and MCM6 (Fig. 6B). Together these data strongly support the conclusion that Mec1 directly targets the S/T-Q sites in Mcm4 and Mcm6, although it is formally possible that Mec1 and Mrc1 activate a different S/T-Q-directed kinase to target Mcm4 and Mcm6.
Checkpoint proteins facilitate growth in the absence of SP phosphorylation
It is possible that eliminating the S/T-P motifs in Mcm4 and Mcm6 causes replication stress that increases the cell’s reliance on a MEC1-dependent S phase checkpoint. We first tested whether any of the replication defects caused by priming site mutations (Fig. 3) conferred sensitivity to hydroxyurea (HU) treatment. Cells lacking the Mcm4 and Mcm6 S/T-P, S-T/Q or both types of priming sites showed no additional growth defects relative to WT cells in the presence of 50 mM HU (Fig. 6C). Thus, neither S/T-Q nor S/T-P sites are required for the checkpoint response to stalled replication forks. Next, we tested if deletion of RAD53, RAD9, MRC1, or TOF1 also inhibited proliferation of the mcm4-AP mcm6-AP cells. These proteins mediate either the DNA-damage (Rad9, Rad53) or the stalled-replication-fork (Mrc1, Tof1, Rad53) checkpoints (Zegerman and Diffley, 2009). Mrc1 and Tof1 have additional functions in replication fork stability/progression (Hodgson et al., 2007; Szyjka et al., 2005; Tourriere et al., 2005). As with MEC1, deletion of each of these genes reduced proliferation of a mcm4-AP mcm6-AP strain but did not affect strains with WT or AQ alleles of MCM4 and MCM6 (Fig. S3). The extent of these defects varied significantly (Figs. 6D and S3). Deletion of RAD53 and RAD9 showed the smallest proliferation defect. Mec1 showed intermediate defects equivalent to a strain lacking both the SP and SQ phosphosites. The RAD53, RAD9 and MEC1 deletion defects suggest a role of the intra-S phase checkpoint in suppressing the proliferation defects of the mcm4-AP mcm6-AP strain. The more severe defect of the MEC1 deletion suggests that Mec1’s role in Mcm4/6 SQ phosphorylation is distinct from the intra-S phase checkpoint.
Unlike the deletions of other checkpoint proteins, deletion of MRC1 in the mcm4-AP mcm6-AP background resulted in a loss of viability (Fig. 6D, the few small colonies do not support further growth). Because loss of MEC1 has a more dramatic effect on SQ phosphorylation than loss of MRC1 (Fig. 6A), the more severe proliferation defect of MRC1 relative to MEC1 deletion suggests that Mrc1 has an additional function unrelated priming phosphorylation and DDK targeting. To address this possibility, we took advantage of strains that eliminated SP and SQ priming phosphorylation and used phospho-mimetic mutations at DDK target sites to rescue the resulting proliferation defect (Fig. 3D). In these strains, eliminating proteins that mediate priming or DDK phosphorylation or that mediate a checkpoint response to incomplete DDK phosphorylation should have no effect on proliferation. Consistent with this logic, deleting either MEC1 or RAD53 in the DDK phosphomimetic background has no effect on proliferation (Fig. 6E compare row 3 and 6). In contrast, deletion of MRC1 in the same background resulted in a loss of viability. This result strongly suggests that the synthetic lethality of the MRC1 deletion with the mcm4-AP mcm6-AP allele is due to a role other than facilitating DDK phosphorylation of Mcm4 and Mcm6.
