Abstract
O6-2′-Deoxyguanosine-alkyl-O6-2′-deoxyguanosine interstrand DNA cross-links (ICL) with a four and seven methylene linkage in a 5′-GNC- motif have been synthesized and their repair by human O6-alkylguanine-DNA alkyltransferase (hAGT) investigated. Duplexes containing 11 base-pairs with the ICL in the center were prepared by automated DNA solid-phase synthesis using a cross-linked 2′-deoxyguanosine dimer phosphoramidite, prepared via a seven step synthesis which employed the Mitsunobu reaction to introduce the alkyl lesion at the O6 atom of guanine. Introduction of the four and seven carbon ICL resulted in no change in duplex stability based on UV thermal denaturation experiments compared to a non-cross-linked control. Circular dichroism spectra of these ICL duplexes exhibited features of a B-form duplex, similar to the control, suggesting that these lesions induce little overall change in structure. The efficiency of repair by hAGT was examined and it was shown that hAGT repairs both ICL containing duplexes, with the heptyl ICL repaired more efficiently relative to the butyl cross-link. These results were reproducible with various hAGT mutants including one that contains a novel V148L mutation. The ICL duplexes displayed similar binding affinities to a C145S hAGT mutant compared to the unmodified duplex with the seven carbon containing ICL displaying slightly higher binding. Experiments with CHO cells to investigate the sensitivity of these cells to busulfan and hepsulfam demonstrate that hAGT reduces the cytotoxicity of hepsulfam suggesting that the O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine interstrand DNA cross-link may account for at least part of the cytotoxicity of this agent.
Introduction
Interstrand cross-links (ICL) that are formed in DNA as a consequence of the action of bifunctional alkylating agents represent some of the most toxic lesions encountered by cells due to the obstruction of unwinding of the two strands, critical to the processes of DNA replication, transcription and recombination.1 Interference with these key cellular processes by the presence of ICL is the basis of the mechanism of action of bifunctional alkylating agents, such as mechlorethamine, that are used as cancer therapeutics.2 However, the potency of these agents is reduced by the ability of cancer cells to repair the lesions resulting in an overall resistance to therapy.
In eukaryotic cells the repair of ICL is a complex process with numerous repair pathways including nucleotide excision repair, homologous recombination and non-homologous end joining implicated in the removal of the damage.3 Understanding the molecular basis of how ICL repair occurs will play an important role towards the development of new chemotherapeutic agents that may evade this process, thus increasing the efficacy of these drugs.
One approach employed in elucidating the roles various DNA repair pathways contribute in removing DNA ICL involves the use of chemically synthesized oligonucleotides that contain representative lesions formed by bifunctional alkylating chemotherapeutics.4,5,6 These oligonucleotides are designed to contain lesions linking specific atoms in DNA in well defined orientations and can be incorporated into plasmids for DNA repair experiments. This approach uses solid-phase synthesis which generates sufficient amounts of ICL DNA to enable structural studies and repair assays.7,8,9,10,11
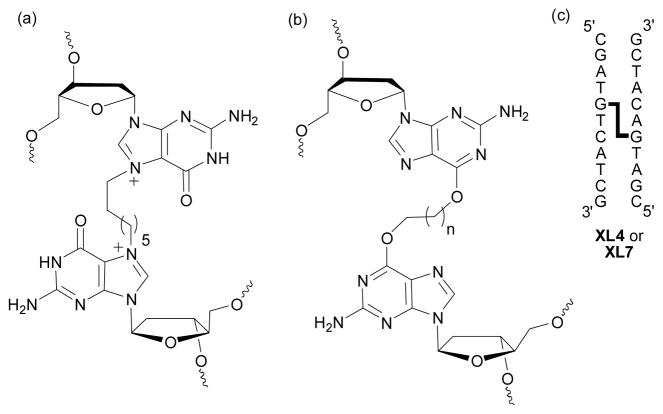
Some of these ICL are challenging to prepare synthetically. For example, the bifunctional alkylating agent hepsulfam (1,7-heptanediol disulfamate) which has been investigated clinically12, has been shown to form a cross-link between the N7 atoms of guanines in 5′-GNC sequences (Figure 1) as demonstrated through the use of mass spectrometry and identification of 1, 7-bis(guanyl)heptane.13 N7-alkylated guanines are chemically unstable, for example the presence of N7-methylguanine can result in an apurinic site in DNA or undergo ring opening to yield the ring opened formamido pyrimidine (Fapy) derivative.14,15
Fig. 1.
Structures of the (a) N7-2′-deoxyguanosine-heptyl-N7-2′-deoxyguanosine cross-link induced by hepsulfam, (b) the O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine cross-link (where n = 3 or 6) and oligomers (c) XL4 and XL7.
In order to prepare oligonucleotides containing a cross-link that models the orientation of the lesion formed by hepsulfam, our group has explored methodologies to prepare ICL DNA that links the O6 atoms of 2′-deoxyguanosine.16 The O6 atom of guanine is a known site of alkylation by chemotherapeutic drugs such as the methylating agent temozolomide and the chloroethylating agent carmustine which proceeds to form a guanine-cytosine ICL.17
Different chemical groups at the O6 position of guanine have been incorporated by organic synthesis for numerous purposes including protection during oligonucleotide synthesis and as substrates for DNA repair studies.18,19 Examples of synthetic methods that have been employed to introduce these groups include the post-synthetic modification of oligonucleotides containing the 2′-deoxyribonucleoside of 2-amino-6-methylsulfonylpurine and the Mitsunobu reaction.19,20
Using a combination of solution and solid-phase synthesis, a 5′-GNC ICL has been prepared (Figure 1) which required a O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine phosphoramidite that contains various protecting groups to permit orthogonal removal, enabling a unit of asymmetry to be engineered in the final ICL DNA duplex to allow for a clinically relevant staggered 1,3 orientation. The synthetic scheme to produce this amidite involved the Mitsunobu reaction which was selected due to its mild conditions, compatibility with all protecting groups employed and direct route to introduce alkyl linkers of various lengths. This method allows for the production of ICL duplexes to explore the influence of linker length in substrates of defined structure and for the systematic investigation of susceptibility to DNA repair mechanisms.
The ability of wild-type human O6-alkylguanine-DNA alkyltransferase (hAGT) and some mutants to repair these ICLs was also investigated. O6-Alkylguanine-DNA alkyltransferases (AGT) are a class of repair proteins that are found in numerous organisms whose role is to repair alkylation at the O6 position of the guanine base and thus they play an important role in maintaining genomic integrity.21 Alkylation at the O6 position of guanine can be mutagenic, disrupting normal Watson-Crick base pairing leading to point mutations which may be fatal. The mechanism by which hAGT repairs alkylated DNA involves flipping the alkylated base out of the duplex and into the active site of the protein where transfer of the alkyl group from the O6 position of guanine to a C145 residue occurs.22 The protein can only act once due to the irreversible alkylation of C145, whereupon the protein is degraded by the ubiquitin pathway.23 hAGT has been shown not only to repair small adducts at the O6 position of guanine such as a methyl group but also larger groups such as benzyl and 4-(3-pyridyl)-4-oxobutyl.24 The study of these novel ICL substrates to undergo repair by hAGT will contribute to the range of DNA substrates that can undergo repair via this pathway.
Results and discussion
Syntheses and characterization of the ICL duplexes
Interstrand cross-linked DNA repair studies require access to substrates of well defined structure. The bifunctional alkylating agent hepsulfam has been shown to alkylate specifically at the N7 atom of 2′-deoxyguanosine in sequences containing a 5′-GNC motif placing a heptyl linkage between the two guanines.13 Because they are synthetically challenging to prepare, we have focused on the synthesis of a cross-link that joins the O6 atoms in this particular sequence motif. It should be emphasized that this specific cross-link has not been identified as a product of alkylation by hepsulfam. The methodology to produce this ICL was used to prepare duplexes containing alkyl linkages of various lengths to investigate the affect of linker length on ICL stability, structure and repair by hAGT. The chemotherapeutic agent busulfan (1,4-butanediol dimethanesulfonate) is another example of a bifunctional alkylating agent that has been used for the treatment of chronic myelogenous leukemia.25 This agent introduces a butyl linkage between the nucleobases with the demonstration that this agent reacts with guanosine to form 1,4-di(7-guanosinyl)butane.26 Mass spectrometry of the reaction products of busulfan with oligonucleotides suggested that this agent forms intrastrand cross-links with 5′-GA-3′ sequences.27
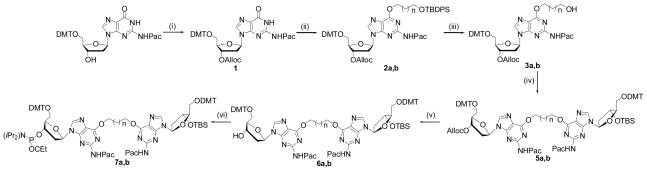
The structure of the O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine cross-link and the sequences containing the butyl and heptyl cross-links (XL4 and XL7) are shown in Figure 1. The synthesis of the cross-linked amidites 7a and 7b were performed according to the synthetic pathway in Scheme 1. Starting from commercially available 5′-O-dimethoxytrityl-N2-phenoxyacetyl-O6-2′-deoxyguanosine the free 3′-alcohol was protected as an allyloxycarbonyl ester 1 using allyl 1-benzotriazoyl carbonate.28 The adducts 2a and 2b were prepared by introduction of 1-tert-butyldiphenylsilyloxy-butan-4-ol or 1-tert-butyldiphenyl silyloxy-heptan-7-ol at the O6 position of 1 via the Mitsunobu reaction.18,20 Removal of the tert-butyldiphenylsilyl protecting group on compounds 2a and 2b was accomplished with TBAF at room temperature for 30 min to yield the adducts 3a and 3b. The formation of dimers 5a or 5b were achieved via a second Mitsunobu reaction using 5′-O-dimethoxytrityl-3′-O-tert-butyldimethyl-silyl-2′-deoxyguanosine (4) in yields of 63 and 71%, respectively. Amidite precursors 6a and 6b required the removal of the 3′-O-alloxycarbonyl group from 5a or 5b with palladium (0) tetrakistriphenylphosphine in THF at room temperature. These were then converted to phosphoramidites 7a and 7b using a slight excess of N,N-diisopropylamino cyanoethyl phosphonamidic chloride and isolated by hexane precipitation. The isolated phosphoramidites 7a and 7b were analyzed by mass spectrometry and were found to have the expected molecular masses. 31P NMR analysis of these phosphoramidites revealed the presence of two signals for 7a (143.92 and 144.14 ppm) and 7b (143.91 and 144.11 ppm) in the region diagnostic for a phosphoramidite.
Scheme 1.
Synthesis of O6G-alkyl-O6G amidite 7a (n=2) and 7b (n=5) (i) Alloc-OBt, CH2Cl2/pyridine (9:1), 12 h; (ii) 4-(tert-butyldiphenylsiloxy) butanol or 7-(tert-butyldiphenylsiloxy) heptanol (1.1 eqv.), PPh3 (1.25 eqv.), DIAD (1.2 eqv.) (iii) TBAF (1 M in THF), 30 min; (iv) 4 (1.01 eqv.), PPh3 (1.25 eqv.), DIAD (1.2 eqv.), 1 h; (v) Pd(PPh3)4 (5mol%), PPh3 (0.25 eqv.), n-butylamine/formic acid (1:1, 2.5 eqv.), THF, 20 min; (vi) N,N-diisopropylamino cyanoethyl phosphonamidic chloride, DIPEA, THF, 30 min.
