Abstract
A profile of transcript abundances from multiple genes constitutes a molecular signature if the expression pattern is unique to one cell type. Here we measure mRNA copy numbers per cell by normalizing per million copies of 18S rRNA and identify 6 genes (TIE1, KDR, CDH5, TIE2, EFNA1 and MYO5C) out of 79 genes tested as excellent molecular signature markers for endothelial cells (ECs) in vitro. The selected genes are uniformly expressed in ECs of 4 different origins but weakly or not expressed in 4 non-EC cell lines. A multi-gene transcriptional profile of these 6 genes clearly distinguishes ECs from non-ECs in vitro. We conclude that (i) a profile of mRNA copy numbers per cell from a well-chosen multi-gene panel can act as a sensitive and accurate cell type signature marker, and (ii) the method described here can be applied to in vivo cell fingerprinting and molecular diagnosis.
Keywords: Molecular signature, Gene expression profiling, mRNA copy numbers, Endothelial cells
Introduction
Cells of a specific lineage but different subtype may share common expression levels of a number of gene products, due to conserved regulatory molecules in the development system (Ivanova et al. 2002; Whitfield et al. 2006). Meanwhile, a significant fraction of the genome is differentially expressed in different lineages, so that the expression patterns of some of those genes may uniquely fingerprint a particular lineage (Deng et al. 2006; Ramalho-Santos et al. 2002). Genes that fit both categories—common and abundant expression in all cells of one lineage and weak expression in cells of other lineages—would be excellent cell signature markers. Examples of such signature markers include CD4, CD3 and CD8 for T-cells (Haynes et al. 1990), and CD19 and CD20 for B-Cells (Kehrl et al. 1994). A method that reliably measures the abundance of a gene product, either protein or transcript, is capable of identifying such signature markers. However, protein quantification can be challenging, since not all proteins have antibodies suitable for quantification (Haab et al. 2001). Transcript quantification is more feasible, depending chiefly on consistent primer amplification during polymerase chain reactions (PCR) (Bustin 2002; Shih and Smith 2005).
Real-time PCR is regarded as the gold standard for nucleic acid quantification (Bustin 2002; Klein 2002). We pioneered a high throughput approach for multi-gene transcriptional profiling (MGTP) using real-time PCR with a master cDNA template to measure mRNA copy numbers from many genes (Shih and Smith 2005). In this report, we investigate the utility of MGTP for the discovery of multi-gene cell signature markers. First, we quantified copy numbers per cell of 18S rRNA, the most uniformly expressed housekeeping gene (Aerts et al. 2004; Bas et al. 2004), enabling translation of MGTP abundances into per cell quantities. We then looked for signature markers of endothelial cells (ECs) from 79 genes and identified 6 genes (TIE1, KDR, CDH5, TIE2, EFNA1 and MYO5C; all genes are identified with NCBI standard symbols) that are uniformly expressed in ECs and whose abundances clearly distinguish ECs from non-ECs. Our results suggest that multi-gene per-cell mRNA profiles will be a valuable tool for the discovery of cell signature markers and for fingerprinting cell lineages.
Materials and methods
Cell lines and cell culture
This study used 6 human primary cell lines: HUVEC (umbilical vein EC), HPAEC (pulmonary artery EC), HCAEC (coronary artery EC), HDMEC (dermal microvascular EC), HCASMC (coronary artery smooth muscle cells), and HDF (dermal fibroblasts); and 13 established cell lines: HEK-293A (embryonic kidney cells), PC3 (prostate cancer), Colo201 (colon carcinoma), MDA-MB231 (breast cancer), M21 (melanoma), HT-1080 (fibrosarcoma), SW620 (colon carcinoma), U937 (leukemic monocyte) and glioblastoma cell lines (U59, U87, U118, U138, and U373). HDMEC and HDF were acquired from our core facility. HUVEC, HPAEC, and HCAEC were purchased from Clonetics (East Rutherford, NJ), HEK-293A from Invitrogen (Bethesda, MD), and the other cell lines from ATCC (Manassa, VA). All ECs and HCASMC were cultured in EC growth medium-2-MV and smooth muscle growth medium (Clonetics) to 85–95% confluency with 5% CO2. All other cell types were cultured in DMEM containing high glucose with 10% FBS and used within the first 3 weeks. At every passage, the cells in each plate were split into five.
RNA isolation and cDNA preparation
Total RNA was prepared using the RNeasy RNA extraction kit with on-column DNase digestion (Qiagen, Chatsworth, CA) following the manufacturer’s instructions. The DNase-treated RNA (100 ng) was then converted into cDNA using random primers and SuperScript III (Invitrogen) and stored at −80 °C.