Discussion
Studies of Mcm2-7 activation via phosphorylation have been complicated by the existence of multiple kinases that phosphorylate multiple Mcm2-7 subunits (Devault et al., 2008; Lei et al., 1997; Masai et al., 2000; Masai et al., 2006; Montagnoli et al., 2006). Our results provide a comprehensive picture of Mcm2-7 phosphorylation and reveal that a series of kinases collaborate in Mcm2-7 activation. We identified two classes of DDK target sites on Mcm4 and Mcm6, intrinsic and phospho-generated (PG), and showed that both classes of target sites are important for Mcm2-7 function. Importantly, DDK phosphorylation required prior phosphorylation of Mcm4 or Mcm6 at either S/T-P or S/T-Q motifs. We demonstrated that Mec1 targets the S/T-Q priming sites and that these modifications positively regulate DNA replication. Finally, our studies suggest that phosphorylation of S/T-P sites in the N-termini of Mcm4 and Mcm6 is important for proper replication fork function. These findings unravel a key step in the initiation of DNA replication and make a new connection between the critical S phase regulator Mec1 and origin firing. Because DDK acts at each origin as it initiates replication (Labib, 2010), our findings identify mechanisms by which Mec1 and other kinases could modulate origin firing throughout S phase by controlling the extent of DDK phosphorylation of Mcm4 and Mcm6.
Priming phosphorylation stimulates DDK targeting by multiple mechanisms
Our findings indicate that priming phosphorylation stimulates DDK phosphorylation of Mcm4 and Mcm6 in two ways. First, at S-S/T-P and S-S/T-Q sites phosphorylation creates an acidic phosphoserine/threonine residue that targets DDK to an adjacent upstream serine residue. By this mechanism, priming phosphorylation approximately doubles the number of potential DDK target sites in Mcm4 and Mcm6. Many of these phospho-generated DDK target sites are preceded by additional serines or threonines, and it is likely that sequential phosphorylation in the C- to N-terminal direction of these S/T strings is responsible for the reported processive phosphorylation of the Mcm4 N-terminus by DDK (Sheu and Stillman, 2006).
Mutation of the priming kinase target sequences also interferes with DDK modification of intrinsic (S/T-D/E) sites such as Mcm4-Ser82 (Fig. 4A and 5A). Because prior phosphorylation is required for DDK association with loaded Mcm2-7 complexes (Francis et al., 2009), we propose that phosphorylation of S-S-P/Q sites by priming kinases (and possibly DDK) creates DDK binding sites that facilitate subsequent DDK phosphorylation of Mcm2-7. The stronger proliferation defect of the priming deficient strain [mcm4 (AP+AQ) mcm6 (AP+AQ) strain, Fig. 3A] relative to a strain lacking only the PG DDK sites (A-S-P/Q mutations, Fig 2A) also supports a role for the priming sites in S/T-D/E site phosphorylation. Importantly, the severe proliferation defect caused by elimination of the priming sites is bypassed by phosphomimetic mutations of the DDK target sites (Fig. 3D), strongly suggesting that the primary role for the S/T-P and S/T-Q priming sites is to promote DDK targeting to Mcm2-7 during origin firing.
Cell cycle regulation of Mcm2-7 phosphorylation
Our analysis of Mcm2-7 phosphorylation indicates that the two types of priming sites are regulated differently. The Mec1-dependent phosphorylation of S/T-Q sites on Mcm4 and Mcm6 is spatially restricted to chromatin-bound Mcm2-7 and temporally restricted to S phase (Fig. 5). Previous studies have found DDK preferentially targets chromatin-associated Mcm2-7 (Sheu and Stillman, 2006). The spatial and temporal regulation of S/T-Q phosphorylation of Mcm4 and Mcm6 is consistent with this modification mediating this preference. In contrast, Mcm4 and Mcm6 S/T-P sites are phosphorylated in G1, S, and G2/M phases, and this phosphorylation is independent of chromatin loading (Fig. 5).