The synthesis of the 1,3-GNC engineered ICL using phosphoramidite dimers 7a or 7b required three different protecting groups around the cross-linked site enabling an orthogonal approach for their removal. This would allow for the synthesis of three different branches in terms of sequences around the site of the ICL. The Mitsunobu reaction was chosen for its specificity at the O6 position of a protected 2′-deoxyguanosine18,20 and the versatility of this method which allows for the synthesis of cross-links of various lengths, depending on the diol linker used. This reaction was employed twice in the synthetic pathway outlined in Scheme 1, first for the synthesis of the monomers 2a and 2b containing the four and seven carbon linkers at the O6 atom and later in the scheme for dimers 5a and 5b. The dimerization reaction to give dimer 5b proceeded in a slightly higher yield relative to 5a (71 versus 63%), presumably due to the reduced steric hindrance of the longer alkyl linker of 3b that reacts with 4.
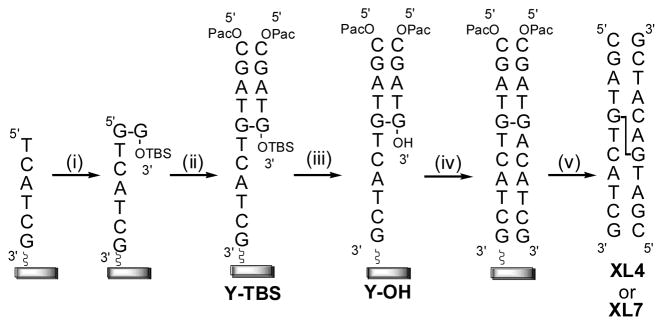
Solid phase synthesis of XL4 and XL7 was performed on a 1 μmol scale using phosphoramidites 7a or 7b according to Figure 2. The first arm of the duplex was synthesized on a polystyrene support using commercially available 3′-O-2′-deoxyphosphoramidites. Coupling of the cross-linked phosphoramidites 7a or 7b introduced the ICL at the 5′-ends to the linear oligomers. The dimethoxytrityl groups of the dimer moiety were removed by brief treatment with 3% TCA in methylene chloride to expose the 5′-OH groups from which the second and third arms of the duplex were then synthesized with a noted increase in the trityl conductivity reading on the synthesizer. Repetitive coupling with protected 3′-O-2′-deoxyphosphoramidites gave, after detritylation and capping (phenoxyacetic anhydride), the branched oligomer Y-TBS (Figure 2). The 3′-O-TBS group was then removed from Y-TBS by treating the support with anhydrous triethylamine for 16 h followed by TEA·3HF twice for 30 min at room temperature. C-18 Reversed-phase HPLC analysis of the deprotected intermediate Y-OH revealed complete removal of the silyl group with the shift of the major peak from 11.2 to 8.2 min in the case of the butyl containing cross-link (see Supplementary Information). Continued synthesis with repetitive coupling of 5′-O-2′-deoxyphosphoramidites at the 3′-end of intermediate Y-OH gave full length ICL duplexes XL4 and XL7.
Fig. 2.
Methodology to construct the cross-linked duplexes XL4 and XL7 via solid phase synthesis: (i) Coupling of amidite 7a or 7b; (ii) extension with 3′-O-2′-deoxyphosphoramidites followed by capping with phenoxyacetic anhydride; (iii) removal of the 3′-O-tert-butyldimethylsilyl group with TEA·3HF; (iv) extension with 5′-O-2′-deoxyphosphoramidites and (v) cleavage from the solid support.
The solid-phase synthesis of the ICL containing oligonucleotides XL4 and XL7 required some changes from the standard synthesis cycle and reagents employed during solid-phase oligonucleotide synthesis. First, synthesis was performed using a polystyrene rather than controlled-pore glass (CPG) solid-support due to the incompatibility of TEA·3HF with the latter.5 As an added precaution, the cyanoethyl protecting groups were removed using TEA as it has been observed that prolonged fluoride treatment using TBAF could lead to chain cleavage.29 Phenoxyacetic anhydride rather than acetic anhydride was used as capping reagent due to a undesirable N-acetylation reaction that has been observed with ‘fast-deprotecting’ amidites.30 Additionally, the protection of the exocyclic amine functionalities of the cross-linked guanosine bases with the phenoxyacetyl group would allow for milder deprotection conditions and ease of its removal at the final stage.
Chain assembly in the 3′ to 5′ direction proceeded smoothly using 3′-O-deoxyphosphoramidites and the cross-linked dimers 7a and 7b with essentially quantitative coupling observed by conductivity measurements to assess the trityl values. A higher concentration (0.15 M) and longer coupling time (10 min) was used for phosphoramidites 7a and 7b to ensure a high coupling efficiency due to their larger size compared to the standard 3′-O-deoxyphosphoramidites. At this point, capping with phenoxyacetyl anhydride gave intermediate Y-TBS (Figure 2) which was desilylated to give the free 3′OH functionality. The efficiency of this step was readily monitored by C-18 reversed phase HPLC (see Supplementary Information). A small amount (approximately 1 mg) of the solid support was deprotected using NH4OH/ethanol (1:3), milder conditions that have been developed for the deprotection of RNA to ensure that the TBS groups are not removed.31 The removal of the TBS group decreases the hydrophobicity of the oligomer and a change in the retention time for the oligomer from 11 to 8 min on the C-18 reverse phase HPLC indicates removal to give the more hydrophilic Y-OH intermediate (Figure 2). The final step in the assembly of XL4 and XL7 is continued chain extension with 5′-O-2′-deoxyphosphoramidites. Cleavage and deprotection of XL4 and XL7 were performed with concentrated ammonium hydroxide/ethanol (3:1) at 55°C for 4 h to yield 141.8 and 108.6 OD of material, respectively (see Table 1). Analysis by SAX HPLC analysis revealed that the major product was the desired ICL whose retention times are shown in Table 1 (see Supplementary Information for HPLC traces of crude and purified material). The combination of mild deprotection conditions and bulky nature of the adduct that the ICL represents at the O6 position may account for the stability under these deprotection conditions. Purification by SAX HPLC, followed by desalting gave XL4 and XL7 in approximately 26 and 36% overall isolated yields.
Table 1.
Amounts, retention times, nucleoside ratios and mass spectral data for cross-linked duplexes XL4 and XL7.
| Duplex | A260 Unitsa | Retention Time | Nucleoside | Ratios | Mass | ||
|---|---|---|---|---|---|---|---|
| exp | obs | exp | obs | ||||
| XL4 | 141.8 (36.9) | 23.7 | dC | 6.0 | 6.1 | 6727.5 | 6726.8 |
| dG | 4.0 | 4.2 | |||||
| dT | 5.0 | 5.0 | |||||
| dA | 5.0 | 5.1 | |||||
| dG-dG | 1.0 | 1.1 | |||||
| XL7 | 108.6 (39.1) | 25.5 | dC | 6.0 | 5.8 | 6769.6 | 6769.0 |
| dG | 4.0 | 4.1 | |||||
| dT | 5.0 | 5.0 | |||||
| dA | 5.0 | 5.1 | |||||
| dG-dG | 1.0 | 1.1 | |||||
Amount of crude ICL duplex purified by SAX HPLC. The numbers in parentheses indicate the amount of pure duplex obtained.
Retention times (min) of ICL duplexes on SAX HPLC using a 0.0–0.5 M linear gradient of sodium chloride.
These ICL duplexes were digested to the constituent nucleosides with a combination of snake venom phosphodiesterase and calf intestinal phosphatase and analyzed by C-18 reversed phase HPLC (see Supporting Information for HPLC traces). In addition to the four standard 2′-deoxynucleosides, one additional peak was observed with a retention time of 18.1 min for XL4 and 25.5 min for XL7. These additional peaks had retention times identical to completely deprotected dimers of 6a and 6b. As shown in Table 1, the ratios of the component 2′-deoxynucleosides and cross-linked nucleosides were consistent with the theoretical composition of ICL duplexes XL4 and XL7. The ICL oligomers XL4 and XL7 required additional time for digestion compared to single stranded oligonucleotides (48 h as opposed to 30 min). ESI mass spectrometry of the duplexes XL4 and XL7 had masses of 6726.8 and 6769.0 Da, consistent with the theoretical values for the cross-linked duplexes.
UV Thermal Denaturation and Circular Dichroism Spectra (CD) of ICL Duplexes
UV denaturation experiments were carried out to assess the effect of the alkyl linkers on duplex stability. The UV thermal denaturation (Tm) curves for the ICL duplexes XL4, XL7, and the corresponding non-cross-linked control duplex all exhibited a sigmoidal denaturation profile with similar hyperchromicities for the transition curves (see Supplementary Information). The melting temperature for XL4, XL7 and the non-cross-linked control were all similar with a value of ~ 48°C.
When this ICL was present as a directly opposed O6-2′-deoxyguanosine-heptyl-O6-2′-deoxyguanosine mismatch in an 11-bp duplex, an increase in Tm of 23°C over the control duplex was observed.16 UV thermal denaturation experiments with oligonucleotides containing an O6-methyl-2′-deoxyguanosine/2′-deoxycytosine base pair have shown that a single O6-methyl substitution reduced duplex stability by at least 18°C per substitution in 1 M NaCl buffer and DNA duplexes containing two of these alkylated lesions were found to have a Tm 40°C lower than that of the unmodified duplex.32,33,34 It is believed that the destabilizing nature of the O6-alkyl lesions are counterbalanced by the preorganization of the covalently linked strands of the duplex resulting in a stability comparable to the non-cross-linked control.5,16
The CD spectra of cross-linked duplexes XL4, XL7 and the non-cross-linked control were recorded at 10°C (spectra are shown in Supplementary Information). In all cases, the CD spectra of the duplexes exhibited signatures characteristic of B-form DNA with a positive maximum peak centered around 275 nm, a negative peak at approximately 250 nm and a cross over around 260 nm.35,36 The CD spectra of non-cross-linked controls revealed some minor differences, particularly a reduction of the signal at 275 nm.
These results suggests that the cross-links had minimal effect on the global B-form structure. NMR studies performed with dodecanucleotides containing a O6meG·C base pair implicate the formation of a wobble base pair, with the methylated guanine sliding towards the major groove and the cytosine oriented towards the minor groove. This base pair is stacked in the duplex between the flanking base pairs inducing only a small conformational change from the wild-type duplex.37 In a standard B-form duplex, the distance between the O6-O6 atoms in a 5′-GNC sequence is approximately 6.4 Å. The O-O distance in the fully extended 1,4-butanediol and 1,7-heptanediol is 6.2 and 9.9 Å, respectively, which is sufficient to span the distance linking the two O6 atoms in a 5′-GNC sequence motif without inducing a significant structural change. Molecular models for the oligomers XL4, XL7 and the non-cross-linked control duplex that were geometry optimized using the AMBER forcefield suggest that the alkyl linkers are oriented towards the major groove and do not greatly distort the duplex (see Supplementary Information).