Primer design and validation
Gene-specific areas of the sequence were selected for primer design based on NCBI Blast searches to eliminate regions with homologies to other genes as described previously (Shih and Smith 2005). For genes that have different transcriptional variants, primers were designed from the common sequence area and synthesized by IDT (Coralville, IA). Primers meet the standard primer design parameters contained in the guidelines of the Primer Express oligo design software (ABI, Foster City, CA). Where feasible, the 5′-end of each primer set does not have A/T or G/C complementarity and the 3′-end of the primers finishes in G or C, but 3 out of 5 of the 3′-end nucleotides are A or T.
Quantitative real-time PCR
The SYBR Green I assay and the ABI 7500 Fast Real-Time PCR System were used for detecting PCR products from cDNA samples as described previously (Shih et al. 2002). PCR reactions for each cDNA sample were performed in duplicate and copy numbers were calculated using standard curves generated from a master template (Shih and Smith 2005). For each data point, mean and standard deviation (SD) were calculated from 3 cDNA samples prepared in 3 separate wells of a 6-well culture plate. In different real-time PCR runs, it is common to obtain gene expression variations within a range of ±20% even after careful monitoring and calibration of each experimental step to ensure RNA integrity and cDNA yield (Shih and Smith 2005). In this paper, we will classify gene expression as uniform if the expression has a coefficient of variation (CV; the ratio of the SD to the mean) less than 30%.
18S rRNA quantification
All cell lines were cultured to approximately 85–90% confluency in a duplicate set of wells where one set was used to extract total RNA and another set was used to calculate total cell numbers. The recovered total RNA was divided by total cell number to acquire total RNA weight/cell. The cDNA derived from 1 ng total RNA was then loaded to each of multiple wells and we measured 18S rRNA copies/1 ng total RNA, enabling calculation of 18S rRNA copies/cell.
Immunocytochemistry
HUVECs were plated on an 8 mm glass disc (Fisher Scientific) and fixed with 4% paraformaldehyde at 25 °C for 5 min, then blocked with PBS solution containing 1% FBS. The cells were stained for 2 h at 25 °C with primary antibodies against TIE1 (Abcam, Cambridge, MA), CDH5 (Santa Cruz Biotechnology, Santa Cruz, CA), KDR (Cell Signaling Technology (Danvers, MA), or EFNA1 (Abcam). Alexa Fluor-594 conjugated anti-rabbit, anti-goat, or anti-mouse antibodies were used as secondary antibodies and applied for 2 h at 25 °C. Rhodamine-conjugated phalloidin for 45 min at 37 °C to stain F-actin. ProLong Gold antifade reagent with DAPI (Invitrogen) was used to mount the slide.
Results and discussion
Measurement of 18S rRNA copy numbers per cell in cultured cell lines in vitro
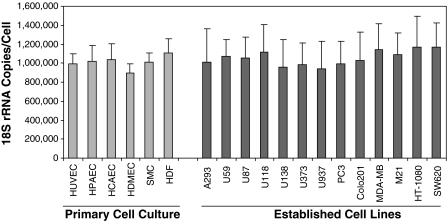
Others have reported that 18S rRNA is uniformly expressed across cell types and exhibits minimal variation in response to stimuli in different cell types; hence, it is a superior normalizer (Aerts et al. 2004; Bas et al. 2004). We quantified 18S rRNA copy numbers per cell in cultured cell lines in vitro. Nineteen cell lines were selected (Fig. 1), 13 of which were established cell lines and 6 of which were passage-5 primary cell cultures. As shown in Fig. 1, all 19 cell lines showed comparable levels of 18S rRNA expression with copy numbers in specific cell lines ranging from 897,858 ± 95,342 to 1,169,507 ± 320,855 per cell and with average copies over all 19 cell lines averaging 1,040,947 ± 223,924 per cell. We conclude that 18S rRNA is expressed uniformly in cultured cells in vitro, regardless of whether they are primary or established cell cultures, at about a million copies of 18S rRNA per cell. As 18S-, 28S-, and 5.8S-rRNA are synthesized by cleavage from a common 13 kb transcription unit, the total number of rRNA copies per cell are therefore at least 3 million copies as 5S rRNA is transcribed separately. This estimate is somewhat lower than that of Sentenac, who estimated that 10 million copies of total rRNA are required in eukaryotes (Sentenac 1985).
Fig. 1.
