Colorectal cancer (CRC) represents a common cancer worldwide and the fourth leading cause of cancer in the US with 150,000 new cases annually in the adult population.1 CRC is ultimately a genetic disease in which damaged DNA and genetic instability initiate malignant transformation. Understanding the potential links between chronic inflammatory processes and the genetic events that drive carcinogenesis is particularly important in designing strategies to prevent CRC. The unrelenting exposure of the colonic mucosa to the microbiome and/or its metabolites has long been proposed to contribute to colon tumorigenesis but without convincing mechanistic links. Enterotoxigenic Bacteroides fragilis (ETBF) emerged over the past 25 years as a global etiology of diarrheal disease in animals and humans that is accompanied by colitis.2 In parallel, it was observed that ~5 to 35% of the studied populations asymptomatically carry ETBF. Central to the pathogenicity of ETBF is their secretion of the B. fragilis toxin (BFT).3 BFT is a 20-kDa zinc-dependent metalloprotease toxin known to bind to colonic epithelial cells (CECs) and to stimulate cleavage of the tumor suppressor protein, E-cadherin (Figure 1). E-cadherin cleavage increases intestinal barrier permeability and augments cell signaling via the β-catenin/Wnt pathway which is constitutively activated in essentially all CRC.2,4 As a result, BFT stimulates proliferation and migration of human colon cancer cells in vitro.4 The ability of BFT to further activate the nuclear factor-kappaB (NFκB) pathway inducing proinflammatory cytokine secretion by CECs and data indicating that specific pools of NFκB foster the initiation and promotion of epithelial tumorigenesis led to the hypothesis that ETBF were pro-inflammatory, oncogenic colonic bacteria. This hypothesis was supported by a recent small study in Turkey suggesting that ETBF colonization is more frequent in CRC patients than in controls without CRC.5
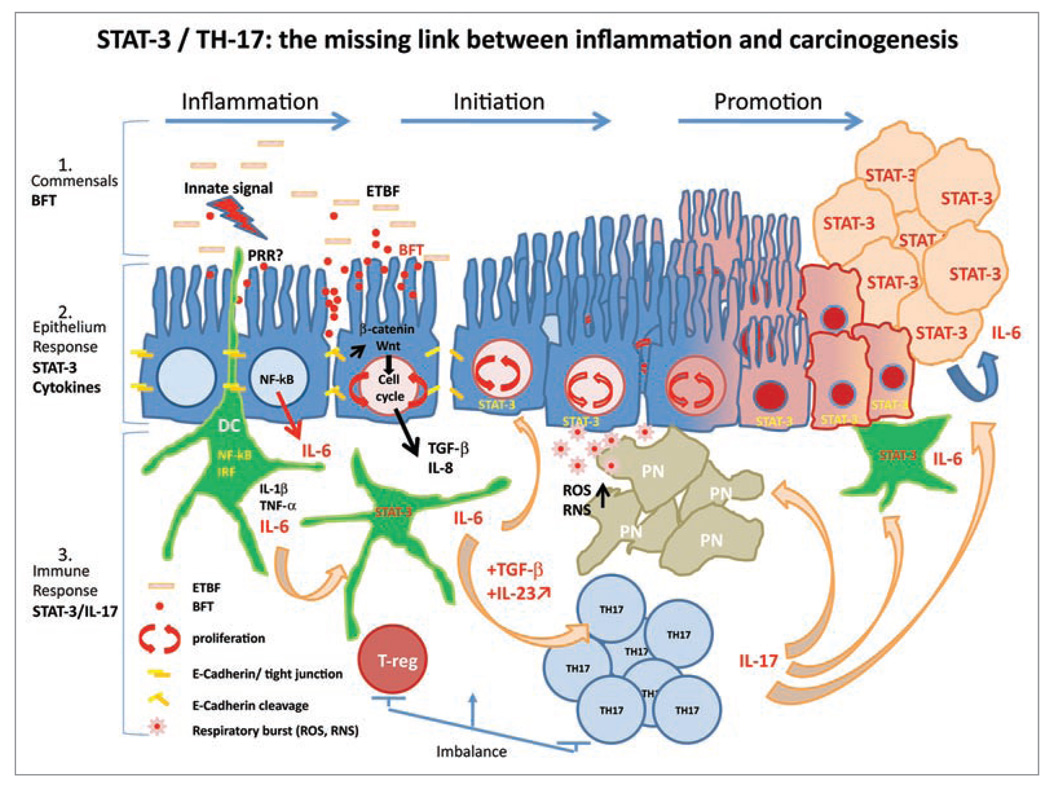
Figure 1.