ICL Repair Assay with hAGT
Repair assays were performed on the ICL containing oligomers XL4 and XL7 with wild-type hAGT and the mutants C145S, P140A and V148L. The C145S mutant was selected as a negative control due to its lack of alkyltransferase activity and for the investigation of protein binding to the O6-dG-alkyl-O6-dG ICLs.38 The P140A mutation has been investigated for the influence of reduced size of the active site pocket on hAGT activity.39,40 In addition, some AGTs such as the E.coli Ada-C, have alanine in place of proline in the equivalent position of hAGT. The V148L mutant was engineered to probe the effect of the side chain of V148 on the repairing ability of the protein. This mutant was selected as little is known about this amino acid other than the carbonyl group of V148 has been shown in wild-type hAGT to accept a H-bond from the N2 atom of a bound guanine base.22,41 We suspected that substituting Val to a larger amino acid would reduce the size of the active site pocket yielding a protein with the same properties as P140A.
All protein (wild-type hAGT, C145S, P140A and V148L) masses were verified by ESI-MS and were in agreement with the calculated values (see Supplementary Information). The secondary structure of the proteins by far UV CD was examined to ensure that the mutations did not compromise the structure. CD scans of the P140A and V148L hAGT mutants revealed similar secondary structures to the wild-type hAGT due the highly conservative nature of these mutations (see Supplementary Information). The C145S mutation had a greater impact on the global secondary structure of the protein. Although the size of the cysteine and serine side groups are relatively similar, the H-bonding capabilities of these two amino acids differ.
The stability of the proteins (wild-type hAGT, C145S, P140A and V148L) were assessed via thermal denaturation studies to ensure that the proteins were properly folded at the assay temperature (37°C) by monitoring the α-helical content of the protein at 222 nm as a function of increasing temperature. The thermal denaturation studies demonstrated that all hAGT proteins were still properly folded at 37°C due to their Tm’s being well above 37°C. Thermal denaturation studies of the wild-type hAGT correlated with values reported of complete inactivation of the protein at 60°C.42 All mutations destabilized the protein based on CD spectra where the stability of the secondary structure of the protein was compromised (see Supplementary Information). Presumably, the presence of an additional methylene group in the V148L mutant affected protein stability negatively while the P140A mutant exhibited a smaller entropy increase upon folding for the alanine versus proline substitution leading to a reduced protein stability.
The intrinsic fluorescence of the hAGT proteins were investigated by monitoring the local environments containing both tryptophan and tyrosine residues using an excitation wavelength of 280 nm while in the case of tryptophan alone the excitation wavelength of 295 nm was used. It is clear from the emission wavelengths and intensities that the mutations had no dramatic affect on the tertiary structure of the protein with V148L having a slight impact on tertiary structure of the protein relative to the other mutants (see Supplementary Information).
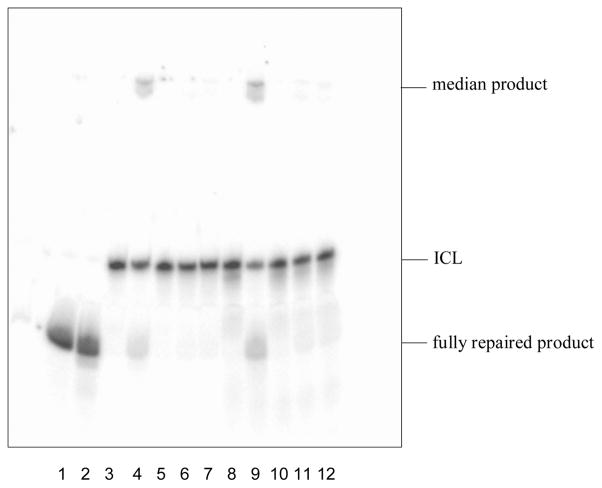
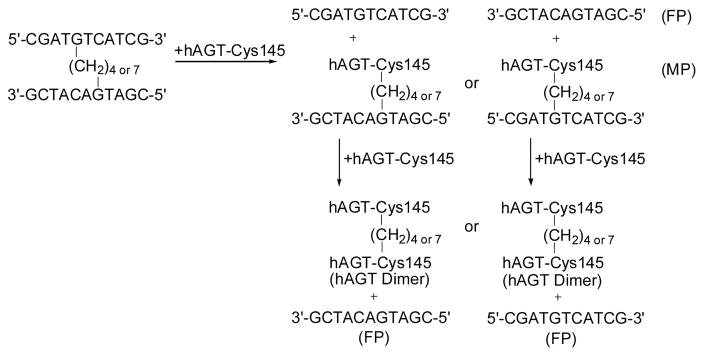
The 5′-32P labeled ICL DNA duplexes XL4 and XL7 (2 pmol) were incubated with 60 pmol of either the wild-type hAGT or the mutants (C145S, P140A and V148L) overnight at 37°C to determine repair. Trials for either 14 or 16 h were performed with virtually identical results obtained, indicating that the reaction was complete as monitored by denaturing PAGE (Figure 3). Lanes 1 and 2 contain 2 pmol of ssDNA corresponding to the sequence of one of the repaired strands with the only difference being the presence of hAGT in lane 2. Lanes 3 and 8 contain 2 pmol of XL4 and XL7, respectively, representing the unrepaired ICL duplexes with a reduced mobility relative to that of completely repaired substrate. Lanes 4 and 9 contain 60 pmol of hAGT with 2 pmol of either XL4 (lane 4) or XL7 (lane 9). In both cases two additional bands are seen, one that migrates more quickly corresponding to the completely repaired single stranded product and the other at the top of the gel with significantly reduced mobility which is likely a single hAGT-bound covalently to a DNA species, a median repair product. For both XL4 and XL7, no repair products are observed with 60 pmol of the mutants C145S (lanes 5 and 10), while P140A (lanes 6 and 11) or V148L (lanes 7 and 12) show very minimal repair of XL7 and no repair of XL4. The repair assays demonstrate that both XL4 and XL7 are repaired by hAGT with XL7 undergoing a greater amount of repair (with 57.0% of ICL repaired) over XL4 (31.4% of ICL repaired) based on quantitation of ICL remaining as measured by the relative counts of the labelled products present on the gel by ImageQuant (see Supplementary Information).
Fig. 3.
Repair of XL4 and XL7 by wild-type hAGT, C145S, P140A and V148L. (A) Denaturing PAGE of repair reactions as described in the experimental section: Lane 1, 2 pmol Control; lane 2, 2 pmol Control + 60 pmol hAGT; lane 3, 2 pmol XL4; lane 4, 2 pmol XL4 + 60 pmol hAGT; lane 5, 2 pmol XL4 + 60 pmol C145S; lane 6, 2 pmol XL4 + 60 pmol P140A; lane 7, 2 pmol XL4 + 60 pmol V148L; lane 8, 2 pmol XL7; lane 9, 2 pmol XL7 + 60 pmol hAGT; lane 10, 2 pmol XL7 + 60 pmol C145S; lane 11, 2 pmol XL7 + 60 pmol P140A; lane 12, 2 pmol XL7 + 60 pmol V148L.
For the repair assays involving XL4 and XL7, the samples were denatured in stop reaction buffer before being loaded on a 7 M urea denaturing gel to prevent self-complexation of the repaired DNA strands which could complicate determination of ratios of the repaired intermediate and fully repaired product versus intact (unrepaired) ICL (Scheme 2).
Scheme 2.
Proposed repair pathway of cross-link species by wild-type hAGT, P140A and V148L (MP - median product; FP- final product).
The inactive C145S mutant was unable to repair both ICL duplexes as expected due to the lack of the active site Cys residue which is known to be the alkyl group acceptor. The other P140A and V148L mutants showed no repair of XL4 and very poor repair for XL7 (4.5% and 3.0%, respectively). The diminished capacity for repair by the P140A mutant has been observed as it is much less active than wild-type hAGT in dealkylating O6-benzylguanine, which has been explained by the reduced size of the binding pocket.43
Time course assays were performed using 60 pmol wild-type hAGT and 2 pmol of the XL4 and XL7 substrates by quantitating the amount of unrepaired, partially and fully repaired species (see Supplementary Information). XL4 was repaired at a much slower rate with 25% repair of the ICL occurring in 8.5 h whereas the same level of repair for XL7 required approximately 1 h.
The results obtained from the time course experiments for the 5′-GNC-3′ ICL for XL4 and XL7 follow similar trends to repair results for the directly opposed O6-dG-alkyl-O6-dG ICL in a mismatch duplex.44 It was shown that 50% final product was formed after 2 h for a directly opposed O6-dG-heptyl-O6-dG mismatch ICL, also observed in the repair of XL7. These results are surprising, as the ICL spanning a 5′-GNC-3′ motif would be expected to be a less flexible substrate relative to directly opposed O6-dG-heptyl-O6-dG mismatch ICL. The mechanism by which hAGT repairs DNA alkylation involves rotation of the alkylated base from the DNA duplex from the minor groove into the active site allowing for transfer of the alkyl group to the residue C145.21 The improved repair of XL7 versus XL4 can be rationalized by the difference in distances of the cross-link. For the four carbon linker of XL4, the O-O distance of the fully extended linker is just sufficient (6.2 Å) to span the distance necessary to link the O6 atoms in a 5′-GNC sequence (6.4 Å). In a geometry optimized model of XL4, the O-O distance was found to be 4.5 Å with some tilting of the guanine bases towards each other (see Supplementary Information). The model of XL7 has an O-O distance of 7.1 Å with the longer alkyl chain protruding into the major groove. This would suggest that rotation of one of the alkylated guanines of XL7 into the active site of hAGT would be less difficult relative to the more strained XL4 ICL. These findings demonstrate that there could have been some distortion in the cross-linked duplexes inherent in the nature of the substrates prior to the repair process and that XL4 is too short to fit optimally in the active site relative to the XL7 counterpart.
The formation of the final product occurs at a faster rate than the formation of the median product for both XL4 and XL7, as the % final product observed is always greater than the % median product. Final and median product formation occur at different rates, suggesting that hAGT preferentially repairs the median product over the initial substrate as indicated by the lack of accumulation of median product at any time. If hAGT preferentially repaired the median product over the initial substrate the median product should not increase with time as the median product would be consumed as it would be formed.
One possible explanation as to why this reaction does not go to completion is that the reacted hAGT binds to another lesion containing ICL or oligonucleotide in our assay. It is known that alkylated hAGT can also bind to DNA.45 In vivo, dissocation is most likely reinforced by ubiquitination resulting in degradation of the alkylated hAGT.46 In addition, it is possible that the cross-link may be oriented in an alternate, less reactive conformation, as has been postulated for O6-(2-hydroxyethyl)guanine and O6-[4-oxo-4-(3-pyridyl)but-1-yl]guanine containing oligonucleotides.47
Binding Studies of C145S hAGT Mutant Using Protein Titration
Binding cooperativity (Hill factor) and monomeric Kd values were determined for the non-cross-linked control duplex, XL4 and XL7 with the C145S hAGT mutant (see Table 2 and Supplementary Information). Similar cooperativity values were obtained for the control duplex (1.41 ± 0.06) and XL7 (1.32 ± 0.06) whereas this value was found to be slightly higher for XL4 (1.75 ± 0.13). All three DNA duplexes showed a similar monomeric dissociation constant ranging from 8.36 for the control duplex to 9.87 μM for XL4 (see Supplementary Information).