It is likely that phosphorylation of S/T-P priming sites is mediated by S-CDKs. A role for S-CDKs in modifying Mcm subunits has been proposed previously (Devault et al., 2008; Masai et al., 2000; Montagnoli et al., 2006; Sheu and Stillman, 2010). If so, our studies provide a clear mechanism for S-CDKs to stimulate DDK targeting of Mcm2-7 (Nougarede et al., 2000). The apparently overlapping function between the S/T-P and S/T-Q sites would explain why Mcm4/6 S/T-P modification is not an essential CDK target (Tanaka et al., 2007; Zegerman and Diffley, 2007). Both our studies and a previous study of S/T-P phosphorylation of human Mcm2 found that these modifications were not cell cycle regulated (Montagnoli et al., 2006). Although it is possible that S-CDKs are solely responsible for the S/T-P phosphorylation and these phosphates are not readily removed outside of S phase, the constitutive nature of these modifications suggests that additional kinases phosphorylate these sites. The S/T-P sites on Mcm4 and Mcm6 also could be targeted by G1- and M-phase CDKs, although G1-CDK activity should be inhibited in α-factor arrested cells. Alternatively, another proline-directed kinase (e.g. Pho85, Huang et al., 2007) could also act as an Mcm2-7 priming kinase.
S-S/T-Q sites are found in the N-termini of Mcm subunits in other yeasts, including S. pombe, but this motif is not present in the N-termini of metazoan Mcm2-7. This suggests that the Mec1/Tel1 homologs ATR/ATM are not involved in Mcm2-7 priming in metazoan cells. Nonetheless, there are an abundance of Ser-Ser-X motifs (including but not limited to S-S/T-P motifs) in metazoan Mcm2-7 amino termini. Such sites would allow CDKs and other kinases to interface with replication by modulating DDK activation of Mcm2-7 in these organisms. Such alternative priming sites may also function in S. cerevisiae as elimination of all S/T-P and S/T-Q sites does not completely eliminate replication (Fig. 3).
Roles for Rad53, Rad9, Mec1 and Mrc1 in suppressing defects in priming phosphorylation
The genetic interactions between mutations that eliminate Mcm4 and Mcm6 S/T-P priming phosphorylation and the deletion of the checkpoint genes RAD53, RAD9, MEC1, and MRC1 suggest that these genes function at three levels to suppress defects caused by the loss of Mcm4/6 S/T-P phosphorylation. First, all of these genes can function in the intra-S phase checkpoint to suppress these replication defects. Second, Mrc1-facilitated Mec1 phosphorylation of Mcm4/6 S/T-Q sites provides a compensatory mechanism to target DDK in the absence of Mcm4/6 S/T-P phosphorylation. Third, in the presence of incomplete priming and DDK phosphorylation, cell proliferation is dependent on Mrc1 (and to a lesser extent Tof1). This dependence is likely related to the role of Mrc1 and Tof1 in replication fork stability and progression (Hodgson et al., 2007). Mrc1 interacts with Mcm6, Cdc45 and Pol ε (Komata et al., 2009; Lou et al., 2008; Naylor et al., 2009) and the synthetic lethality between MRC1 deletion and impaired Mcm2-7 priming is consistent with the requirement of DDK phosphorylation of Mcm2-7 for Cdc45 recruitment to the origins (Labib, 2010). Notably, the severity of the proliferation defects caused by elimination of each protein in the mcm4-AP mcm6-AP strain corresponds to the participation of each protein in these three functions: Rad53 and Rad9 are expected to be exclusive to the intra-S phase checkpoint response, Mec1 is implicated both in the checkpoint and in Mcm4/6 S/T-Q phosphorylation, and Mrc1 is important for these functions and fork stability/progression.
These findings have implications for the function of Mcm2-7 phosphorylation. The finding that elimination of checkpoint proteins is deleterious only in the S/T-P background suggests that loss of these sites is more deleterious than loss of S/T-Q sites. It is noteworthy, however, that the specific interaction of MEC1 and MRC1 deletion with the mcm4/6-AP background is very likely influenced by the role of these proteins in S/T-Q phosphorylation (Fig. 6A) and the overlapping functions of the S/T-P and S/T-Q modifications (Fig. 3A). The lethality observed when MRC1 is deleted in a strain lacking Mcm4/6 priming phosphosites but including DDK phosphomimetic mutations suggests that priming phosphorylation also contributes to the recruitment of Cdc45 or another downstream factor stabilized at the origin by Mrc1 (Fig. 6E). This result is consistent with a previous finding that S/T-P mutations in the Mcm4 N-terminus inhibit growth of a DDK bypass mutant (Sheu and Stillman, 2010). The finding that a deletion of the Mcm4 N-terminus bypasses DDK function (Sheu and Stillman, 2010) suggests that phosphorylation of these regions alleviates an inhibitory function of the Mcm4 N-terminus. Our findings indicate that this mechanism must be dependent on phosphorylation of specific residues rather than the total level of phosphorylation, as only certain subsets of phosphomimetic mutations bypass DDK activity (Fig. 2B). Finally, our findings suggest that strains engineered to replicate their DNA in the absence of CDK (Tanaka et al., 2007; Zegerman and Diffley, 2007) are likely to be dependent on MRC1 for replication.
