Abstract
Molecular mechanisms triggered by high dietary beta-carotene (BC) intake in lung are largely unknown. We performed microarray gene expression analysis on lung tissue of BC supplemented beta-carotene 15,15′-monooxygenase 1 knockout (Bcmo1 −/−) mice, which are—like humans—able to accumulate BC. Our main observation was that the genes were regulated in an opposite direction in male and female Bcmo1 −/− mice by BC. The steroid biosynthetic pathway was overrepresented in BC-supplemented male Bcmo1 −/− mice. Testosterone levels were higher after BC supplementation only in Bcmo1 −/− mice, which had, unlike wild-type (Bcmo1 +/+) mice, large variations. We hypothesize that BC possibly affects hormone synthesis or metabolism. Since sex hormones influence lung cancer risk, these data might contribute to an explanation for the previously found increased lung cancer risk after BC supplementation (ATBC and CARET studies). Moreover, effects of BC may depend on the presence of frequent human BCMO1 polymorphisms, since these effects were not found in wild-type mice.
Keywords: Sex-hormones; Mouse whole genome microarray gene expression analysis; Beta-carotene 15,15′-monooxygenase 1; Steroids; Retinol; Retinoic acid; Gender effect; Transcriptome
Introduction
Beta-carotene (BC) is a lipid-soluble provitamin, and a major dietary source of vitamin A. BC itself is thought to behave as a health-promoting agent. This has been attributed to its ability to efficiently scavenge radicals and thereby prevent macromolecular damage [1]. Furthermore, epidemiological studies focusing on dietary intakes of BC have indicated that a high BC intake is associated with a reduced risk for the development of cardiovascular diseases and several types of cancer [2, 3]. Moreover, BC is a provitamin A, and therefore its consumption prevents vitamin A deficiency, which is of importance even in Western society. For example, pregnant and lactating women are at risk for vitamin A deficiency in the Western society due to the discouragement of the consumption of food containing high amounts of vitamin A, in combination with their increased requirements [4].
Although BC is mainly considered to be a health-promoting agent, large-scale intervention trials failed to confirm its beneficial properties. Two intervention trials even showed an increased lung cancer risk and increased mortality because of lung cancer in smokers after BC supplementation (CARET study and ATBC study) [5, 6]. A 6-year follow-up of the CARET study showed differences in lung cancer risk in response to BC supplementation between male and female volunteers, while the ATBC study was only executed in male volunteers. In the CARET study, male subjects had an increased lung cancer risk upon BC supplementation, but only for the duration of the BC intervention. On the other hand, BC-supplemented females had an increased lung cancer risk that persisted even after the intervention had stopped, and it took several years before the lung cancer risk in females reached the levels observed in the controls [7]. The BC-induced increase in lung cancer risk is thought to occur only in smokers or asbestos-exposed subjects since one large-scale intervention trial with mainly non-smokers showed no effect of BC on lung cancer [8, 9]. Although the effect of BC on lung carcinogenesis has been studied extensively, the mechanism by which BC increases lung cancer risk is still not precisely known. The main reason for this is that research on molecular pathways influenced by BC is hampered by the lack of an appropriate animal model that also allows for the use of state-of-the-art functional genomic tools, such as commercially available microarrays. Mice and rats are animal models for which these tools are commercially available, but they differ greatly in BC metabolizing activity compared to humans. The main cause for this is the presence of a more active beta-carotene 15, 15′-monooxygenase 1 (Bcmo1) enzyme variant in rodents [10]. Bcmo1 is the key enzyme in BC metabolism, and in rodents, Bcmo1 cleaves almost all absorbed BC. Since rodents metabolize BC to a high degree, BC supplementation results in an increase in BC metabolites rather than the accumulation of BC, and rodents are therefore inadequate animals to study the effects of BC in humans [10].
Not only inter-species differences in Bcmo1 activity have been described, but also large inter-individual differences in BCMO1 activity are present between humans. Around 25–45% of the volunteers participating in BC conversion studies have been classified as poor converters [11–13]. These inter-individual differences in BC metabolism are at least partly related to polymorphisms in the BCMO1 gene, which results in a reduced BCMO1 activity [14, 15]. Of the reported polymorphisms, there are two frequently occurring polymorphisms with variant allele frequencies of 42 and 24%. As a result, individuals with these polymorphisms have relatively low vitamin A and high BC plasma concentrations compared to their wild-type counterparts [14]. Since differences in activity of BCMO1 result in different BC metabolite concentrations, the activity of BCMO1 might therefore have an effect on previously reported BC-induced effects.