Table 2.
Hill factor and monomeric Kd values of C145S hAGT-DNA complexes
| DNA | Cooperativity Factor | Kd (μM) |
|---|---|---|
| Control | 1.41 ± 0.06 | 8.36 ± 1.11 |
| XL4 | 1.75 ± 0.13 | 9.87 ± 1.22 |
| XL7 | 1.32 ± 0.06 | 9.67 ± 1.12 |
Experiments to measure cooperativity and dissociation constants for XL4 and XL7 showed only two bands by EMSA corresponding to the free and bound ICL DNA. The absence of an intermediate band on the native gel indicates that there is cooperativity in the binding of hAGT for the ICL DNA. XL7 and the control duplex showed similar cooperativity and dissociation constants indicating that the presence of the 7-methylene cross-link had little influence on hAGT interaction with the DNA. XL4 had a slightly higher cooperativity of interaction with hAGT, however, all the measured Hill factors for ICL duplexes XL4 and XL7 were between 1 and 2 in accordance with previous work which showed that 1 hAGT protein binds every 4 nucleotides.48 A stoichiometry value of 2 indicates perfect cooperativity, as previously shown in the case of hAGT for many oligonucleotide sequences under different assay conditions whereas a value of 1 indicates no cooperativity.48
In the absence of NMR or X-ray structural data for XL4 or XL7, it can only be speculated that the 4-methylene cross-link induces a structural change in the DNA that has an influence on the interaction of the ICL with hAGT contributing to the slight increase in cooperativity that is observed. It is known that binding of hAGT to DNA causes structural changes in the DNA, which may play a role in aiding the rotation of the damage base from the DNA double helix and subsequently promote hAGT cooperative binding.22,49 However, the shorter alkyl linker of XL4 versus XL7 may hinder rotation of the alkylated guanine into the active site contributing to a reduction in repair by hAGT.
Colony Forming Assay in CHO Cells Treated with Busulfan and Hepsulfam and Relevance with O6-dG-alkyl-O6-dG ICL
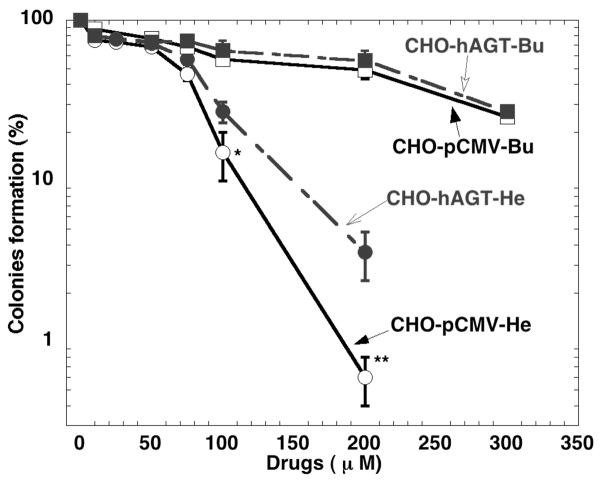
CHO cells that do not express AGT were very sensitive to killing by hepsulfam (Figure 4). Expression of hAGT provided a significant but not complete protection from this agent. This is consistent with the concept that an O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine cross-link accounts for at least part of the cytotoxicity of this agent and that the efficient repair of this 7 carbon adduct by hAGT prevents this killing. There was no protection from busulfan, which is in accord with the poor repair of the 4 carbon adduct produced by this agent.
Fig. 4.
The effect of the expression of hAGT on sensitivity of CHO cells to busulfan and hepsulfam. Cell survival was measured with a colony forming assay as described in the Experimental Section. * p<0.05, Compared to cells expressing hAGT; ** p<0.01, Compared to cells expressing hAGT.
Extensive study of the hAGT reaction has shown that DNA repair by this protein is specific for adducts on the O6-position with a minor capability to repair adducts on the O4-positon of thymine. The results shown here using in vitro assays show that a DNA interstrand cross-link containing 7 carbon atoms linking two O6-2′-deoxyguanosine residues is efficiently repaired by hAGT. Such a cross-link would be expected to be formed in treated cells by hepsulfam and, although it has not yet been characterized, the finding that hAGT reduces the cytotoxicity of hepsulfam is strong evidence that it does occur. Unrepaired DNA interstrand cross-links are very toxic and only a small number of such cross-links may be formed. The results also suggest that a high level of hAGT expression would render tumor cells resistant to therapy with hepsulfam.
Conclusions
The synthesis of nucleoside dimers containing a O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine cross-link that enables the construction of a 1,3-staggered 5′-GNC motif by solid-phase synthesis using phosphoramidite chemistry and an orthogonal approach to removing protecting groups is described. These ICL were obtained in high yield and purity that is required for DNA repair studies. ICL duplexes containing a four and seven methylene linkage were found to have similar thermal stability and structure relative to a non-cross-linked control. Both were also repaired by hAGT, with efficiency higher for the ICL containing the seven methylene linkage. Binding studies suggested similar affinity of hAGT for both ICL. Studies with cells treated with hepsulfam demonstrate that hAGT reduces the cytotoxicity this bifunctional alkylating agent induces and serves as evidence that the O6-2′-deoxyguanosine-alkyl-O6-2′-deoxyguanosine cross-link may account for at least part of the cytotoxicity of this agent.
Experimental
5′-O-Dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanosine, 3′-O-dimethoxytrityl-2′-deoxyribonucleoside-5′-O-(β-cyanoethyl-N,N′-diisopropyl) phosphoramidites and N,N-diisopropylamino cyanoethyl phosphonamidic chloride were purchased from ChemGenes Inc. (Wilmington, MA). 5′-O-Dimethoxytrityl-2′-deoxyribonucleoside-3′-O-(β-cyanoethyl-N,N′-diisopropyl)phosphoramidites and protected 2′-deoxyribonucleoside-polystyrene supports were purchased from Glen Research (Sterling, Virginia). All other chemicals and solvents were purchased from the Aldrich Chemical Company (Milwaukee, WI) or EMD Chemicals Inc. (Gibbstown, NJ). Flash column chromatography was performed using silica gel 60 (230–400 mesh) obtained from Silicycle (Quebec City, QC). Thin layer chromatography (TLC) was performed using precoated TLC plates (Merck, Kieselgel 60 F254, 0.25 mm) purchased from EMD Chemicals Inc. (Gibbstown, NJ). NMR spectra were recorded on a Varian INOVA 300 MHz NMR spectrometer at room temperature. 1H NMR spectra were recorded at a frequency of 300.0 MHz and chemical shifts were reported in parts per million downfield from tetramethylsilane. 31P NMR spectra (1H decoupled) were recorded at a frequency of 121.5 MHz with H3PO4 used as an external standard. ESI mass spectra for oligonucleotides were obtained at the Concordia University Centre for Biological Applications of Mass Spectrometry (CBAMS) using a Micromass Qtof2 mass spectrometer (Waters) equipped with a nanospray ion source. The mass spectrometer was operated in full scan, negative ion detection mode. ESI mass spectra for small molecules were recorded at the McGill University Department of Chemistry Mass Spectrometry Facility with a Finnigan LCQ DUO mass spectrometer in methanol or acetone. Ampicillin, isopropyl β-D-thiogalactopyranoside (IPTG), and most other biochemical reagents as well as polyacrylamide gel materials were purchased from Bioshop Canada Inc (Burlington, ON). Ni-NTA Superflow Resin was purchased from Qiagen (Mississauga, ON). Complete, Mini, EDTA-free Protease Inhibitor Cocktail Tablets were obtained from Roche (Laval, QC) Nitrocellulose filters (0.20 μm) were obtained from Millipore. XL-10 Gold E. coli cells were obtained from Stratagene (Cedar Creek, TX). DpnI, T4 polynucleotide kinase (PNK), Unstained Protein Molecular Weight Marker and restriction enzymes EcoRI and KpnI were obtained from Fermentas (Burlington, ON). [γ-32P]ATP was purchased from Amersham Canada Ltd. (Oakville, ON).
3′-O-alloxycarbonyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanosine (1)
To a solution of 5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanosine (2.15 g, 3.06 mmol) in anhydrous THF/pyridine (9:1) was added a catalytic amount of dimethylaminopyridine (~ 1 mg) followed by allyl 1-benzotriazoyl carbonate (0.88 g, 4.05 mmol) and the mixture allowed to stir at room temperature for 24 h. The solvent was removed in vacuo, the crude taken up in CH2Cl2 (50 mL) and washed twice with 5% sodium bicarbonate (50 mL). The organic layer was then dried over sodium sulfate and evaporated to near dryness. The crude compound was then purified by silica gel column chromatography using a gradient of CH2Cl2/methanol (100:0.5 → 100:1.4) to yield 1.84 g (76.2%) of 1 as a colorless foam. Rf (SiO2): 0.37 in CH2Cl2/MeOH (20:1). 1H NMR (300 MHz, CDCl3, ppm): 11.79 (s, 1H, NH), 8.99 (s, 1H, NH), 7.85 (s, 1H, H8), 7.35–7.44 (m, 4H, Ar), 7.18–7.35 (m, 9H, Ar), 7.12 (t, 1H, Ar), 7.02 (dd, 2H, Ar), 6.82 (dd, 4H, Ar), 6.32 (dd, 1H, H1′, J = 6.0), 5.90–6.05 (m, 1H, allyl), 5.31–5.40 (m, 2H, allyl), 4.44–4.50 (m, 1H, H3′), 4.69 (d, 2H, PhOCH2CO), 4.67 (d, 2H, vinylCH2CO), 4.31–4.36 (m, 1H, H4′), 3.79 (s, 6H, OCH3), 3.35–3.51 (dd, 2H, H5′, H5″), 2.83–2.88 (m, 1H, H2′), 2.64–2.70 (m, 1H, H2″). MS (ESI): m/z calcd for C43H42N5O10[M+H]+, 788.3; found, 788.3.
3′-O-alloxycarbonyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-O6-(4-tert-butyldiphenylsiloxybutyl)-2′-deoxyguanosine (2a)
To a solution of 1 (5.37 g, 6.82 mmol) in anhydrous dioxane (40 mL) was added 1-tert-butyldiphenylsilylbutanol (1.97 g, 9.63 mmol) and triphenylphosphine (2.55 g, 9.72 mmol) followed by the dropwise addition of diisopropylazodicarboxylate (1.89 g, 9.34 mmol) introduced in dioxane (4.4 mL). The solvent was evaporated after 1 h and the crude was taken up in CH2Cl2 (50 mL) and washed twice with 5% sodium bicarbonate (50 mL). The organic layer was then dried over magnesium sulfate, filtered and evaporated. The crude compound was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (19:1 → 13:7). Triphenylphosphine oxide was precipitated from contaminated fractions with diethyl ether and separated by filtration. Evaporation of the solvent afforded 5.16 g (78.8 %) of the compound as a colorless foam. Rf (SiO2): 0.67, hexanes/ethylacetate 2:3. 1H NMR (300 MHz, DMSO-d6, ppm): 10.52 (s, 1H, NH), 8.36 (s, 1H, H8); 7.34–7.64 (m, 11H, Ar); 7.11–7.29 (m, 11H, Ar); 6.89–6.95 (m, 3H, Ar); 6.65–6.74 (dd, 4H, Ar); 6.40 (dd, 1H, H1′, J = 7.2); 5.84–5.98 (m, 1H, allyl); 5.20–5.35 (m, 3H, allyl, H3′); 4.97–5.06 (m, 2H, CH2OAr); 4.50–4.64 (m, 4H, vinylCH2CO, PhOCH2CO); 4.18–4.24 (m, 1H, H4′); 3.70 (t, 2H, CH2OSi); 3.67 (s, 3H, OCH3); 3.66 (s, 3H, OCH3); 3.46 (dd, 1H, H5′); 3.23–3.27 (m, 1H, H2′); 3.15 (dd, 1H, H5″); 2.58–2.63 (m, 1H, H2″); 1.90 (q, 2H, CH2); 1.67–1.76 (m, 2H, CH2); 0.94 (s, 9H, SiC(CH3)3). MS (ESI): m/z calcd for C63H67N5O11SiNa [M+Na]+, 1062.5, found 1062.4.