Measurements of 18S rRNA copy numbers per cell in cultured cell lines in vitro. Six primary cell lines (passage-5) and thirteen established cell lines were used to measure 18S rRNA copies/cell (see “Materials and methods”). Three individual RNA samples prepared from three different cultures of each cell line were used to calculate mean ± SD. All nineteen in vitro cultured cells expressed about one million copies of 18S rRNA/cell
To obtain target gene abundances per cell, we measure both 18S rRNA and target gene copy numbers in a given sample and normalize target gene abundance per million copies of 18S rRNA. With this approach, we typically obtain approximately 1,000 β-actin mRNA copies per cell for in vitro cultured cells, which is similar to the number per cell reported by others (Bjarnadottir and Jonsson 2005), and frequently obtain 1–15 mRNA copies/cell for many biologically active genes. Hereafter, we will regard expression levels greater than 15 copies/cell as a high expression level, expression levels between 1–15 copies/cell as a moderate expression level, and expression levels less than 1 copy/cell as a low expression level.
Based on unpublished data from simultaneous MGTP, flow cytometry, and Western blot studies of over 60 genes, we find that samples with more than 15 mRNA copies/cell of a membrane bound gene are consistently measurable by flow cytometry, whereas samples with of less than 1 copy/cell are generally detectable only by Western blot. Western blot can typically detect proteins when the mRNA abundance is above 0.1 copies/cell, an mRNA level that indicates that about 10% of the cells express a single copy of the transcript of interest at any given moment. Poor correlation between mRNA abundances and protein abundances measured by flow cytometry is rare but can occur; for membrane bound proteins this usually indicates cleavage from membrane surface as in the case of CD23 (Lemieux et al. 2007) but could indicate protein internalization as in the case of KDR (Dougher and Terman 1999).
A search for endothelial cells (ECs) EC cell signature markers using multi-gene transcriptional profiling (MGTP)
ECs are heterogeneous, encompassing a variety of phenotypes that are differentially regulated in space and time (Aird 2005). We searched for excellent EC signature markers by looking for genes that are uniformly (coefficient of variation CV less than 30%; see “Quantitative real-time PCR” in “Materials and methods”) and moderately to highly (>1 mRNA copy/cell) expressed in ECs of different origin [artery (HPAEC and HCAEC), vein (HUVEC), and microvasculature (HDMEC)] but weakly or not expressed (<1 mRNA copy/cell) in all four types of non-ECs we selected (HDF, HCASMC, 293A, and U937).
We selected 79 marker candidates, consisting of (i) 16 genes whose proteins have been widely used to isolate ECs (ENG, MCAM, and PECAM1; NCBI, National Center for Biotechnology Information, symbols are used throughout the paper), or are widely studied for their molecular functions in ECs (CD34, ESM1, FLT1, KDR, ICAM1, CDH5, SELE, SELP, THBD, TIE1, TIE2, VCAM1, and VWF), and (ii) 63 genes that were identified through a gene chip study of cultured ECs of different origin (Chi et al. 2003).
We measured mRNA copy numbers per cell for all 79 genes in 24 EC samples (6 total RNA samples from different cultures of each of HUVEC, HPAEC, HCAEC and HDMEC). Results were divided into 3 categories based on mean copy number in EC subtypes (Table 1): (i) 46 genes had mean expression >1 copy/cell in all four EC subtypes; (ii) 20 genes had mean expression >1 copy/cell in at least one EC subtype; and (iii) 13 genes had mean expression <1 copy/cell in all four EC subtypes. This last group was excluded from further study.