The aim of our study was to investigate gene expression changes induced by dietary BC to try to explain the previously found harmful effects of BC in the lung. For this purpose we used a recently described mouse model, which has no functional Bcmo1 enzyme and which therefore displays increased BC plasma concentrations upon BC supplementation [16]. We performed whole-genome microarray gene expression analysis on lung tissue of both female and male wild-type (Bcmo1 +/+) and Bcmol knockout (Bcmo1 −/−) mice with or without 14 weeks of 150 mg/kg BC supplementation. We previously analyzed the effects of BC in female Bcmo1 −/− mice, and the major effect observed was that BC supplementation counteracted inflammation associated gene expression effects, which were most likely caused by an increased vitamin A demand in Bcmo1 −/− mice [17]. This effect was not observed in male Bcmo1 −/− mice, where two genes, Frizzled 6 (Fzd6) and Collagen triple helix receptor containing 1 (Cthrc1) were strongly downregulated by BC supplementation, which was not observed in female mice [18]. Thus, the major BC-affected processes seemed different between both genders. However, both females and males displayed increased cancer risk after BC supplementation during the intervention of the CARET study. Therefore, we aimed to specifically focus on those genes that are commonly regulated by BC supplementation in both genders, i.e., in male as well as female Bcmo1 −/− mice.
Materials and methods
Animals and treatment
The experimental setup has been described previously (females, [17] and males [18]). Twelve female (fe) and 12 male (ma) B6129SF1 (Bcmo1 +/+) and 12 female and 12 male B6;129S-Bcmo1 tm1dnp (Bcmo1 −/−) mice [16] were used for the dietary intervention, which was conducted in accordance with the German animal protection laws by the guidelines of the local veterinary authorities. During the breeding and weaning periods of the mice, mothers were maintained on KLIBA 3430 chow containing 14,000 IU vitamin A/kg diet (Provima Kliba AG, Kaiseraugst, Switzerland). Five-week-old female and male Bcmo1 +/+ and Bcmo1 −/− mice were caged in groups containing two to four siblings per group and were maintained under environmentally controlled conditions (temperature 24°C, 12 h/12 h light/dark cycle). Mice had ad libitum access to feed and water. Basic feed consisted of the pelletized diet D12450B (Research Diets Inc, USA) with a fat content of 10%. The diet was modified to contain 1,500 IU vitamin A/kg of diet, which is a vitamin A-sufficient diet, and the control diet (control) was supplemented with water-soluble vehicle beadlets (DSM Nutritional Products Ltd., Basel, Switzerland) containing dl-alpha-tocopherol and ascorbyl palmitate as stabilizers, as well as carriers such as gelatin, corn oil sucrose and starch. The BC diet (BC) was supplemented with identical water-soluble beadlets containing BC (DSM Nutritional Products Ltd., Basel, Switzerland) to generate 150 mg BC/kg diet. Beadlets were added by the manufacturer before low temperature pelletting. Feed pellets were color marked and stored at 4°C in the dark.
After 14 weeks of dietary intervention, six female and six male Bcmo1 +/+ mice on the control diet (Bcmo1 +/+ Co), six female and six male Bcmo1 +/+ mice on the BC diet (Bcmo1 +/+ BC), three female and six male Bcmo1 −/− mice on the control diet (Bcmo1 −/− Co), and three female and six male Bcmo1 −/− mice on the BC diet (Bcmo1 −/− BC) were randomly killed on three subsequent mornings. Due to an insufficient number of female Bcmo1 −/− mice in the original breeding pool, six additional female Bcmo1 −/− mice were used that were born 2 weeks later from an identical experiment, three were fed the control diet and three the BC diet, to generate n = 6 for each group. These mice were killed 2 weeks after the first group of mice. Blood was collected from the vena cava after isoflurane and ketamin anesthesia. Blood was coagulated for at least 20 min at room temperature, cooled to 4°C and centrifuged. Lung tissue was removed, rinsed in phosphate-buffered saline (PBS) and snap frozen in liquid nitrogen. The lung tissues were stored at −80°C.
RNA isolation
Left lung lobes were homogenized in liquid nitrogen using a cooled mortar and pestle. Total RNA was isolated using TRIzol reagent (Invitrogen, Breda, The Netherlands) followed by purification using RNeasy columns (Qiagen, Venlo, The Netherlands) using the instructions of the manufacturer. RNA concentration and purity were measured using the Nanodrop system (IsoGen Life Science, Maarsen, The Netherlands). Approximately 30 μg of total RNA was isolated with A260/A280 ratios above 2 and A260/A230 ratios above 1.9 for all samples, indicating good RNA purity. RNA degradation was checked on the Experion (Bio-Rad, Veenendaal, The Netherlands) using Experion StdSense chips (Bio-Rad). Two samples did not meet RNA quality (female Bcmo1 +/+ mice, one on BC diet and one on control diet) and were omitted from the experiment.