A model for Mcm2-7 priming by Mec1 and other kinases
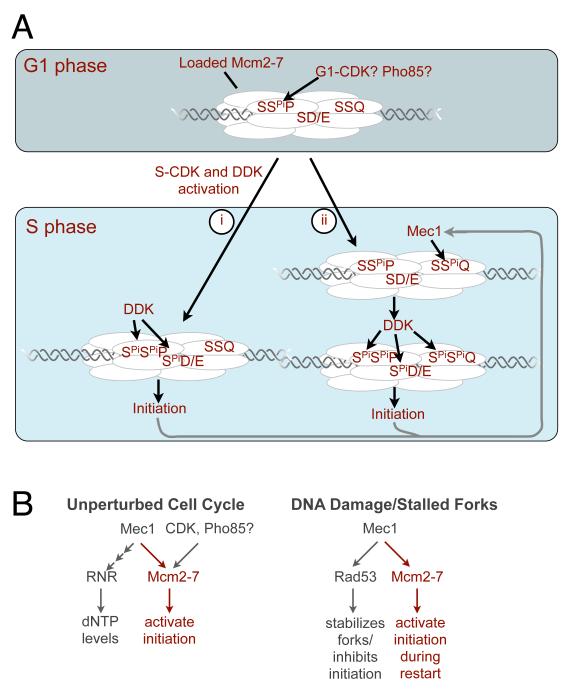
Based on our findings, we propose the following model for the cell cycle regulation of Mcm2-7 phosphorylation (Fig. 7A). As cells exit G1, DDK is activated by the accumulation of Dbf4 (Sclafani and Holzen, 2007). We propose that phosphorylation of Mcm4/6 S/T-P sites, which are already phosphorylated in G1, allows initial Mcm2-7 phosphorylation by DDK and initiation from the first origins of replication (Fig. 7Ai). Because we observe S-phase-dependent S/T-Q phosphorylation in an unperturbed cell cycle (Fig. 5), we propose that the same mechanism that activates Mec1 to regulate dNTP synthesis after cells enter S phase also results in Mec1 phosphorylation of chromatin-associated Mcm4 and Mcm6. Based on our finding that S/T-P and S/T-Q sites both contribute to full DDK phosphorylation of Mcm2-7 (Figure 4), we propose that the resulting Mec1 modification of Mcm4 and Mcm6 further activates DDK phosphorylation of Mcm2-7 (Fig. 7Aii). Our findings suggest that priming and DDK phosphorylation of Mcm4 and Mcm6 both facilitate the subsequent recruitment of Cdc45 or another critical replication factor stabilized at the replication fork by Mrc1. Thus, in this model Mec1 functions as a coordinator of two key S phase events: origin initiation and dNTP synthesis (Fig. 7B, “unperturbed”). Such an activity could facilitate the completion of DNA replication by increasing the probability of origin firing as S phase progresses, and is consistent with recent data that links DDK activity to the efficient completion of DNA synthesis (Patel et al., 2008).
Figure 7. A model for phospho-regulation of Mcm2-7.