3′-O-alloxycarbonyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-O6-(7-tert-butyldiphenylsiloxyheptyl)-2′-deoxyguanosine (2b)
To a solution of 1 (3.48 g, 4.41 mmol) in anhydrous dioxane (20 mL) was added 1-tert-butyldiphenylsilyloxyheptanol (1.80 g, 4.86 mmol) and triphenylphosphine (1.76 g, 6.71 mmol) followed by diisopropylazodicarboxylate (1.37 g, 6.46 mmol) introduced dropwise in dioxane (1.6 mL). The solvent was evaporated after 1 h and the crude was taken up in CH2Cl2 (50 mL) and washed twice with 5% sodium bicarbonate (50 mL). The organic layer was then dried over magnesium sulfate, filtered and evaporated to a solid. The crude compound was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (19:1 → 13:7). Triphenylphosphine oxide was precipitated from contaminated fractions with diethyl ether and separated by filtration. Evaporation of the solvent afforded 3.08 g (61.9%) of the product as a colorless foam. Rf (SiO2): 0.67, hexanes/ethylacetate 2:3. 1H NMR (300 MHz, CDCl3, ppm): 8.68 (s, 1H, NH); 8.10 (s, 1H, H8); 7.66–7.71 (m, 4H, Ar); 7.16–7.46 (m, 18H, Ar); 7.01–7.10 (m, 3H, Ar); 6.79 (dd, 4H, Ar); 6.48 (dd, 1H, H1′, J = 5.7); 5.89–6.04 (m, 1H, allyl); 5.30–5.46 (m, 2H, allyl); 4.47–4.52 (m, 1H, H3′); 4.78 (s, 2H, CH2OAr); 4.67 (dt, 2H, vinylCH2CO); 4.59 (d, 2H, PhOCH2CO); 4.35–4.39 (m, 1H, H4′); 3.78 (s, 6H, OCH3); 3.67 (t, 2H, CH2OSi); 3.52 (dd, 1H, H5′); 3.41 (dd, 1H, H5″); 3.18–3.23 (m, 1H, H2′); 2.71–2.76 (m, 1H, H2″); 1.89 (q, 2H, CH2); 1.33–1.62 (m, 6H, (CH2)3); 1.06 (s, 9H, SiC(CH3)3). MS (ESI): m/z calcd C66H73N5O11SiNa [M+Na]+, 1120.5, found 1120.4.
3′-O-alloxycarbonyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-O6-(4-hydroxybutyl)-2′-deoxyguanosine (3a)
To a solution of 2a (5.15 g, 5.38 mmol) in anhydrous THF (10 mL) was added dropwise tetrabutylammonium fluoride (1 M in THF, 645 μL, 6.45 mmol) and the solution allowed to stir at room temperature for 30 min. On completion the reaction was concentrated, taken up in CH2Cl2 (50 mL), washed twice with 5% sodium bicarbonate (50 mL), the organic layer dried over sodium sulfate and evaporated to dryness. The crude was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (60:40 → 20:80) to yield 3.25 g (70.3 %) of product as a colorless foam. Rf (SiO2): 0.50, CH2Cl2/MeOH 10:1. 1H NMR (300 MHz, DMSO-d6, ppm): 10.58 (s, 1H, NH), 8.41 (s, 1H, H8); 7.31–7.36 (m, 4H, Ar); 7.18–7.21 (m, 7H, Ar); 6.95–7.02 (m, 3H, Ar); 6.71–6.81 (dd, 4H, Ar); 6.45 (dd, 1H, H1′, J = 6.9); 5.89–6.04 (m, 1H, allyl); 5.27–5.41 (m, 3H, H3′, allyl); 4.87–5.15 (m, 2H, CH2OAr); 4.63–4.67 (dt, 2H, vinylCH2CO); 4.59 (d, 2H, PhOCH2CO); 4.53 (t, 1H, OH); 4.23–4.30 (m, 1H, H4′); 3.74 (s, 3H, OCH3); 3.73 (s, 3H, OCH3); 3.44–3.55 (m, 3H, CH2OH, H5′); 3.26–3.35 (m, 1H, H5″); 3.20 (dd, 1H, H2′); 2.57–2.69 (m, 1H, H2″); 1.90 (q, 2H, CH2); 1.56–1.68 (m, 2H, CH2). MS (ESI): m/z calcd for C47H49N5O11Na [M+Na]+, 882.3, found 882.2.
3′-O-alloxycarbonyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-O6-(7-hydroxyheptyl)-2′-deoxyguanosine (3b)
To a solution of 2b (1.35 g, 1.20 mmol) in anhydrous THF (10 mL) was added dropwise tetrabutylammonium fluoride (1 M in THF, 135 μL, 1.35 mmol) and the solution was allowed to stir at room temperature for 30 min. On completion the reaction was concentrated, taken up in CH2Cl2 (50 mL) and washed twice with 5% sodium bicarbonate (50 mL). The organic layer was then dried over sodium sulfate, evaporated and the crude was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (60:40 → 20:80) to yield 0.949 g (89.6%) of a colorless foam. Rf (SiO2): 0.50, CH2Cl2/MeOH 10:1. 1H NMR (300 MHz, DMSO-d6, ppm): 10.55 (s, 1H, NH), 8.36 (s, 1H, H8); 7.23–7.33 (m, 4H, Ar); 7.10–7.20 (m, 7H, Ar); 6.88–6.98 (m, 3H, Ar); 6.71 (dd, 4H, Ar); 6.40 (dd, 1H, H1′, J = 5.7); 5.85–6.00 (m, 1H, allyl); 5.31–5.39 (m, 1H, H3′); 5.22–5.35 (m, 2H, allyl); 5.00 (t, 2H, CH2OAr); 4.60 (d, 2H, vinylCH2CO); 4.52 (d, 2H, PhOCH2CO); 4.34 (s, 1H, CH2OH); 4.18–4.26 (m, 1H, H4′); 3.69 (s, 6H, OCH3); 3.42 (dd, 1H, H5′); 3.30–3.41 (m, 2H, CH2OH); 3.19–3.31 (m, 1H, H2′); 3.14 (dd, 1H, H5″); 2.56–2.64 (m, 1H, H2″); 1.80 (q, 2H, CH2CH2OAr); 1.21–1.47 (m, 6H, (CH2)3). MS (ESI): m/z calcd for C50H55N5O11Na [M+Na]+, 924.4, found 924.4.
3′-O-tert-butyldimethylsilyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanosine (4)
To a solution of 5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanosine (1.73 g, 2.46 mmol) in anhydrous N,N-dimethylformamide was added imidazole (1.13 g, 16.64 mmol) followed by tert-butyldimethylsilyl chloride (1.24 g, 8.23 mmol) and the mixture was allowed to stir at room temperature for 16 h. The solvent was removed, and the crude taken up in CH2Cl2 (25 mL) and washed twice with 5% sodium bicarbonate (25 mL). The organic layer was then dried over sodium sulfate and evaporated. The crude was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (5:1 → 1:3) to yield 1.58 g (83.2%) of the product which was a colorless foam. Rf (SiO2): 0.55, CH2Cl2/MeOH 10:0.8. 1H NMR (300 MHz, DMSO-d6, ppm): 11.85 (s, 1H, NH), 11.78 (s, 1H, NH), 8.19 (s, 1H, H8); 7.10–7.36 (m, 12H, Ar); 6.92–7.02 (m, 3H, Ar); 6.74–6.84 (t, 3H, Ar); 6.21 (dd, 1H, H1′, J = 6.9); 4.85 (d, 2H, PhOCH2CO); 4.49–4.54 (m, 1H, H3′); 3.81–3.86 (m, 1H, H4′); 3.70 (s, 6H, OCH3); 3.05–3.23 (dd, 2H, H5′, H5″); 2.72–2.77 (m, 1H, H2′); 2.32–2.37 (m, 1H, H2″); 0.79 (s, 9H, SiC(CH3)3); 0.03 (s, 3H, SiCH3); −0.04 (s, 3H, SiCH3). MS (ESI): m/z calcd for C45H51N5O8SiNa [M+Na]+, 840.3, found 840.2.
1-{O6-[3′-O-alloxycarbonyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-4-{O6-[3′-O-tert-butyldimethylsilyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-butane (5a)
To a solution of compound 3a (1.70 g, 1.98 mmol) in anhydrous dioxane (15 mL) was added triphenylphosphine (0.81 g, 3.09 mmol) and compound 4 (1.56 g, 1.93 mmol). Then, diisopropylazodicarboxylate (0.64 g, 3.02 mmol) was introduced in a solution of dioxane (5 mL). The solvent was evaporated after 14 h and the crude was taken up in CH2Cl2 (50 mL) and washed twice with 5% sodium bicarbonate (50 mL). The organic layer was then dried over magnesium sulfate and evaporated to dryness, which was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (1:0 → 1:5) to yield 2.07 g (63.1%) of product which was a colorless foam. Rf (SiO2): 0.45, CH2Cl2/MeOH 20:1. 1H NMR (300 MHz, CDCl3, ppm): 8.82 (s, 1H, NH); 8.78 (s, 1H, NH); 8.06 (s, 1H, H8); 8.00 (s, 1H, H8); 6.67–7.45 (m, 22H, Ar); 6.94-6.09 (m, 6H, Ar); 6.64–6.80 (m, 8H, Ar); 6.42 (m, 2H, 2 x H1′); 5.86–6.02 (m, 1H, allyl); 5.43–5.51 (m, 1H, H3′); 5.27–5.44 (m, 2H, allyl); 4.78 (s, 4H, CH2OAr); 4.71 (d, 4H, PhOCH2CO); 4.57–4.69 (m, 2H, vinylCH2CO); 4.52–4.64 (m, 1H, H3′); 4.31–4.39 (m, 1H, H4′); 4.03–4.11 (m, 1H, H4′); 3.75 (s, 12H, OCH3); 3.30–3.56 (m, 4H, 2 x H5′, 2 x H5″); 3.00 (m, 1H, H2′); 2.62–2.76 (m, 2H, H2′, H2″); 2.38–2.49 (m, 1H, H2″); 2.13 (m, 4H, (CH2)2); 0.85 (s, 9H, SiC(CH3)3); 0.04 (s, 3H, SiCH3); 0.00 (s, 3H, SiCH3). MS (ESI): m/z calcd for C92H98N10O18SiNa [M+Na]+, 1681.7, found 1681.5.