Microarray hybridization procedure
The 4 × 44 k Agilent whole mouse genome microarrays (G4122F, Agilent Technologies, Inc. Santa Clara, CA) were used. Preparation of the sample and the microarray hybridization were carried out according to the manufacturer’s protocol with a few exceptions as described previously [19, 20]. In brief, cDNA was synthesized from 1 μg lung RNA using the Agilent Low RNA Input Fluorescent Linear Amplification Kit for each animal without addition of spikes. Thereafter, samples were split in two equal amounts to synthesize Cyanine 3-CTP (Cy3) and Cyanine 5-CTP (Cy5) labeled cRNA using half the amounts per dye as indicated by the manufacturer (Agilent Technologies). Labeled cRNA was purified using RNeasy columns (Qiagen). Yield, A260/A280 ratio, and Cy3 or Cy5 incorporation were examined for every sample using the nanodrop. All samples met the criteria of a cRNA yield higher than 825 ng and a specific activity of at least 8.0 pmol Cy3 or Cy5. Then 1,200 ng of every Cy3-labeled cRNA sample was pooled and used as a common reference pool. Individual 825 ng Cy5-labeled cRNA and 825 ng pooled Cy3-labeled cRNA were fragmented in 1× fragmentation and 1× blocking agent (Agilent Technologies) at 60°C for 30 min and thereafter mixed with GEx Hybridization Buffer HI-RPM (Agilent Technologies) and hybridized in a 1:1 ratio at 65°C for 17 h in an Agilent Microarray hybridization Chamber rotating at 4 rpm. After hybridization, slides were washed according to the wash protocol with Stabilization and Drying solution (Agilent Technologies). Arrays were scanned with an Agilent scanner with 10 and 100% laser power intensities (Agilent Technologies).
Data analyses of microarray results
Signal intensities for each spot were quantified using Feature Extraction 9.1 (Agilent Technologies). Median density values and background values of each spot were extracted for both the experimental samples (Cy5) and the reference samples (Cy3). Quality control for every microarray was performed visually, by using quality control graphs from Feature Extraction, M-A plots and boxplots that were made using limmaGUI in R (Bioconductor) [21]. Data were imported into GeneMaths XT 2.0 (Applied Maths, Sint-Martens-Latem, Belgium).
The median Cy5 signal intensity per spot per array was used for the individual gene expression, while the median Cy3 signal intensity was used for normalization. Since values close to the background may give aberrant fold changes between groups, spots with an average Cy5 or Cy3 median density value below two times the average of the background density were discarded. Spots with an average of the median Cy5 and Cy3 densities twice above the average of the background density were selected and log transformed. Thereafter, the log transformed Cy5 median density value was normalized against the fold change in the log-transformed Cy3 median density value compared to the average log-transformed Cy3 median density value for each spot as described before [22]. Pathway analysis was performed using Gene-Ontology (GO) overrepresentation analysis (ErmineJ) [23], and classification of genes was based on OMIM (www.ncbi.nlm.nih.gov), genecards (www.genecards.org) and literature mining.