(A) Prior to replication we propose that SP sites in Mcm4 and Mcm6 are phosphorylated either by G1-CDKs or Pho85. It is also possible that this phosphorylation occurs earlier (e.g. by S-CDKs) and is not readily removed during G1. Upon entry into S phase we envision two possible mechanisms of DDK targeting: (i) DDK targets S-SPi-P and S-D/E sites to trigger helicase/origin activation; or (ii) Mec1 targets S-S-Q sites followed by DDK targeting of S-SPi-P, S-SPi-Q and S-D/E sites to trigger initiation. Based on our cell cycle studies, we propose that Mec1 is activated initially as a consequence of initiation events mediated by type (i) DDK targeting. Additional activation of Mec1 could be mediated by replication intermediates derived from type (ii) DDK driven initiation or checkpoint signaling.
(B) Mec1 coordinates multiple events in both unperturbed and perturbed cell cycles. In a normal S phase, Mec1 activates both dNTP synthesis and DDK targeting to Mcm2-7. Under conditions of DNA damage or stalled replication forks, Mec1 activates a checkpoint pathway that stabilizes stalled forks and inhibits further origin firing. We propose that Mec1 activation could also facilitate firing of additional origins upon checkpoint recovery.
The HU-insensitivity of the mcm4(AP+AQ) mcm6(AP+AQ) strain (Fig. 6C) and the weaker effects of deleting RAD53 or RAD9 relative to MEC1 in the absence of Mcm4/6 S/T-P sites suggest that Mec1 phosphorylation of Mcm2-7 is not essential for the intra S-phase checkpoint. It remains possible that Mrc1 and Mec1 promote the activation of additional replication origins during checkpoint recovery (Fig. 7B, “DNA damage/stalled forks”). Elevated Mec1 activity during replication stress could increase Mcm2-7 S/T-Q phosphorylation (as we observe after HU treatment; Fig. 6A) and thus subsequent Mcm2-7 phosphorylation by DDK. Such a mechanism would be distinct from Mec1-dependent inhibition of initiation after fork stalling or DNA damage (Zegerman and Diffley, 2009). In this context, we propose that that S/T-Q phosphorylation acts to rescue Mcm2-7 complexes that have not been activated, either due to mutation (e.g. mcm4-AP mcm6-AP) or those that are normally dormant but need activation under conditions of replication stress. Although these potential stress-related functions are intriguing, we stress that S/T-Q phosphorylation of Mcm4 and Mcm6 is observed in the absence of replication stress (Fig. 5), supporting a function for Mec1-dependent Mcm2-7 modification in normal S phase.
Experimental Procedures
Identification of phosphorylation sites by mass spectrometry
Pre-RC samples for mass spectrometry were generated in a 1 ml in vitro reaction using recombinant Cdc6 and extracts from α-factor-arrested cells overexpressing ORC. This reaction corresponds to a 25-fold scale-up of the standard pre-RC assembly reaction described in (Randell et al., 2006). Where indicated, pre-RCs were phosphorylated by recombinant DDK as described previously (Francis et al., 2009). Pre-RCs were concentrated by TCA precipitation and the entire sample was loaded on an 8% SDS/PAGE gel. Bands corresponding to Mcm2-7 proteins were excised from the gel and digested with trypsin or Asp-N. The resulting peptides were analyzed by LC/MS/MS to identify phosphorylated residues (Taplin Mass Spectrometry Facility, Harvard Medical School). Peptides covering more than 80% of each Mcm2-7 protein were identified. Table S1 lists of all the Mcm2-7 phosphopeptides identified.
Generation of phosphospecific antisera
Phosphospecific antisera were generated by Abgent (San Diego, CA) and were used in immunoblotting assays at a dilution of 1:400.
Generation and analysis of Mcm2/4/6 mutations
Mutant alleles of the MCM2, MCM4, and MCM6 N-termini were synthesized as 712, 602, or 430 bp DNAs by gene synthesis (BioBasic, Inc.). These synthetic constructs were cloned into TRP+ ARS/CEN vectors containing MCM2, MCM4, or MCM6 under the control of the MCM5 promoter (Schwacha and Bell, 2001). The resultant constructs were then tested for MCM2, MCM4, or MCM6 function by a plasmid shuffle assay (Schwacha and Bell, 2001). The precise sites mutated in the different alleles are indicated in Fig. S1 and further details of strain construction are in the Supplementary Methods. The genotypes of all the strains used is presented in table S2.