Table 1.
Expression abundances of seventy-nine genes in ECs
| (i) Mean > 1 for all ECs | (ii) Mean < 1 for some ECs | (iii) Mean < 1 for all ECs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Symbol | Mean | SD | Symbol | Mean | SD | Symbol | Mean | SD | Symbol | Mean | SD |
| SPARC | 379.23 | 121.19 | PGF | 11.28 | 5.46 | CD36 | 6.39 | 11.49 | ROBO1 | 0.62 | 0.38 |
| CLU | 151.66 | 84.23 | EVA1 | 10.58 | 4.84 | GMFG | 5.37 | 6.83 | GDF | 0.57 | 0.10 |
| PECAM | 109.10 | 15.60 | JAM3 | 9.59 | 3.89 | ITM2A | 5.22 | 3.55 | EGFR | 0.38 | 0.31 |
| FABP4 | 90.11 | 189.49 | FLT1 | 8.38 | 3.58 | EDNRB | 3.01 | 5.19 | ITGA4 | 0.29 | 0.32 |
| MCAM | 73.31 | 34.02 | SORBS2 | 6.80 | 4.53 | CETP | 2.83 | 4.84 | SFRP1 | 0.26 | 0.24 |
| ALDH1A1 | 56.90 | 21.47 | DHCR24 | 5.75 | 2.10 | APOD | 2.67 | 1.86 | GAL | 0.21 | 0.11 |
| CDH5 | 56.64 | 9.21 | ANGPT2 | 5.25 | 5.25 | NOTCH4 | 2.32 | 1.35 | MYO7A | 0.11 | 0.17 |
| FABP5 | 53.83 | 36.02 | NAV1 | 4.89 | 1.49 | ITGA1 | 2.05 | 3.19 | FABP7 | 0.10 | 0.21 |
| VWF | 41.65 | 21.27 | TIE2 | 4.66 | 0.95 | SELE | 1.96 | 1.00 | SIX3 | 0.04 | 0.07 |
| CD34 | 37.04 | 39.10 | THBD | 4.61 | 1.80 | CEACAM1 | 1.75 | 2.88 | OGN | 0.02 | 0.01 |
| COL4A1 | 36.51 | 16.13 | BTD | 4.51 | 0.84 | HEY2 | 1.55 | 1.53 | LEFTY2 | 0.01 | 0.01 |
| COL4A2 | 35.71 | 13.21 | IL1R1 | 4.51 | 2.32 | ITGB4 | 1.52 | 1.78 | CALCR | 0.01 | 0.01 |
| ESM1 | 33.64 | 11.53 | FGF2 | 4.34 | 0.77 | SMO | 1.45 | 1.29 | KRT7 | 0.00 | 0.01 |
| ENG | 33.39 | 4.37 | SQLE | 4.29 | 1.37 | NAV3 | 1.25 | 0.39 | |||
| KDR | 22.75 | 3.39 | SSH1 | 4.24 | 1.32 | LIPG | 1.24 | 0.92 | |||
| COL5A2 | 21.35 | 6.91 | CD44 | 4.01 | 1.99 | MYLK | 1.11 | 0.54 | |||
| TIE1 | 20.83 | 4.08 | MYO1B | 3.98 | 1.93 | SELP | 0.86 | 0.81 | |||
| EFNA1 | 18.22 | 1.85 | HMGCS1 | 3.27 | 1.72 | TGFA | 0.79 | 1.27 | |||
| COL5A1 | 16.24 | 10.05 | VCAM1 | 2.60 | 1.27 | VAV3 | 0.71 | 1.23 | |||
| ICAM1 | 14.20 | 10.30 | MYO5C | 2.16 | 0.62 | IL6R | 0.63 | 0.36 | |||
| NRP2 | 12.35 | 3.60 | VEGFC | 1.86 | 0.92 | ||||||
| EPHB4 | 12.23 | 2.56 | PDGFD | 1.82 | 0.65 | ||||||
| ABLIM1 | 11.54 | 4.71 | PLA2G12A | 1.74 | 0.26 | ||||||
The seventy-nine genes were divided into three categories based on whether expression was greater or less than one copy per cell in all, some, or non-EC samples. The sixteen genes whose proteins are known to be expressed in ECs are highlighted in bold
We measured expression of the remaining 66 genes in 24 non-EC samples (6 total RNA samples from different cultures of each of HCASMC, U937, HEK293, and HDF) and compared the expression level with ECs. Thirty-one genes met two requirements for possible use as EC lineage markers: a p-value <0.00075 in the student t-test to distinguish ECs from non-ECs, which corresponds to a p-value of 0.05 subject to the Bonferroni correction for 66 genes; and mean expression in ECs greater than mean expression in non-ECs. These 31 genes are listed in Table 2. Fourteen were from the pre-selected genes whose proteins are known to be expressed in ECs (highlighted in bold in Table 2) and seventeen were from the Chi et al. (2003) gene chip study. The two eliminated known EC genes, ICAM1 and VCAM1, were more abundantly expressed in some non-ECs than in ECs, notably in smooth muscle cells (data not shown).
Table 2.