Analysis of mRNA expression by real-time Q-PCR
Differential gene expression of lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) (Lrat), glutamate receptor, ionotropic, AMPA1 (alpha 1) (Gria1) and endothelin 1 (Edn1) was analyzed using quantitative PCR (Q-PCR) to validate microarray results. Then 1 μg RNA isolated from lung of individual animals was converted into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). One sample was taken along without reverse transcriptase to examine the presence of DNA (without RT reaction). An equal amount of cDNA of all individual samples was pooled and diluted (10×, 31.6× 100×, 316× 1,000×, 3,160×, 10,000×) for the generation of a calibration curve to test the efficiency of the Q-PCR reaction. Each PCR reaction (25 μl) contained 1× iQ SYBR green supermix (Bio-Rad), 10 μM sense primer, 10 μM antisense primer and 2 μl 100 times diluted cDNA. Primers for Lrat were sense 5′-GCTCTCGGATCAGTCCACAGGC-3′ and antisense 5′-TCCCAAGACAGCCGAAGCAAGA-3′, for Gria1 sense 5′-TGGTGGTGGTGGACTGTGAATC-3′ and antisense 5′-CAGGTTGGCGAGGATGTAGTGG-3′ and for Edn1 sense 5′-AGACCAGGCAGTTAGATGTCAGTG and antisense 5′-TCGTAGTTTCCTTCCTTCCACCAG. For the amplification of the reference genes we used syntaxin 5a (Stx5a) with sense 5′-TTAAAGAACAGGAGGAAACGATTCAGAG-3′ and antisense 5′-CAGGCAAGGAAGACCACAAAGATG-3′, and ring finger protein 130 (Rnf130) with sense 5′-ACAGGAACCAGCGTCGTCTTG-3′ and antisense 5′-ACCCGAACAACATCATTCTGCTTATAG-3′, which showed equal gene expression levels for all individual animals on the microarray. These intron-spanning primers were designed using Beacon designer 7.00 (Premier Biosoft International, Palo Alto, CA) or using primer-BLAST (http://www.ncbi.nlm.nih.gov/). Amplification was performed in duplicate using the MyIQ single-color real-time PCR detection system (Bio-Rad) using the following temperature cycles: 1× 3 min at 95°C, 40× 2-step amplification (15 s 95°C, 45 s 60°C), 1× 1 min 95°C, and 1× 1 min 65°C followed by melting curve analysis (60× 10 s 65°C with an increase of 0.5°C/10 s. A negative control without cDNA template and the minus reverse transcriptase sample were taken along with every assay. Using the standard curves for every gene, the relative level of expression of all genes was calculated. Normalized Gene Expression (ΔΔCt) analysis was calculated using the IQ5 software version 2.0 (Bio-Rad).
Testosterone measurement
Testosterone concentrations were measured using radio immunoassays (DSL 4100, Diagnostic Systems Laboratories, Beckman Coulter, Assendelft, The Netherlands) and were performed according to the manufacturer’s protocol using quarters of all volumes as has been described previously [24]. Gamma-radiation was measured on a Wallac 1470 Wizard Gamma Counter (Perkin-Elmer). The recovery of testosterone spikes and linearity of the dilution were tested before use.
Statistical analysis
General effects of diet, gender and genotype on concentrations of BC, retinol and retinyl esters in lung and serum were analyzed using 2 × 2 × 2 factorial univariate ANOVA (SPSS version 15.0) and considered statistically significant when p < 0.05. Effects of BC compared to control diet were tested using a Student’s t-test and considered statistically significant at p < 0.05). Fold changes for both microarray gene expression and Q-PCR gene expression were calculated using mean log signal intensities. P-values for differential expressions were calculated between two groups using two-tailed Student’s t-test statistics on log intensity values. Changes were considered statistically different at p < 0.05. Regression analysis of gene expression in males versus females was performed in GraphPad Prism software (version 5.0; GraphPad Software, Inc, San Diego, CA) using nonlinear regression analysis.
Results
BC supplementation and knockout of Bcmo1 results in changes in BC and BC metabolites
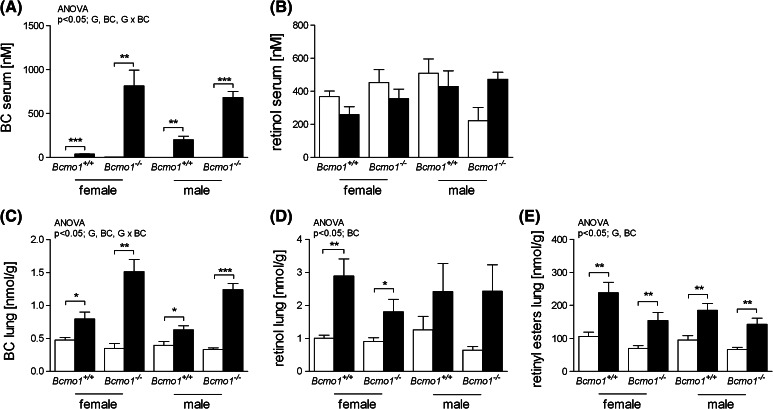
BC concentrations in lung and serum were increased after 14 weeks of BC treatment compared to control diet-fed mice, and as expected, to a higher level in Bcmo1 −/− mice as in Bcmo1 +/+ mice (as has been described separately for female [17] and male [18] mice). There were no significant differences in retinol serum concentrations between the different groups. In lung tissue, retinol and retinyl ester concentrations were increased upon BC supplementation compared to the control diet fed mice, but there was no interaction between BC and the Bcmo1 genotype concerning the concentration of these metabolites (Fig. 1).
Fig. 1.