Preparation of phosphorylated pre-RC complexes
Pre-RC assembly assays, pre-RC dephosphorylation, purification of DDK, and DDK kinase assays were performed as described (Francis et al., 2009).
Supplementary Material
Acknowledgements
We thank Angelika Amon, Hannah Blitzblau and Andreas Hochwagen for comments on the manuscript. This work was supported by the Howard Hughes Medical Institute. S.P.B. was an employee of the Howard Hughes Medical Institute. J.C.W.R. was a Special Fellow of the Leukemia and Lymphoma Society. K.G. was a Fellow of the Leukemia and Lymphoma Society. R.H. was supported by a fellowship from the Damon Runyon Cancer Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bruck I, Kaplan D. Dbf4-Cdc7 phosphorylation of Mcm2 is required for cell growth. J Biol Chem. 2009;284:28823–28831. doi: 10.1074/jbc.M109.039123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charych DH, Coyne M, Yabannavar A, Narberes J, Chow S, Wallroth M, Shafer C, Walter AO. Inhibition of Cdc7/Dbf4 kinase activity affects specific phosphorylation sites on MCM2 in cancer cells. J Cell Biochem. 2008;104:1075–1086. doi: 10.1002/jcb.21698. [DOI] [PubMed] [Google Scholar]
- Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci U S A. 2006;103:11521–11526. doi: 10.1073/pnas.0604990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devault A, Gueydon E, Schwob E. Interplay between S-cyclin-dependent kinase and Dbf4-dependent kinase in controlling DNA replication through phosphorylation of yeast Mcm4 N-terminal domain. Mol Biol Cell. 2008;19:2267–2277. doi: 10.1091/mbc.E07-06-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell SJ, Romanowski P, Diffley JF. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci U S A. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B, Calzada A, Labib K. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell. 2007;18:3894–3902. doi: 10.1091/mbc.E07-05-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Friesen H, Andrews B. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol Microbiol. 2007;66:303–314. doi: 10.1111/j.1365-2958.2007.05914.x. [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komata M, Bando M, Araki H, Shirahige K. The direct binding of Mrc1, a checkpoint mediator, to Mcm6, a replication helicase, is essential for the replication checkpoint against methyl methanesulfonate-induced stress. Mol Cell Biol. 2009;29:5008–5019. doi: 10.1128/MCB.01934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liku ME, Nguyen VQ, Rosales AW, Irie K, Li JJ. CDK phosphorylation of a novel NLS-NES module distributed between two subunits of the Mcm2-7 complex prevents chromosomal rereplication. Mol Biol Cell. 2005;16:5026–5039. doi: 10.1091/mbc.E05-05-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell. 2008;32:106–117. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 bY Cdks. J Biol Chem. 2000;275:29042–29052. doi: 10.1074/jbc.M002713200. [DOI] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, Molinari A, Santocanale C. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem. 2006;281:10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci U S A. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor ML, Li JM, Osborn AJ, Elledge SJ. Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc Natl Acad Sci U S A. 2009;106:12765–12770. doi: 10.1073/pnas.0904623106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougarede R, Seta F. Della, Zarzov P, Schwob E. Hierarchy of S-phase-promoting factors: yeast Dbf4-Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol Cell Biol. 2000;20:3795–3806. doi: 10.1128/mcb.20.11.3795-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Hirota K, Fukuda T, Kakusho N, Kugou K, Kawasaki Y, Shibata T, Masai H, Ohta K. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 2008;22:398–410. doi: 10.1101/gad.1626608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyjka SJ, Viggiani CJ, Aparicio OM. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol Cell. 2005;19:691–697. doi: 10.1016/j.molcel.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Wan L, Niu H, Futcher B, Zhang C, Shokat KM, Boulton SJ, Hollingsworth NM. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 2008;22:386–397. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 2009;8:1077–1088. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20:3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.