EC signature marker candidates
| EC | Non-EC | EC versus non-EC | ||||||
|---|---|---|---|---|---|---|---|---|
| Symbol | Mean | SD | CV | Mean | SD | p value | Fold | |
| (A) Markers expressed at less than 1 copy per cell in all non-EC | ||||||||
| Unform (CV < 30%) | CDH5 | 56.6 | 9.2 | 16.3 | 0.3 | 0.4 | 8.0E-32 | 203.1 |
| KDR | 22.6 | 3.4 | 15.1 | 0.1 | 0.1 | 2.6E-33 | 310.2 | |
| TIE1 | 20.8 | 4.1 | 19.6 | 0.0 | 0.1 | 2.2E-28 | 650.3 | |
| EFNA1 | 18.2 | 1.8 | 10.2 | 0.6 | 0.4 | 1.4E-39 | 27.0 | |
| TIE2 | 4.7 | 1.0 | 20.5 | 0.1 | 0.2 | 1.2E-26 | 31.9 | |
| MYO5C | 2.2 | 0.6 | 28.6 | 0.2 | 0.2 | 2.8E-19 | 10.8 | |
| Variable (CV > 30%) | VWF | 41.6 | 21.3 | 51.1 | 0.0 | 0.0 | 1.2E-13 | 3,338.1 |
| CD34 | 39.2 | 32.3 | 82.4 | 0.2 | 0.3 | 3.8E-07 | 217.1 | |
| PGF | 11.7 | 6.0 | 51.6 | 0.2 | 0.2 | 4.1E-12 | 47.0 | |
| EVA1 | 10.6 | 4.8 | 45.7 | 0.2 | 0.3 | 8.5E-14 | 54.3 | |
| SORBS2 | 6.8 | 4.5 | 66.6 | 0.2 | 0.2 | 5.6E-09 | 36.0 | |
| ANGPT2 | 5.2 | 5.3 | 100.1 | 0.0 | 0.0 | 1.3E-05 | 354.5 | |
| ITM2A | 5.2 | 3.5 | 67.9 | 0.2 | 0.3 | 1.3E-08 | 24.0 | |
| APOD | 2.7 | 1.9 | 69.7 | 0.0 | 0.0 | 1.1E-08 | 81.7 | |
| NOTCH4 | 2.32 | 1.35 | 58.3 | 0.02 | 0.02 | 9.8E-11 | 120.7 | |
| SELE | 2.0 | 1.0 | 50.9 | 0.1 | 0.1 | 4.8E-12 | 30.2 | |
| ITGB4 | 1.5 | 1.8 | 116.9 | 0.2 | 0.3 | 5.4E-04 | 9.7 | |
| SELP | 0.9 | 0.8 | 93.3 | 0.0 | 0.1 | 7.0E-06 | 31.1 | |
| (B) Markers expressed at greater than 1 copy per cell in some non-EC | ||||||||
| Unform (CV < 30%) | PECAM1 | 109.1 | 15.6 | 14.3 | 1.7 | 2.6 | 8.1E-34 | 64.2 |
| ENG | 33.4 | 4.4 | 13.1 | 5.0 | 3.6 | 3.6E-28 | 6.6 | |
| ESM1 | 32.6 | 11.3 | 34.6 | 1.0 | 1.1 | 7.9E-18 | 34.4 | |
| NRP2 | 12.4 | 3.6 | 29.2 | 4.3 | 4.3 | 9.1E-09 | 2.9 | |
| EPHB4 | 12.2 | 2.6 | 20.9 | 2.4 | 1.4 | 4.0E-21 | 5.1 | |
| BTD | 4.5 | 0.8 | 18.6 | 2.4 | 2.1 | 4.7E-05 | 1.9 | |
| Variable (CV > 30%) | CLU | 151.7 | 84.2 | 55.5 | 35.4 | 42.9 | 2.7E-07 | 4.3 |
| MCAM | 73.3 | 34.0 | 46.4 | 1.6 | 1.5 | 1.5E-13 | 45.9 | |
| ALDH1A1 | 56.9 | 21.5 | 37.7 | 2.1 | 2.7 | 2.7E-16 | 27.3 | |
| FABP5 | 53.8 | 36.0 | 66.9 | 15.9 | 16.3 | 2.4E-05 | 3.4 | |
| ABLIM1 | 11.5 | 4.7 | 40.9 | 2.1 | 1.5 | 3.4E-12 | 5.4 | |
| THBD | 8.5 | 7.8 | 92.4 | 1.8 | 2.0 | 1.8E-04 | 4.8 | |
| FLT1 | 5.5 | 3.4 | 62.2 | 1.8 | 2.0 | 3.1E-05 | 3.1 | |
The 31 EC signature marker candidates were divided into two sub-tables: markers with gene expression in non-ECs always less than one copy/cell were gathered in sub-table (A), and the rest gathered in sub-table (B). In each sub-table, genes are ordered by mean expression in ECs. For both sub-tables, the selected genes are further divided into two groups, the top group having CV less than 30% and bottom group CV greater than 30%
SD standard deviation, CV coefficient of variation in ECs, p-value from student t-test, fold ratio of mean EC expression to mean non-EC expression
The 33 genes from Chi et al. (2003) which were disqualified as good EC signature marker candidates fell into three categories of exclusion: (i) the mean expression of 15 genes (CD44, DHCR24, FGF2, HMGCS1, IL6R, MYLK, MYO1B, PLA2G12A, NAV3, SMO, SQLE, SSH1, TGFA, VAV3, and VEGFC) was greater in non-ECs than in ECs; (ii) p-value of 10 genes (CD36, COL5A1, COL5A2, COL4A2, GMFG, IL1R1, ITGA1, NAV1, PDGFD, and SPARC,) was greater than 0.05 between ECs and non-ECs; and (iii) p-value of 8 genes (CEACAM1, CETP, COL4A1, EDNRB, FABP4, HEY2, JAM3, and LIPG) was between 0.05 and 0.00075, making them unable to adequately distinguish ECs from non-ECs after Bonferroni correction.