Beta-carotene (BC) (a, c), retinol (b, d) concentrations in serum and lung tissue, respectively, and retinyl ester concentrations in lung tissue (e) of female and male Bcmo1 +/+ mice and Bcmo1 −/− mice fed a control diet (white bars) or a BC diet (black bars). Data are expressed as mean ± SEM; significance was tested for diet (BC), gender (S) and genotype (G) using ANOVA and considered significant at p < 0.05. Significant effects for the different factors or any interaction between those factors are displayed on top of each figure. Student’s t-test was used between BC and control groups when there was a significant diet effect by ANOVA and considered significant when p < 0.05. Using Student’s t-test statistics: *p < 0.05, **p < 0.01 and ***p < 0.001
Gene expression of all groups was analyzed, and of the 43,379 spots (number of spots minus control spots) on the whole genome microarrays, 31,128 spots had an average signal twice above background and were regarded as positive spots and included in the analysis. There were two genes involved in downstream BC metabolism that were significantly regulated by BC in Bcmo1 +/+ mice (Table 1). The biggest fold change was found for lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase (Lrat; 1.6 fold increase after BC supplementation). Five genes involved in downstream BC metabolism were regulated because of the knock-out of Bcmo1. Most striking was the high fold change up regulation of Lrat (1.7–2.1 fold increase in Bcmo1 −/− mice) and the high fold change down regulation of alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide (Adh7; 1.6–2.7 fold decrease in Bcmo1 −/− mice).
Table 1.
Genes involved in BC metabolism or retinoic acid catabolism that were significantly regulated after BC supplementation or Bcmo1 knockout
| Symbol | Name | Probe name | Systematic name | Fold change induced by BC supplementation | Fold change induced by Bcmo1 knockout | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Female | Male | ||||||||
| Bcmo1 +/+ | Bcmo1 −/− | Bcmo1 +/+ | Bcmo1 −/− | Co | BC | Co | BC | ||||
| Aldh1a2 | Aldehyde dehydrogenase family 1, subfamily A2 | A_52_P58145 | NM_009022 | −1.04 | −1.03 | −1.02 | −1.02 | 1.25* | 1.25** | 1.35*** | 1.35** |
| Aldh1a3 | Aldehyde dehydrogenase family 1, subfamily A3 | A_52_P87843 | NM_053080 | −1.25** | 1.00 | −1.11 | −1.12 | −1.43*** | −1.14 | −1.14 | −1.14 |
| Aldh2 | Aldehyde dehydrogenase 2, mitochondrial | A_52_P116134 | NM_009656 | 1.05 | 1.17* | −1.03 | 1.01 | −1.18* | −1.07 | −1.10 | −1.05 |
| Lrat | Lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) | A_52_P669005 | NM_023624 | 1.58** | 1.34 | 1.57* | 1.49 | 2.58* | 1.88* | 1.80* | 1.71** |
| Adh7 | Alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | A_51_P233797 | NM_009626 | −1.11 | 1.23 | 1.01 | 1.13 | −2.67*** | −1.91*** | −1.83*** | −1.64** |