STAT-3/TH-17: the missing link between inflammation and carcinogenesis. ETBF is a gram negative bacterium, which is thought to stimulate pattern recognition receptors (PRR ) expressed at the surface of colonic epithelial cells (CEC) and/or immune cells such as interdigiting dendritic cells (DC) present in the lamina propria (LP) of colonic epithelium. This innate signal triggers the inflammatory response to the pathogenic commensal colonization, including the downstream NFκB cascade and the production of pro-inflammatory cytokines such as IL-1β, TNFα and IL-6. ET BF colonization of CECs is accompanied by the production of the Bacteroides fragilis toxin (BFT). BFT triggers the cleavage of E-cadherin and complex signal transduction in CECs involving the β-catenin/Wnt pathway leading to the production of c-Myc and CEC proliferation as well as the release from CECs of cytokines/chemokines including IL-8 and TGFβ.2 Decreased barrier function due to E-cadherin cleavage facilitates exposure of ETBF to mucosal immune cells such as DCs. BFT is thought to, in part, initiate oncogenesis via specific mechanisms yet to be defined. The paracrine/autocrine expression of IL-6 and STAT-3 activation may play central roles in promoting tumorigenesis via their pro-proliferative, anti-apoptotic and/or pro-angiogenesis properties. IL-6 secretion by and STAT-3 activation in epithelial and immune cells also contribute to diverting local T cell differentiation from a homeostatic regulatory pathway (T-reg) under TGFβ control to a Th17 pro-inflammatory response sustained by the production of key cytokines such as IL-23. IL-17 recruits polymorphonuclear leukocytes (PN; neutrophils) and may promote CEC proliferation through IL-6-dependent activation of STAT-3. The respiratory burst originating from the anti-bacterial activity of PN is known to induce DNA damage and genetic instability, which can also initiate oncogenesis. Chronic asymptomatic ETBF colonization is proposed to cause a persistent Th17 inflammatory colonic response, which promotes CRC genesis, at least in part, through the actions of STAT-3 and IL-6.
Both mechanistic and translational insights into the link between infection, colitis and CRC are limited by the nonphysiologic nature of current established murine colitis-tumorigenesis models which include adoptive transfer of CD45RBhi T cells into RAG-deficient mice, immune gene knockout (KO) mice or chemically induced colitis-tumorigenesis [e.g., azoxymethane (AOM)/dextran-sulfate sodium (DSS)]. Development of a mouse model of human commensal-driven CRC has the potential to facilitate identification of critical environmental (colonic luminal) components contributing to malignant transformation and to permit studies of attractive targets for therapeutic or preventive intervention. Thus, we sought to establish the role of ETBF-driven colitis in colonic tumorigenesis using multiple intestinal neoplasia (Min) mice a point mutation in the adenomatous polyposis coli (Apc) gene.6 Min mice provide a valuable in vivo system to model human CRC since mutations in Apc are responsible for the familial adenomatous polyposis syndrome and occur in the vast majority of sporadic CRC.7,8 Min mice, however, develop predominantly small intestinal adenomas after ~2–3 months, contrasting with human disease in which adenomas occur predominantly in the colon, especially the distal colon, and develop into progressive CRC. The usual tumorigenesis pattern observed in Min mice contrasts dramatically with the results when Min mice are orally colonized with ETBF that trigger an inflammatory colitis with rapid promotion of predominantly distal colonic tumors. In contrast, nontoxigenic B. fragilis (NTBF) similarly colonizes only the colon but does not induce colonic tumors in excess of sham-inoculated Min mice. Colon tumors in ETBF-colonized Min mice are visible as early as 4 weeks when the usual small bowel adenomas are not yet detectable and, even more striking, is the induction of microadenomas (aka GIN, gastrointestinal intraepithelial neoplasia) as early as one week after ETBF Min mouse colonization. At one week, ETBF, but not NTBF, induce a strong infiltration of the lamina propria with IL-17-producing CD4+ T cells (Th17) and γδ-T cells with a dominant signal transducer activator of transcription-3 (STAT-3) pathway (absolutely required for Th17 cell differentiation) response in both hyperplastic and adenomatous CECs as well as a subset of infiltrating immune cells. The role of the IL-17 in ETBF tumorigenesis is demonstrated by decreased colonic tumors when ETBF-colonized Min mice are treated with blocking antibodies to