We further segregated the 31 genes listed in Table 2 into two groups based on their expression in non-ECs. Of the 31 genes, 18 genes are expressed at low levels (<1 copy/cell) in all non-ECs tested; these are listed in Table 2a. Thirteen genes were expressed at moderate levels (>1 copy/cell) in at least one non-EC line; these are listed in Table 2b and are eliminated as the candidates for the search of excellent EC signature markers. PECAM1 and ESM1 can be good EC markers as their expression difference between EC to non-EC are greater than 34-fold.
Finally, we further segregated eighteen genes in Table 2a into two groups based on their uniformity of expression in ECs (CV < 30%). Six genes, 4 pre-selected EC genes (TIE1, KDR, CDH5, and TIE2) and 2 from the Chi el al gene chip study (EFNA1 and MYO5C), met all four criteria of EC signature markers. The criteria were: (i) A high fold difference in ECs over non-ECs. Mean expression was at least 200-fold higher in ECs for TIE1, KDR, and CDH5 and at least ten-fold higher in ECs for TIE2, EFNA1, and MYO5C. (ii) A low p-value to distinguish ECs from non-ECs. All 6 genes distinguished ECs from non-ECs with a p-value of less than 10−19. (iii) Uniform expression in ECs: the CV was less than 30% among ECs. (iv) Low expression in all non-EC samples tested: expression in all of the non-EC lines was less than 1 copy/cell.
Two well-known EC markers, PECAM1 and ENG, were uniformly expressed among the 4 EC subtypes, but were excluded as EC signature markers because they were moderately expressed in at least one non-EC cell line: PECAM1 in U937 cells, ENG in U937, HCASMC, and HDF cells.
Our finding of variable expression in ECs or positive expression in non-ECs for many of the EC markers listed in Table 2 are consistent with others’ proteomic results: (i) ENG is strongly expressed on vascular ECs but weakly expressed on hematopoietic cells, fibroblasts, and smooth muscle cells (Fonsatti et al. 2001), (ii) PECAM1 is expressed on endothelia and also a variety of mature hemopoietic cell types (Davies et al. 1993; Watt et al. 1995), (iii) intact endothelium expresses undetectable or low levels of SELE and VCAM1 (Eppihimer et al. 1996; Kuzu et al. 1993), (iv) ICAM1 is expressed in smooth muscle cells (Couffinhal et al. 1994) and in ECs the expression level varies between different vascular beds and different segments of the vascular loop (Henninger et al. 1997; Kuzu et al. 1993), and (v) VWF is known to be differentially regulated by vascular bed-specific pathways in response to signals derived from the local microenvironment (Aird et al. 1997). This consistency with the results of proteomic studies suggests that measuring mRNA copy numbers per cell by MGTP will facilitate the identification of cell signature biomarkers.
Figure 2a shows an MGTP profile for the 6 EC signature markers, illustrating their ability to distinguish ECs from non-ECs. When HUVEC were mixed with HEK293 cells in varying percentages, as expected, mRNA copy numbers were directly proportional to HUVEC cell percentage verifying that expression is specific to the ECs and unregulated by the presence of the non-ECs (data not shown).
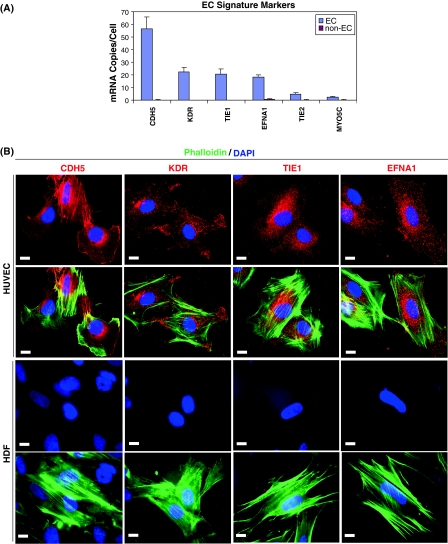
Fig. 2.
EC signature marker expression in endothelial cells. a MGTP profile of the 6 excellent EC signature markers, showing a clear difference in expression in ECs compared to non-ECs. CDH5, KDR, TIE1, and EFNA1 are expressed at high levels (>15 mRNA copies/cell) in all EC subtypes, while TIE2 and MYO5C are expressed at moderate levels (1–10 mRNA copies/cell). b ICC staining of TIE1, KDR, CDH5, and EFNA1 in red, with phalloidin stained F-actin in green and DAPI stained nucleus in blue, in HUVEC and HDF. The bar embedded in each picture indicates a length of 10 μm
Four of the EC signature markers, TIE1, KDR, CDH5, and EFNA1, are highly expressed (>15 copies/cell); they were selected for protein staining in HUVEC and HDF using immunocytochemistry (ICC). As shown in Fig. 2b, all four EC signature markers showed positive ICC staining in HUVEC whereas none showed a positive signal in HDF, as expected given their low mRNA copy numbers in those cells.