* Significant difference (p < 0.05)
** Significant difference (p < 0.01)
*** Significant difference (p < 0.001)
Gene expression is changed in the opposite direction by BC in male and female Bcmo1−/− mice
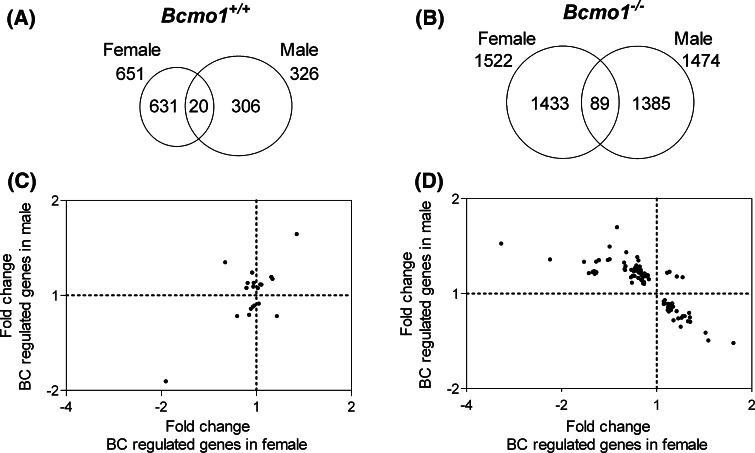
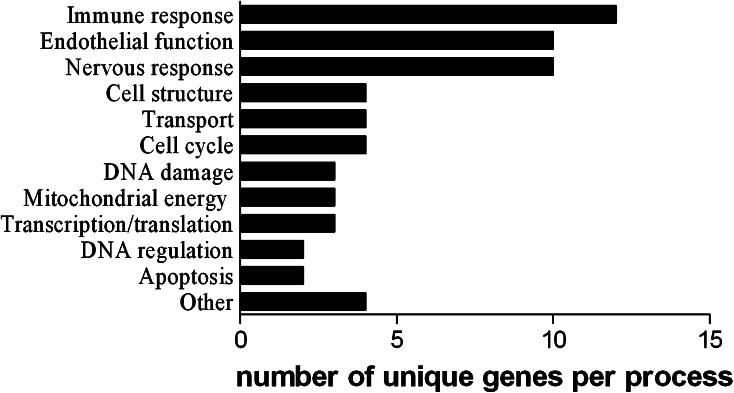
BC supplementation resulted in a considerably higher number of significantly regulated spots in the Bcmo1 −/− mice than in the Bcmo1 +/+ mice (Fig. 2a, b). Only 20 regulated genes overlapped in male and female Bcmo1 +/+ mice and 89 genes in Bcmo1 −/− mice. These two groups of regulated genes commonly affected in both genders showed no overlap between Bcmo1 −/− and Bcmo1 +/+ mice. To further determine whether these genes were similarly regulated by BC in both genders, fold changes of gender-independent BC-regulated genes were plotted for female mice against the fold changes in male mice. The 20 BC-regulated genes in male and female Bcmo1 +/+ mice were distributed in all four quadrants of the plot (Fig. 2c). Regression analysis resulted in a significant correlation [R = 0.66; slope = 0.787; 95% CI slope = (0.2988–1.276)]. This correlation was however based on only two data points (−1.93; −1.87) and (1.34; 1.56). Without these two points, there was no significant correlation [R = 0.14; slope = −0.193; 95% CI slope = (−0.8832; 0.4960)]. To our surprise, genes regulated by BC in female Bcmo1 −/− were regulated in opposite directions compared to BC-regulated genes in male Bcmo1 −/− mice (upwards regulated genes in males were down in females and vice versa), with only 4 out of the 89 probes being regulated in the same direction (in italics in Table 3). We found >95% of the probes being regulated in the opposite direction by BC between males and females, suggesting that this is a real effect of BC. Randomly affected gene expression would theoretically result in 50% of the probes being regulated in a similar direction and 50% in opposite directions. Regression analysis resulted in a significant negative regression [R = 0.76; slope = −0.436; 95% CI slope = (−0.5144; −0.3582)]. Genes regulated by BC in both male and female Bcmo1 +/+ mice (Table 2) or Bcmo1 −/− mice (Table 3) were categorized into biological processes, with endothelial functioning, immune response and nervous response being most prominently regulated by BC in Bcmo1 −/− mice (Fig. 3).
Fig. 2.
Venn diagrams representing the number of genes significantly (p < 0.05) regulated by BC in female and male mice and the number of genes regulated by BC in both females as well as males (overlap) in Bcmo1 +/+ (a) and Bcmo1 −/− (b) mice. The scatter plots represent the gene expression fold changes of the genes regulated by BC independent of gender in females (x-axis) and males (y-axis) in Bcmo1 +/+ (c) and in Bcmo1 −/− (d) mice
Table 3.