1-{O6-[3′-O-alloxycarbonyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-7-{O6-[3′-O-tert-butyldimethylsilyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-heptane (5b)
To a solution of 3b (1.73 g, 1.92 mmol) in anhydrous dioxane (10.0 mL) was added triphenylphosphine (0.77 g, 2.91 mmol) and compound 4 (1.54 g, 1.88 mmol). Then diisopropylazodicarboxylate (0.59 g, 2.78 mmol) was introduced dropwise in a solution of dioxane (3 mL). After 30 min the solvent was evaporated, the crude was taken up in CH2Cl2 (50 mL) and washed twice with 5% sodium bicarbonate (50 mL). The organic layer was then dried over sodium sulfate and evaporated to dryness. The crude was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (1:0 → 1:4) to yield 2.32 g (71.1%) of a colorless foam. Rf (SiO2): 0.70, CH2Cl2/MeOH 10:1. 1H NMR (300 MHz, DMSO-d6, ppm): 10.58 (s, 1H, NH); 10.53 (s, 1H, NH); 8.39 (s, 1H, H8); 8.35 (s, 1H, H8); 7.10–7.34 (m, 22H, Ar); 6.88–6.97 (m, 6H, Ar); 6.64–6.80 (m, 8H, Ar); 6.40 (dd, 1H, H1′, J = 6.0); 6.34 (dd, 1H, H1′, J = 6.6); 5.85–6.00 (m, 1H, allyl); 5.31–5.39 (m, 1H, H3′); 5.21–5.35 (m, 2H, allyl); 4.97–5.03 (m, 4H, PhOCH2CO); 4.61–4.72 (m, 1H, H3′); 4.60 (d, 2H, vinylCH2CO); 4.34 (d, 4H, CH2OAr); 4.17–4.26 (m, 1H, H4′); 3.69–3.77 (m, 1H, H4′); 3.68 (s, 12H, OCH3); 3.46 (dd, 1H, H5′); 3.10–3.32 (m, 4H, H2′, H5′, 2 x H5″); 2.86–2.98 (m, 1H, H2′); 2.54–2.65 (m, 1H, H2″); 2.27–2.38 (m, 1H, H2′); 1.80 (m, 4H, CH2CH2OAr); 1.41; (m, 6H, (CH2)3); 0.78 (s, 9H, SiC(CH3)3); 0.00 (s, 3H, SiCH3); −0.05 (s, 3H, SiCH3). MS (ESI): m/z calcd for C94H104N10O18SiNa [M+Na]+, 1723.7, found 1724.5.
1-{O6-[3′-O-tert-butyldimethylsilyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-4-{O6-[5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-butane (6a)
To a solution of 5a (0.97 g, 0.59 mmol) in anhydrous THF (3 mL) was added triphenylphosphine (23.0 mg, 0.09 mmol), palladium (0) tetrakistriphenylphosphine (34.0 mg, 0.03 mmol) and a butylamine/formic acid solution (1:1, 1.17 mmol) in THF (1 mL). After stirring the mixture for 30 min the reaction was concentrated in vacuo and the crude taken up in CH2Cl2 (25 mL) and washed twice with 5% sodium bicarbonate (25 mL). The organic layer was dried over sodium sulfate, filtered and evaporated. The crude was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (3:7 → 1:9) to give 0.87 g (94.4%) of colorless product. Rf (SiO2): 0.41 hexanes/ethyl acetate (1:9). 1H NMR (300 MHz, DMSO-d6, ppm): 10.65 (s, 2H, NH); 8.45 (s, 1H, H8); 8.41 (s, 1H, H8); 7.21–7.38 (m, 22H, Ar); 7.03-6.96 (m, 6H, Ar); 6.76–6.85 (m, 8H, Ar); 6.40–6.49 (m, 2H, 2 x H1′); 5.40 (d, 1H, 3′OH); 5.07 (s, 4H, 2 x PhOCH2CO); 4.64–4.76 (m, 5H, H3′, 2 x CH2CH2OAr); 4.52–4.60 (m, 1H, H3′); 4.02–4.07 (m, 1H, H4′); 3.80–3.93 (m, 1H, H4′); 3.76 (s, 12H, OCH3); 3.26–3.39 (m, 3H, 2 x H5′, H5″); 3.13–3.17 (m, 1H, H5″); 2.90–3.03 (m, 3H, 2 x H2′, H2″); 2.40–2.44 (m, 1H, H2″); 2.03–2.10 (m, 4H, CH2CH2OAr); 0.85 (s, 9H, SiC(CH3)3); 0.07 (s, 3H, SiCH3); 0.00 (s, 3H, SiCH3). MS (ESI): m/z calcd for C88H95N10O16Si [M+H]+, 1575.7 found 1575.7.
1-{O6-[3′-O-tert-butyldimethylsilyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-7-{O6-[5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-heptane (6b)
To a solution of compound 5b (0.52 g, 0.31 mmol) in anhydrous THF (3 mL) was added triphenylphosphine (24.4 mg, 0.09 mmol), palladium (0) tetrakistriphenylphosphine (35.2 mg, 0.03 mmol) and a butylamine/formic acid solution (1:1, 0.66 mmol) in THF (1 mL). After stirring at room temperature for 40 min the reaction was concentrated, taken up in CH2Cl2 (25 mL) and washed twice with 5% sodium bicarbonate (25 mL). The organic layer was then dried over sodium sulfate and evaporated. The crude was purified by silica gel column chromatography using a gradient of hexanes/ethyl acetate (1:0 → 1:4) to yield 0.376 g (76.0%) of a colorless foam. Rf (SiO2): 0.33 hexanes/ethyl acetate (1:4). 1H NMR (300 MHz, CDCl3, ppm): 8.76 (s, 1H, NH); 8.67 (s, 1H, NH); 8.03 (s, 1H, H8); 8.01 (s, 1H, H8); 7.14–7.44 (m, 22H, Ar); 6.97–7.09 (m, 6H, Ar); 6.74–6.84 (m, 8H, Ar); 6.60 (dd, 1H, H1′, J = 6.6); 6.43 (dd, 1H, H1′, J = 6.4); 4.40–4.85 (m, 10H, 2 x H3′, 2 x PhOCH2CO, 2 x CH2OAr); 4.17–4.26 (m, 1H, H4′); 4.05–4.14 (m, 1H, H4′); 3.77 (s, 6H, OCH3); 3.75 (s, 6H, OCH3); 3.45 (dd, 1H, H5′); 3.00–3.38 (m, 3H, H5′, 2 x H5″); 3.21 (s, 1H, 3′OH); 2.56–2.83 (m, 3H, 2 x H2′, H2″); 2.40–2.51 (m, 1H, H2″); 1.90 (m, 4H, CH2CH2OAr); 1.52; (s, 6H, (CH2)3); 0.86 (s, 9H, SiC(CH3)3); 0.00 (s, 3H, SiCH3); −0.05 (s, 3H, SiCH3). MS (ESI): m/z calcd for C91H100N10O16SiNa [M+Na]+, 1639.7, found 1639.5.
1-{O6-[3′-O-tert-butyldimethylsilyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-4-{O6-[5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl-3′-O-(β-2-cyanoethyl-N,N′-diisopropyl)phosphoramidite]}-butane (7a)
Compound 6a (0.45 g, 0.29 mmol) was dissolved in THF (1.4 mL) and diisopropylethylamine (0.055 g, 0.43 mmol) followed by N, N-diisopropylamino cyanoethyl phosphonamidic chloride (0.081 g, 0.343 mmol). The reaction was allowed to stir at room temperature for 2 h after which it was quenched by the addition of ethyl acetate (50 mL) and the solution washed twice with 5% sodium bicarbonate (50 mL). The organic layer was dried over sodium sulfate, filtered and evaporated. The product was precipitated from vigorously stirring hexanes to give 0.43 g (86%) of a colorless powder. Rf (SiO2): 0.64, 0.70 in ethyl acetate. 31P NMR (121.5 MHz, acetone-d6, ppm): 143.92, 144.14. MS (ESI): m/z calcd for C97H111N12O17PSiNa [M+Na]+, 1797.8, found 1797.6.
1-{O6-[3′-O-tert-butyldimethylsilyl-5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl]}-7-{O6-[5′-O-dimethoxytrityl-N2-phenoxyacetyl-2′-deoxyguanidyl-3′-O-(β-2-cyanoethyl-N,N′-diisopropyl)phosphoramidite]}-heptane (7b)
Compound 6b (0.35 g, 0.22 mmol) was dissolved in THF (1.1 mL) and diisopropylethylamine (0.042 g, 0.32 mmol) followed by N, N-diisopropylamino cyanoethyl phosphonamidic chloride (0.061 g, 0.26 mmol). The reaction was allowed to stir at room temperature for 2 h after which it was quenched by the addition of ethyl acetate (40 mL) and the solution washed twice with 5% sodium bicarbonate (50 mL). The organic layer was dried over sodium sulfate, filtered and evaporated. The product was precipitated from vigorously stirring hexanes to give 0.28 g (71%) of a colorless powder. Rf (SiO2): 0.71, 0.82 in ethyl acetate. 31P NMR (121.5 MHz, acetone- d6, ppm): 143.91, 144.11. MS (ESI): m/z calcd for C100H117N12O17PSiNa [M+Na]+, 1839.8, found 1839.5.
Oligonucleotide Synthesis, Purification and Characterization
The cross-linked duplexes XL4 and XL7, whose sequences are shown in Figure 1, were assembled using an Applied Biosystems Model 3400 synthesizer on a 1 μmole scale employing standard β-cyanoethylphosphoramidite cycles supplied by the manufacturer with slight modifications to coupling times described below. The nucleoside phosphoramidites containing standard protecting groups were prepared in anhydrous acetonitrile at a concentration of 0.1 M for the 3′-O-2′-deoxyphosphoramidites, 0.15 M for the cross-linked 3′-O-2′-deoxyphosphoramidites (7a and 7b) and 0.2 M for the 5′-O-2′-deoxyphosphoramidites. Assembly of sequences first involved detritylation (3% trichloroacetic acid [TCA] in CH2Cl2), followed by nucleoside phosphoramidite coupling with commercial 3′-O-2′-deoxyphosphoramidites (2 min), 5′-O-2′-deoxyphosphoramidites (3 min) or cross-linked phosphoramidites 7a or 7b (10 min); Subsequent capping with phenoxyacetic anhydride/pyridine/tetrahydrofuran (1:1:8, v/v/v; solution A, and 1-methyl-1 H-imidazole/tetrahydrofuran 16:84 w/v; solution B) and oxidation (0.02 M iodine in tetrahydrofuran/water/pyridine 2.5:2:1) followed every coupling. To complete assembly of the cross-linked duplexes, the cyanoethyl groups were removed from the polystyrene-linked oligomers by treating the support with 1 mL of anhydrous triethylamine (TEA) for at least 12 h. The support was then washed with 30 mL of anhydrous acetonitrile (ACN) followed by anhydrous THF. The tert-butyldimethylsilyl (TBS) group was removed from the partial duplex by treating the support with 2 × 1 mL triethylamine trihydrofluoride (TEA·3HF) for a total of 1 h. The support was then washed with 30 mL each of anhydrous THF and ACN followed by drying via high vacuum (20 min). The final extension of the cross-linked duplex was then achieved using 5′-O-2′-deoxyphosphoramidites with a total detritylation exposure of 130 seconds and removal of the 3′-terminal trityl group by the synthesizer to yield duplexes XL4 and XL7 on the solid support.