Expression of these 6 EC signature markers was consistent over different experimental batches and different donors, passages, and serum starvation (data not shown), suggesting that they are likely to be epigenetically regulated. Such EC-specific gene expression makes these markers ideal for applications like EC specific gene expression control. Indeed, promoters of CDH5 (Gory et al. 1999), KDR (Fadel et al. 1998), TIE1 (Boutet et al. 2001), and TIE2 (Patterson et al. 1995) have already been used for EC-specific gene expression regulation in vivo. In future work, it will be desirable to address whether the various EC signature markers identified here consistently mark the EC lineage in various in vivo microenvironments.
Conclusion
MGTP, a method we developed to quantify mRNA copy numbers (Shih and Smith 2005), enables creation of quantitative gene expression profiles and facilitates comparison of mRNA abundances across genes, samples, and experiments. By normalizing per million copies of 18S rRNA, results can be interpreted as mRNA copies/cell.
This study identified 6 genes (CDH5, KDR, TIE1, TIE2, EFNA1, and MYO5C) out of 79 candidates as excellent EC signature markers based on the following criteria: (1) They distinguished ECs from non-ECs with a p-value in the student t-test of less than 0.00075. (2) Expression was at least 1 mRNA copy/cell in ECs; we regard this as an appropriate minimum abundance for a useful marker, as it is in our experience the mRNA level at which protein products become detectable by flow cytometry. (3) Expression was uniform in ECs (CV < 30%) and below 1 copy/cell in non-ECs; these criteria minimize uncertainty over the source of expression and permit ready estimation of the EC percentage in samples with mixed cell populations.
Multi-gene abundance profiles, such as those provided by MGTP, have several advantages over single-gene marker measurements, especially measurements of fold-change: (1) The statistical reliability of a multi-gene signature increases with the number of markers, and improved statistics may be critical in clinical applications due to the heterogeneity of genomic expression in human populations; (2) Abundance information facilitates elimination of weakly expressed genes from marker panels; and (3) Inter-gene abundance ratios rather than the abundance of single genes may be of critical biological importance. In combination with DNA microarrays, which screen the entire genome, MGTP will facilitate the discovery of signature marker panels whose expression profile is unique to a specific normal or pathological cell type. Such marker panels are likely to be valuable for molecular diagnosis.
Acknowledgments
Grants Supported in part by US Public Health Service grants HL-64402 and P01 CA92644.
References
- Aerts JL, Gonzales MI, Topalian SL. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. Biotechniques. 2004;36:84–86. doi: 10.2144/04361ST04. [DOI] [PubMed] [Google Scholar]
- Aird WC. Spatial and temporal dynamics of the endothelium. J Thromb Haemost. 2005;3:1392–1406. doi: 10.1111/j.1538-7836.2005.01328.x. [DOI] [PubMed] [Google Scholar]
- Aird WC, Edelberg JM, Weiler-Guettler H, Simmons WW, Smith TW, Rosenberg RD. Vascular bed-specific expression of an endothelial cell gene is programmed by the tissue microenvironment. J Cell Biol. 1997;138:1117–1124. doi: 10.1083/jcb.138.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bas A, Forsberg G, Hammarstrom S, Hammarstrom ML. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol. 2004;59:566–573. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- Bjarnadottir H, Jonsson JJ. A rapid real-time qRT-PCR assay for ovine beta-actin mRNA. J Biotechnol. 2005;117:173–182. doi: 10.1016/j.jbiotec.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Boutet SC, Quertermous T, Fadel BM. Identification of an octamer element required for in vivo expression of the TIE1 gene in endothelial cells. Biochem J. 2001;360:23–29. doi: 10.1042/0264-6021:3600023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Chi JT, Chang HY, Haraldsen G, Jahnsen FL, Troyanskaya OG, Chang DS, Wang Z, Rockson SG, et al. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci USA. 2003;100:10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couffinhal T, Duplaa C, Moreau C, Lamaziere JM, Bonnet J. Regulation of vascular cell adhesion molecule-1 and intercellular adhesion molecule-1 in human vascular smooth muscle cells. Circ Res. 1994;74:225–234. doi: 10.1161/01.res.74.2.225. [DOI] [PubMed] [Google Scholar]