IL-17 alone or together with antibodies to the receptor for IL-23, the major cytokine that maintains Th17 cells. Unexpectedly, only the depletion of IL-17-producing CD4+ αβ-T cells, but not the innate γδ-T cells, results in decreased colon tumor numbers in ETBF-colonized Min mice. The role of IL-17 in tumorigenesis is supported by other recent data showing that IL-17 promotes tumor growth in vitro and in vivo through the production of IL-6 by IL-17 receptor-bearing tumor cell lines.9 ETBF-induced colon tumors in the Min mouse model represents the first in vivo demonstration of the direct role of a pathogenic human commensal triggering lymphoid and/or colonic epithelium-associated STAT-3 signaling to hijack the adaptive immunity towards a pro-carcinogenic Th17 differentiation pathway. The distinct distal tumor distribution in ETBF-colonized Min mice suggests an opportunity to understand mechanisms by which different colon regions may be susceptible to malignant transformation.
Although these results are provocative, much remains to be learned about the molecular mechanisms by which ETBF and Th17 adaptive immunity promote carcinogenesis. Both NFκB10 and STAT-3,11 have assumed center stage roles as mediators of inflammation-driven carcinogenesis through putatively direct antiapoptotic and cell cycle activity in CECs and promotion of pro-carcinogenic mediators by immune cells. Critical questions remain to be answered about how and if NFκB and activated Stat3 co-regulate oncogenic vs. homeostatic balance in the colonic epithelium and the contributions of other drivers of Th17 differentiation to oncogenesis and, specifically, putative microbiome-induced neoplastic transformation in the colon. We postulate that the composition of an individual’s colonic microbiota and the repeated gastrointestinal insults associated with life (~1–2 episodes of diarrhea/person/year) combined with the individual’s proclivity, likely genetically determined, to respond with a Th17 colonic mucosal response may determine the individual risk for developing the critical CEC mutations that define colorectal carcinogenesis. ETBF is likely just one example of a microbiota organism armed with an oncogenic protease that can trigger a mucosal response contributing to malignant transformation. The observations that Stat3 and IL-23 receptor polymorphisms enhance risk for chronic inflammatory bowel diseases that are, in turn, inextricably connected to CRC risk further support this hypothesis.12 Identifying the molecular mechanisms by which ETBF, a proposed pathogenic human commensal, and Th17 colonic mucosal immune responses result in colon tumorigenesis in the Min mouse model will enhance our ability to translate these findings to understand the early inciting events in the pathogenesis of human CRC. Because ETBF is difficult to detect and/or quantify in stools, translation to studies of human disease remains rudimentary at present but the persistence of ETBF colonization and whether ETBF colonization induces chronic, asymptomatic colonic inflammation should be scrutinized as a potential etiologic component of CRC. Studies of H. pylori without doubt and perhaps ETBF provide excellent opportunities to investigate how certain microbes can upset the often symbiotic relationship with the host to yield epithelial disease and cancers, at least 10% of which are estimated as infectious in origin.13
Acknowledgements
We thank Drew M. Pardoll for his helpful comments. C.L.S. is supported by R01 DK45496 and R01 DK080817.
References
- 1.Jemal A, et al. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Sears CL. Clin Microbiol Rev. 2009;22:349–369. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee KJ, et al. Infect Immun. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S, et al. Gastroenterology. 2003;124:392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- 5.Toprak NU, et al. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 6.Wu S, et al. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCart AE, et al. Pathol Res Pract. 2008;204:479–490. doi: 10.1016/j.prp.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Nathke IS. Annu Rev Cell Dev Biol. 2004;20:337–366. doi: 10.1146/annurev.cellbio.20.012103.094541. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, et al. J Exp Med. 2009;206:1457–1464. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karin M. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, et al. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 12.Cho JH. Nature Rev. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 13.Parkin DM. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]