The oligomer-derivatized polystyrene beads were transferred from the reaction column to screw cap microfuge tubes fitted with teflon lined caps and the oligomer released from the support and protecting groups removed by treatment with a mixture of concentrated ammonium hydroxide/ethanol (0.3 mL:0.1 mL) for 4 h at 55°C. The cross-linked final products were separated from pre-terminated products by strong anion exchange (SAX) HPLC using a Dionex DNAPAC PA-100 column (0.4 cm × 25 cm) purchased from Dionex Corp, (Sunnyvale, CA) with a linear gradient of 0–50% buffer B over 30 min (buffer A: 100 mM Tris HCl, pH 7.5, 10% acetonitrile and buffer B: 100 mM Tris HCl, pH 7.5, 10% acetonitrile, 1 M NaCl) at 40°C. The columns were monitored at 260 nm for analytical runs or 280 nm for preparative runs. The purified oligomers were desalted using C-18 SEP PAK cartridges (Waters Inc.) as previously described.5 The amounts of purified oligomers obtained are shown in Table 1.
The cross-linked oligomers (0.1 A260 units) were characterized by enzymatic digestion (snake venom phosphodiesterase: 0.28 units and calf intestinal phosphatase: 5 units, in 10 mM Tris, pH 8.1 and 2 mM magnesium chloride) for a minimum of 36 h at 37°C as previously described.5 The resulting mixture of nucleosides was analyzed by reversed phase HPLC carried out using a Symmetry® C-18 5μm column (0.46 × 15 cm) purchased from Waters Inc, Milford, MA. The C-18 column was eluted with a linear gradient of 0–60% buffer B over 30 min (buffer A, 50 mM sodium phosphate, pH 5.8, 2% acetonitrile and buffer B, 50 mM sodium phosphate, pH 5.8, 50% acetonitrile). The resulting peaks were identified by co-injection with the corresponding standards and eluted at the following times: dC (4.6 min), dG (7.9 min), dT (8.5 min), dA (9.6 min), cross-link dimers (18.1 for the four carbon and 26.2 min for the seven carbon cross-link), and the ratio of nucleosides was determined. The results are given in Table 1. The molecular weights of the cross-linked oligomers were determined by ESI-MS and these were in agreement with the calculated values (see Supplementary Information for mass spectra).
UV Thermal Denaturation Studies
Molar extinction coefficients for the oligonucleotides and cross-linked duplexes were calculated from those of the mononucleotides and dinucleotides according to nearest-neighbour approximations (M−1 cm−1). Non-cross-linked duplexes were prepared by mixing equimolar amounts of the interacting strands and lyophilizing the mixture to dryness. The resulting pellet was then re-dissolved in 90 mM sodium chloride, 10 mM sodium phosphate, 1 mM EDTA buffer (pH 7.0) to give a final concentration of 2.8 μM duplex. The cross-linked duplexes were dissolved in the same buffer to give a final concentration of 2.8 μM. The solutions were then heated to 90°C for 10 min, cooled slowly to room temperature, and stored at 4°C overnight before measurements. Prior to the thermal run, samples were degassed by placing them in a speed-vac concentrator for 2 min. Denaturation curves were acquired at 260 nm at a rate of heating of 0.5°C/min, using a Varian CARY Model 3E spectrophotometer fitted with a 6-sample thermostated cell block and a temperature controller. The data were analyzed in accordance with the convention of Puglisi and Tinoco50 and transferred to Microsoft Excel™.
Circular Dichroism (CD) Spectroscopy
Circular dichroism spectra were obtained on a Jasco J-815 spectropolarimeter equipped with a Julaba F25 circulating bath. Samples were allowed to equilibrate for 5–10 min at 10°C in 90 mM sodium chloride, 10 mM sodium phosphate, 1 mM EDTA (pH 7.0), at a final concentration of 2.8 μM for the cross-linked duplexes and ca. 2.8 μM for control duplexes. Each spectrum was an average of 5 scans. Spectra were collected at a rate of 100 nm/min, with a bandwidth of 1 nm and sampling wavelength of 0.2 nm using fused quartz cells (Starna 29-Q-10). The CD spectra were recorded from 350 to 200 nm at 10°C. The molar ellipticity was calculated from the equation [φ] = ε/Cl, where ε is the relative ellipticity (mdeg), C is the molar concentration of oligonucleotides (moles/L), and l is the path length of the cell (cm). The data were processed on a PC computer using Windows™ based software supplied by the manufacturer (JASCO, Inc.) and transferred into Microsoft Excel™ for presentation.
Molecular Modeling of ICL Duplexes
The DNA duplex (5′-dCGATGTCATCG)/(5′-CGATGACATCG) and cross-linked duplexes XL4 and XL7 were built using HyperChem™ molecular modeling software. All duplexes were geometry optimized using the AMBER forcefield.
Protein Expression and Purification
Site directed mutagenesis as well as transformation into XL-10 E. coli cells were performed as directed by the Stratagene manual. Cells containing either a plasmid coding for wild-type or mutant hAGT were grown in a 1 L cultures of LB broth + 100 μg/mL ampicillin until an OD600=0.6 was reached. The cells were induced with 0.3 mM IPTG, incubated at 37°C for 4 h with shaking at 225 rpm and harvested by centrifugation at 6000 x g at 4°C for 30 min. Pellets were weighed and resuspended in 5 mL of buffer [20 mM Tris HCl (pH 8.0), 250 mM NaCl, 20 mM β-mercaptoethanol supplemented with Complete, Mini, EDTA-free Protease Inhibitor Cocktail Tablets] for each gram of pellet. The cells were homogenized by using approximately 20 strokes in a dounce homogenizer, lysed using two rounds of French press, centrifuged at 17000 x g for 45 min at 4°C. The lysate was applied to a pre-equilibrated Ni-NTA Superflow column (100 mL resuspension buffer) containing 3.5 mL of resin. The lysate was run twice through the column at 1 mL/min to ensure complete binding. The column was washed with 200 mL resuspension buffer and 20 mM imidazole at 1 mL/min, until the OD280 of the eluant was constant. The protein eluted with 50 mL resuspension buffer supplemented with 200 mM imidazole at 1 mL/min. 1 mL fractions were collected and fractions that displayed protein content (generally fractions 4–11) were pooled followed by dialysis versus 4 L of dialysis buffer [50 mM Tris HCl (pH 7.6), 250 mM NaCl, 20 mM β-mercaptoethanol and 0.1 mM EDTA] using 8000 Da cutoff dialysis tubing. Typically a yield of 8–10 mg of purified protein was obtained per liter of culture inoculated.
Protein Characterization
All proteins were analyzed by ESI-MS and were prepared by precipitation using one volume of TCA for every four volumes of protein. Samples were incubated for 10 min on ice, centrifuged at 4°C for 5 min at 14 krpm on a table-top centrifuge, the supernatant removed, the pellet washed with 200 μL of cold acetone and washing repeated. The protein pellet was dried at 95°C for 5 min, resuspended in a 50% acetonitrile and 1% formic acid solution at a protein concentration of 10 μM. The samples were analyzed with a Waters Micromass Q-ToF-2 mass spectrometer operating in positive-ion mode.
Far-UV circular dichroism (CD) spectra were obtained on a JASCO 815 spectropolarimeter using a 2 mm path length cell. The scans were performed at 20°C, by averaging 5 wavelength scans from 260 to 200 nm (1 nm bandwidth) in 0.2 nm steps at a rate of 20 nm/min, and 1 sec response at a protein concentration of 5 μM in CD buffer [50 mM potassium phosphate (pH 7.5), 75 mM NaCl and 2 mM dithiothreitol (DTT)].
Thermal denaturation curves were obtained by monitoring the change in mdeg at 222 nm (corresponding to the α-helical content of the protein). The heating rate was set at 15°C/h using a Peltier-type temperature controller, which ranged from 40–70°C. The Tm was obtained by noting the temperature at which the 1st derivative of the denaturation plot was the highest.
Fluorescence spectra were obtained using a Varian Cary Eclipse Fluorescence Spectrophotometer in a 10 mm quartz cuvette with 3.5 μM protein in CD buffer at room temperature. Excitation and emission slits were set at 5 nm with a detector voltage of 650 and the spectra were the average of 10 accumulations. Tyrosine and tryptophan fluorescence were obtained using an excitation wavelength of 280 nm and recording the emission spectrum between 300 and 400 nm. To monitor the tryptophan fluorescence solely an excitation wavelength of 295 nm was used and the emission spectrum was monitored between 300 and 400 nm.
Denaturing Polyacrylamide Gel Electrophoresis Assay of O6-dG-Alkyl-O6-dG Interstrand Cross-Link Repair
The cross-linked oligonucleotides XL4 and XL7 as well as the control DNA (non-cross-linked duplex) were labeled at the 5′-end using [γ-32P]ATP. 10 μL reaction volumes were used for the labeling of the DNA. The reaction mixture was comprised of 1x T4 PNK buffer (Fermentas), 21 μM oligonucleotides, 10 μM cold-ATP, 1 μL [γ-32P]ATP (10 μCi/μL) and 5 units of T4 PNK. The reaction mixture was incubated at 37°C for 1 h and terminated by placing the reaction in boiling water for 10 min. 60 pmol of wild-type and mutant hAGT protein was incubated with 2 pmol of labeled DNA in a total reaction volume of 15 μL comprised of hAGT buffer [80 mM Tris HCl (pH 7.6), 0.1 mM EDTA and 5 mM DTT]. The reactions were incubated at 37°C for 14 to 16 h with similar results indicating that the reaction was complete by 14 h. Denaturing PAGE in 7 M urea was used to analyze the products. The reaction was terminated by adding 18.2 μL of stop reaction buffer [81 mM Tris-HCl, 81 mM boric acid, 1.8 mM EDTA and 1% SDS (pH 8.0) in 80% formamide] to the reaction tube and heating as above. The samples were loaded onto 14 cm × 16 cm, 17% polyacrylamide gels (19:1) in the presence of 7 M urea. The gels were run using 1x TBE [89 mM Tris-HCl, 89 mM boric acid, 2 mM EDTA (pH 8.0)] for 1.5 h at 700 V, which heated the gels to 40°C. After electrophoresis, the gels were covered in Saran wrap and exposed to a storage phosphor screen. The image was captured using a Typhoon 9400 (GE Healthcare, Piscataway, NJ) and the counts of the radiolabelled products quantified using ImageQuant™ (Amersham Biosciences). A control lane was used as a standard in order to quantitate the other bands on the gel. For the repair time course assay of the ICL a 150 μL master mix composed of 600 pmol of wild-type hAGT and 20 pmol of DNA substrate in hAGT buffer was prepared. The sample was placed at 37°C. At every time point 7.5 μL (1 pmol of DNA) was removed from the master mix and placed in a tube with 9.1 μL of stop reaction buffer and terminated as above.