Classification of genes that were significantly regulated by BC in both female and male Bcmo1 −/− mice
| Gene symbol | Gene name | Probe name | Systematic name | BC-induced fold change | |||
|---|---|---|---|---|---|---|---|
| Female | Male | ||||||
| Bcmo1 +/+ | Bcmo1 −/− | Bcmo1 +/+ | Bcmo1 −/− | ||||
| Immune response | |||||||
| B2 m | Beta-2 microglobulin | A_51_P129006 | NM_009735 | 1.00 (ns) | −1.48* | −1.00 (ns) | 1.17* |
| B2 m | Beta-2 microglobulin | A_51_P129006 | NM_009735 | 1.01 (ns) | −1.50* | 1.01 (ns) | 1.18* |
| B2 m | Beta-2 microglobulin | A_51_P129006 | NM_009735 | 1.01 (ns) | −1.48* | −1.02 (ns) | 1.18* |
| B2 m | Beta-2 microglobulin | A_51_P129006 | NM_009735 | 1.02 (ns) | −1.47* | 1.02 (ns) | 1.17* |
| B2 m | Beta-2 microglobulin | A_51_P129006 | NM_009735 | 1.01 (ns) | −1.48* | 1.03 (ns) | 1.16* |
| Cd247 | CD247 antigen | A_52_P173555 | NM_031162 | −1.06 (ns) | −1.14* | 1.13 (ns) | 1.12* |
| Dapp1 | Dual adaptor for phosphotyrosine and 3-phosphoinositides 1 | A_52_P422172 | NM_011932 | −1.03 (ns) | −1.11* | −1.05 (ns) | 1.14* |
| Gja5 | Gap junction membrane channel protein alpha 5 | A_52_P612636 | AK082993 | 1.04 (ns) | −1.17* | 1.21 (ns) | 1.14* |
| Gvin1 | GTPase, very large interferon inducible 1 | A_52_P535484 | NM_029000 | −1.14 (ns) | −1.56* | 1.01 (ns) | 1.16* |
| H2-Q8 | Histocompatibility 2, Q region locus 8 | A_52_P152133 | NM_023124 | −1.01 (ns) | −1.56* | −1.11* | 1.26** |
| Il18 bp | Interleukin 18 binding protein | A_52_P577384 | NM_010531 | 1.11 (ns) | −1.49* | −1.04 (ns) | 1.18* |
| Ifi44 | Interferon-induced protein 44 | A_52_P550858 | AK085407 | 1.25 (ns) | −2.36* | −1.03 (ns) | 1.47* |
| Nfam1 | Nfat activating molecule with ITAM motif 1 | A_52_P686701 | NM_028728 | −1.02 (ns) | −1.13*** | 1.09 (ns) | 1.21* |
| LOC668139 | Similar to very large inducible GTPase 1 isoform A | A_52_P666442 | XR_001627 | −1.10 (ns) | −2.14** | 1.00 (ns) | 1.31* |
| LOC675624 | Similar to very large inducible GTPase 1 isoform A | A_52_P764477 | XM_983705 | −1.14 (ns) | −1.53** | −1.05 (ns) | 1.26* |
| Oas1d | 2′-5′ oligoadenylate synthetase 1D | A_51_P183025 | NM_133893 | −1.10 (ns) | −1.48* | 1.05 (ns) | 1.27*** |
| Psme2 | Proteasome (prosome, macropain) 28 subunit, beta | A_52_P305289 | NM_011190 | 1.04 (ns) | −1.19* | −1.02 (ns) | 1.08* |
| Endothelial functioning | |||||||
| Aebp1 | AE binding protein 1 | A_51_P336770 | NM_009636 | −1.04 (ns) | 1.12* | −1.05 (ns) | −1.07* |
| Col25a1 | Procollagen, type XXV, alpha 1 | A_51_P333929 | NM_029838 | −1.09 | −1.14* | −1.07 (ns) | 1.16* |
| Darc | Duffy blood group, chemokine receptor | A_52_P110052 | NM_010045 | −1.16 (ns) | −1.36* | 1.05 (ns) | 1.29* |
| Ecgf1 | Endothelial cell growth factor 1 (platelet-derived) | A_51_P267080 | NM_138302 | 1.11 (ns) | 1.11* | −1.03 (ns) | −1.11* |
| Edn1 | Endothelin 1 | A_51_P115005 | NM_010104 | −1.33 (ns) | 1.44* | 1.22 (ns) | −1.33* |
| Gata1 | GATA binding protein 1 | A_52_P670095 | NM_008089 | −1.03 (ns) | −1.26* | 1.02 (ns) | 1.25*** |
| Gata1 | GATA binding protein 1 | A_52_P670095 | NM_008089 | −1.07 (ns) | −1.17* | 1.06 (ns) | 1.18* |
| Habp4 | Hyaluronic acid binding protein 4 | A_51_P269078 | NM_019986 | 1.03 | 1.10* | 1.01 (ns) | -1.10* |
| Pthlh | Parathyroid hormone-like peptide | A_51_P129363 | NM_008970 | −1.15 | −1.15** | 1.01 (ns) | 1.20* |