- Davies MJ, Gordon JL, Gearing AJ, Pigott R, Woolf N, Katz D, Kyriakopoulos A. The expression of the adhesion molecules ICAM-1, VCAM-1, PECAM, and E-selectin in human atherosclerosis. J Pathol. 1993;171:223–229. doi: 10.1002/path.1711710311. [DOI] [PubMed] [Google Scholar]
- Deng DX, Spin JM, Tsalenko A, Vailaya A, Ben-Dor A, Yakhini Z, Tsao P, Bruhn L, et al. Molecular signatures determining coronary artery and saphenous vein smooth muscle cell phenotypes: distinct responses to stimuli. Arterioscler Thromb Vasc Biol. 2006;26:1058–1065. doi: 10.1161/01.ATV.0000208185.16371.97. [DOI] [PubMed] [Google Scholar]
- Dougher M, Terman BI. Autophosphorylation of KDR in the kinase domain is required for maximal VEGF-stimulated kinase activity and receptor internalization. Oncogene. 1999;18:1619–1627. doi: 10.1038/sj.onc.1202478. [DOI] [PubMed] [Google Scholar]
- Eppihimer MJ, Wolitzky B, Anderson DC, Labow MA, Granger DN. Heterogeneity of expression of E- and P-selectins in vivo. Circ Res. 1996;79:560–569. doi: 10.1161/01.res.79.3.560. [DOI] [PubMed] [Google Scholar]
- Fadel BM, Boutet SC, Quertermous T. Functional analysis of the endothelial cell-specific Tie2/Tek promoter identifies unique protein-binding elements. Biochem J. 1998;330(Pt 1):335–343. doi: 10.1042/bj3300335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonsatti E, Del Vecchio L, Altomonte M, Sigalotti L, Nicotra MR, Coral S, Natali PG, Maio M. Endoglin: an accessory component of the TGF-beta-binding receptor-complex with diagnostic, prognostic, and bioimmunotherapeutic potential in human malignancies. J Cell Physiol. 2001;188:1–7. doi: 10.1002/jcp.1095. [DOI] [PubMed] [Google Scholar]
- Gory S, Vernet M, Laurent M, Dejana E, Dalmon J, Huber P. The vascular endothelial-cadherin promoter directs endothelial-specific expression in transgenic mice. Blood. 1999;93:184–192. [PubMed] [Google Scholar]
- Haab BB, Dunham MJ, Brown PO (2001) Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol 2:RESEARCH0004 [DOI] [PMC free article] [PubMed]
- Haynes BF, Denning SM, Le PT, Singer KH. Human intrathymic T cell differentiation. Semin Immunol. 1990;2:67–77. [PubMed] [Google Scholar]
- Henninger DD, Panes J, Eppihimer M, Russell J, Gerritsen M, Anderson DC, Granger DN. Cytokine-induced VCAM-1 and ICAM-1 expression in different organs of the mouse. J Immunol. 1997;158:1825–1832. [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Kehrl JH, Riva A, Wilson GL, Thevenin C. Molecular mechanisms regulating CD19, CD20 and CD22 gene expression. Immunol Today. 1994;15:432–436. doi: 10.1016/0167-5699(94)90273-9. [DOI] [PubMed] [Google Scholar]
- Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med. 2002;8:257–260. doi: 10.1016/S1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- Kuzu I, Bicknell R, Fletcher CD, Gatter KC. Expression of adhesion molecules on the endothelium of normal tissue vessels and vascular tumors. Lab Invest. 1993;69:322–328. [PubMed] [Google Scholar]
- Lemieux GA, Blumenkron F, Yeung N, Zhou P, Williams J, Grammer AC, Petrovich R, Lipsky PE, et al. The low affinity IgE receptor (CD23) is cleaved by the metalloproteinase ADAM10. J Biol Chem. 2007;282:14836–14844. doi: 10.1074/jbc.M608414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson C, Perrella MA, Hsieh CM, Yoshizumi M, Lee ME, Haber E. Cloning and functional analysis of the promoter for KDR/flk-1, a receptor for vascular endothelial growth factor. J Biol Chem. 1995;270:23111–23118. doi: 10.1074/jbc.270.39.23111. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18:31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Shih SC, Smith LE. Quantitative multi-gene transcriptional profiling using real-time PCR with a master template. Exp Mol Pathol. 2005;79:14–22. doi: 10.1016/j.yexmp.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Shih SC, Robinson GS, Perruzzi P, Calvo A, Desai K, Green J, Ali I, Smith LE, et al. Molecular profiling of angiogenesis markers. Am J Pathol. 2002;161:35–41. doi: 10.1016/S0002-9440(10)64154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt SM, Gschmeissner SE, Bates PA. PECAM-1: its expression and function as a cell adhesion molecule on hemopoietic and endothelial cells. Leuk Lymphoma. 1995;17:229–244. doi: 10.3109/10428199509056827. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]