Binding Studies of C145S hAGT Using Protein Titration
Reaction tubes consisting of 5 nM DNA and increasing C145S hAGT concentrations (ranging from 1 to 35.69 μM) were prepared in a total solution volume of 20 μl of binding buffer [10 mM Tris-HCl (pH 7.6), 5 mM DTT and 0.1 mM EDTA]. The samples were allowed to equilibrate on ice for 30 min. Triplicate samples were incubated for longer (45 min and 2 h) and yielded similar results indicating that equilibrium had been attained. Each sample (0.1 pmol) was loaded on 14 cm × 16 cm, 4°C, preequilibrated native 6% polyacrylamide (75:1) gels. The gels were electrophoresed in 10 mM Tris acetate (pH 7.6) and 0.25 mM EDTA buffer at 125 V for 1 h. On completion, the gels were covered and processed as described above.
Estimates of the monomeric dissociation constant (Kd) and the cooperativity factor (n) were obtained from the electrophoretic mobility shift assays as explained previously.48 The binding of n moles of hAGT protein [P] to 1 mole of cross-linked DNA [D] follows the equation: nP + D ↔ PnD. Isolating the variables and taking the logarithm of the equation yields:
Plotting log [PnD]/[P] as a function of log [P] gives a slope equal to n. At half-saturation: log [PnD]/[P] = 0 and observed Kd can be obtained from the relation observed Kd = −n ln [P]. The monomeric Kd can be obtained by taking the nth root of the observed Kd.
Colony forming assay in CHO cells treated with busulfan and hepsulfam
CHO cells transfected with either the empty pCMV vector or pCMV-hAGT were grown in α-MEM media (Gibco-Invitrogen, Carlsbad, CA) containing 26 mM NaHCO3, 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, CA), 100 U/ml penicillin and 100μg/ml streptomycin. The cells were maintained by seeding at 1×105 cells/25 cm2 flask at weekly intervals. For colony forming assays, cells were plated at a density of 106 per 25 cm2 flask and 24 h later were treated with different concentrations of busulfan (Sigma, St Louis, MO) or hepsulfam (NCI, Frederick, MD) for 4 h. The cells were washed with phosphate buffered saline (PBS) twice, and then the medium was replaced with fresh medium and the cells allowed to grow for another 16–18 h. They were then replated at densities of 250–1000 cells per 25 cm2 flask and grown for 6–8 days until discrete colonies had formed. The colonies were washed with 0.9% saline solution, stained with 0.5% crystal violet and counted.
Supplementary Material
Acknowledgments
We thank the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canada Foundation for Innovation (CFI), the Canada Research Chair (CRC) program (to CJW) and grant CA-018137 from the NCI, US Public Health Service (to AEP) for financial support of this project. FPM is the recipient of graduate fellowships from NSERC, the Fonds Québécois de la Recherche sur la Nature et les Technologies (FQRNT) and Groupe de Recherche Axé sur la Structure des Protéines (GRASP). We also thank Mr. Nadim Saadeh and Dr. Bruce Lennox of McGill University for ESI-MS analysis.
Footnotes
Electronic Supplementary Information (ESI) available: [1H NMR and 31P NMR of compounds 6a, 6b, 7a and 7b, HPLC chromatorgraphs and MS spectra for oligonucleotide characterization and repair and binding data of hAGT to the ICL]. See DOI: 10.1039/b000000x/
References
- 1.Dronkert ML, Kanaar R. Mutat Res. 2001;486:217. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 2.Rajski SR, Williams RM. Chem Rev. 1998;98:2723. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 3.Noll DM, Mason TM, Miller PS. Chem Rev. 2006;106:277. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alzeer J, Schärer OD. Nucleic Acids Res. 2006;34:4458. doi: 10.1093/nar/gkl587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilds CJ, Xu F, Noronha AM. Chem Res Toxicol. 2008;21:686. doi: 10.1021/tx700422h. [DOI] [PubMed] [Google Scholar]
- 6.Angelov T, Guainazzi A, Schärer OD. Org Lett. 2009;11:661. doi: 10.1021/ol802719a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens K, Madder A. Nucleic Acids Res. 2009;37:1555. doi: 10.1093/nar/gkn1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali MM, Imoto S, Li Y, Sasaki S, Nagatsugi F. Bioorg Med Chem. 2009;17:2859. doi: 10.1016/j.bmc.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Taniguchi Y, Kurose Y, Nishioka T, Nagatsugi F, Sasaki S. Bioorg Med Chem. 2010;18:2894. doi: 10.1016/j.bmc.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Huang H, Kim HY, Kozekov ID, Cho YJ, Wang H, Kozekova A, Harris TM, Rizzo CJ, Stone MP. J Am Chem Soc. 2009;131:8416. doi: 10.1021/ja809543j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noll DM, Webba da Silva M, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS. Biochemistry. 2006;44:6764. doi: 10.1021/bi050014n. [DOI] [PubMed] [Google Scholar]
- 12.Ravdin PM, Havlin KA, Marshall MV, Brown TD, Koeller JM, Kuhn JG, Rodriguez G, Von Hoff DD. Cancer Res. 1991;51:6268. [PubMed] [Google Scholar]
- 13.Streeper RT, Cotter RJ, Colvin ME, Hilton J, Colvin OM. Cancer Res. 1995;55:1491. [PubMed] [Google Scholar]
- 14.Haines JA, Reese CB, Lord T. J Chem Soc. 1962;25:5281. [Google Scholar]
- 15.Robins RK, Townsend LB. J Am Chem Soc. 1963;85:252. [Google Scholar]
- 16.Wilds CJ, Booth JD, Noronha AM. Tetrahedron Letters. 2006;47:9125. [Google Scholar]
- 17.Khan O, Middleton MR. Expert Opin Investig Drugs. 2007;16:1573. doi: 10.1517/13543784.16.10.1573. [DOI] [PubMed] [Google Scholar]
- 18.Himmelsbach F, Schulz BS, Trichtinger T, Charubala R, Pfleiderer W. Tetrahedron. 1984;40:59. [Google Scholar]
- 19.Shibata T, Glynn N, McMurry TBH, McElhinney RS, Margison GP, Williams DM. Nucleic Acids Res. 2006;34:1884. doi: 10.1093/nar/gkl117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitsunobu O. Synthesis. 1981;1:1. [Google Scholar]
- 21.Pegg AE. Mutation Research. 2000;462:83. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 22.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. Nature Structural & Molecular Biology. 2004;11:714. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 23.Srivenugopal KS, Yuan XH, Friedman HS, Ali-Osman F. Biochemistry. 1996;35:1328. doi: 10.1021/bi9518205. [DOI] [PubMed] [Google Scholar]
- 24.Coulter R, Blandino M, Tomlinson JM, Pauly GT, Krajewska M, Moschel RC, Peterson LA, Pegg AE, Spratt TE. Chem Res Toxicol. 2007;20:1966. doi: 10.1021/tx700271j. [DOI] [PubMed] [Google Scholar]
- 25.Buggia I, Locatelli F, Regazzi MB, Zecca M. Ann Pharmacother. 1994;28:1055. doi: 10.1177/106002809402800911. [DOI] [PubMed] [Google Scholar]
- 26.Tong WP, Ludlum DB. Biochim Biophys Acta. 1980;608:174. doi: 10.1016/0005-2787(80)90145-8. [DOI] [PubMed] [Google Scholar]
- 27.Iwamoto T, Hiraku Y, Oikawa S, Mizutani H, Kojima M, Kawanishi S. Cancer Sci. 2004;95:454. doi: 10.1111/j.1349-7006.2004.tb03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayakawa Y, Kato H, Uchiyama M, Kajino H, Noyori R. J Org Chem. 1986;51:2400. [Google Scholar]
- 29.Braich RS, Damha MJ. Bioconjugate Chem. 1997;8:370. doi: 10.1021/bc9700300. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Q, Delaney MO, Greenberg MM. Bioorg Med Chem Lett. 2001;11:1105. doi: 10.1016/s0960-894x(01)00161-5. [DOI] [PubMed] [Google Scholar]
- 31.Damha MJ, Ogilvie KK. In: Methods in Molecular Biology. Agrawal S, editor. Vol. 20. The Humana Press, Inc; Totowa, NJ: 1993. pp. 81–114. [DOI] [PubMed] [Google Scholar]
- 32.Gaffney BL, Marky LA, Jones RA. Biochemistry. 1984;23:5686. doi: 10.1021/bi00319a004. [DOI] [PubMed] [Google Scholar]
- 33.Gaffney BL, Jones RA. Biochemistry. 1989;28:5881. doi: 10.1021/bi00440a026. [DOI] [PubMed] [Google Scholar]
- 34.Kuzmich S, Marky LA, Jones RA. Nucleic Acids Res. 1993;11:3393. doi: 10.1093/nar/11.10.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson WC., Jr . In: Methods of Biochemical Analysis. Glick D, editor. Vol. 31. Wiley; New York: 1985. p. 61. [DOI] [PubMed] [Google Scholar]
- 36.Johnson WC., Jr . In: Circular Dichroism and the Conformational Analysis of Biomolecules. Fasman GD, editor. Plenum Press; New York: 1996. p. 433. [Google Scholar]
- 37.Patel DJ, Shapiro L, Kozlowski SA, Gaffney BL, Jones RA. Biochemistry. 1986;25:1027. doi: 10.1021/bi00353a012. [DOI] [PubMed] [Google Scholar]
- 38.Rasimas JJ, Pegg AE, Fried MG. J Biol Chem. 2003;278:7973. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- 39.Crone TM, Pegg AE. Cancer Res. 1993;53:4750. [PubMed] [Google Scholar]
- 40.Crone TM, Goodtzova K, Edara S, Pegg AE. Cancer Res. 1994;54:6221. [PubMed] [Google Scholar]
- 41.Daniels DS, Mol CD, Arvai AS, Kanugula S, Pegg AE, Tainer JA. EMBO J. 2000;19:1719. doi: 10.1093/emboj/19.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanugula S, Pegg AE. Biochem J. 2003;375:449. doi: 10.1042/BJ20030809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodtzova K, Crone TM, Pegg AE. Biochemistry. 1994;33:8385. doi: 10.1021/bi00194a001. [DOI] [PubMed] [Google Scholar]
- 44.Fang Q, Noronha AM, Murphy SP, Wilds CJ, Tubbs JL, Tainer JA, Chowdhury G, Guengerich FP, Pegg AE. Biochemistry. 2008;47:10892. doi: 10.1021/bi8008664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasimas JJ, Pegg AE, Fried MG. J Biol Chem. 2003;278:7973. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- 46.Rasimas JJ, Dalessio PA, Ropson IJ, Pegg AE, Fried MG. Protein Sci. 2004;13:301. doi: 10.1110/ps.03319404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coulter R, Blandino M, Tomlinson JM, Pauly GT, Krajewska M, Moschel RC, Peterson LA, Pegg AE, Spratt TE. Chem Res Toxicol. 2007;20:1966. doi: 10.1021/tx700271j. [DOI] [PubMed] [Google Scholar]
- 48.Fried MG, Kanugula S, Bromberg JL, Pegg AE. Biochemistry. 1996;35:15295. doi: 10.1021/bi960971k. [DOI] [PubMed] [Google Scholar]
- 49.Duguid EM, Rice PA, He C. J Mol Biol. 2005;350:657. doi: 10.1016/j.jmb.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 50.Puglisi JD, Tinoco IJ. Methods in Enzymology. 1989;180:304. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.