| Selp | Selectin, platelet | A_51_P211854 | NM_011347 | −1.03 (ns) | −1.14** | −1.01 (ns) | 1.17* |
| Serpinb2 | Serine (or cysteine) peptidase inhibitor, clade B, member 2 | A_52_P639522 | NM_011111 | −1.37 (ns) | −1.25** | −1.06 (ns) | 1.23* |
| Serpinb2 | Serine (or cysteine) peptidase inhibitor, clade B, member 2 | A_51_P101146 | NM_011111 | −1.46 (ns) | −1.39* | 1.06 (ns) | 1.38* |
| Nervous response | |||||||
| Ahnak | AHNAK nucleoprotein (desmoyokin) | A_51_P224534 | NM_009643 | 1.01 (ns) | 1.16* | −1.03 (ns) | −1.14* |
| Atxn7l2 | Ataxin 7-like 2 | A_52_P287195 | NM_175183 | −1.00 (ns) | 1.05* | 1.00 (ns) | −1.06* |
| Dyrk4 | Dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 4 | A_51_P307463 | NM_207210 | −1.01 (ns) | −1.25* | 1.02 (ns) | 1.18* |
| Gad1 | Glutamic acid decarboxylase 1 | A_51_P282538 | NM_008077 | −1.12 (ns) | −1.17** | −1.01 (ns) | 1.21** |
| Gria1 | Glutamate receptor, ionotropic, AMPA1 (alpha 1) | A_52_P39575 | NM_008165 | −1.03 (ns) | 1.28* | −1.02 (ns) | −1.24** |
| Kcnn2 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 | A_51_P309854 | NM_080465 | 1.06 | 1.22* | −1.03 (ns) | 1.12* |
| Olfml1 | Olfactomedin-like 1 | A_51_P476879 | NM_172907 | 1.00 (ns) | −1.20* | −1.01 (ns) | 1.19*** |
| Plcl1 | Phospholipase C-like 1 | A_52_P191567 | AK140530 | 1.01 (ns) | 1.06* | 1.00 (ns) | −1.07* |
| Reln | Reelin | A_51_P365369 | NM_011261 | 1.15* | 1.20* | −1.05 (ns) | −1.20* |
| Reln | Reelin | A_51_P365369 | NM_011261 | 1.09* | 1.21* | 1.02 (ns) | −1.17* |
| Reln | Reelin | A_51_P365369 | NM_011261 | 1.08* | 1.27* | −1.06 (ns) | −1.15* |
| Reln | Reelin | A_51_P365369 | NM_011261 | 1.13** | 1.24* | −1.00 (ns) | −1.17* |
| Reln | Reelin | A_51_P365369 | NM_011261 | 1.09 (ns) | 1.26** | 1.03 (ns) | −1.22* |
| Slc6a17 | Solute carrier family 6 (neurotransmitter transporter), member 17 | A_52_P373637 | NM_172271 | −1.07 (ns) | −1.12* | 1.07 (ns) | 1.19* |
| Cell structure | |||||||
| Elmo1 | Engulfment and cell motility 1, ced-12 homolog (C. elegans) | A_51_P306129 | NM_080288 | 1.00 | −1.06* | 1.02 (ns) | 1.14* |
| Mmp13 | Matrix metallopeptidase 13 | A_51_P184484 | NM_008607 | 1.06 | −1.40* | −1.06 (ns) | 1.28* |
| Mmp24 | Matrix metallopeptidase 24 | A_52_P605500 | NM_010808 | −1.20 | 1.10** | 1.04 (ns) | 1.17* |
| Twf1 | Twinfilin, actin-binding protein, homolog 1 (Drosophila) | A_52_P463365 | NM_008971 | 1.01 | −1.06* | −1.00 (ns) | 1.11* |
| Transport | |||||||
| Slc12a7 | Solute carrier family 12, member 7 | A_51_P421734 | NM_011390 | 1.07 | 1.09** | 1.02 (ns) | −1.08* |
| Slc35a5 | Solute carrier family 35, member A5 | A_52_P618379 | NM_028756 | −1.02 | −1.08* | 1.01 (ns) | 1.15* |
| Slc5a11 | Solute carrier family 5 (sodium/glucose cotransporter), member 11 | A_52_P271085 | NM_146198 | −1.20 | −1.14* | 1.04 (ns) | 1.21* |
| Rab6 | RAB6, member RAS oncogene family | A_51_P209873 | NM_024287 | 1.11 | 1.77** | 1.08 (ns) | −1.39* |
| Cell cycle | |||||||
| Ccnb1 | Cyclin B1 | A_52_P558401 | NM_172301 | −1.02 (ns) | −1.15* | 1.01 (ns) | 1.18* |
| Esco2 | Establishment of cohesion 1 homolog 2 (S. cerevisiae) | A_51_P195034 | NM_028039 | 1.11 (ns) | −1.15* | −1.04 (ns) | 1.28* |
| Fbxw2 | F-box and WD-40 domain protein 2 | A_52_P643001 | BC003834 | 1.26 (ns) | −1.11* | −1.04 (ns) | 1.16*** |
| Hspa2 | Heat shock protein 2 | A_51_P453149 | NM_008301 | −1.08 (ns) | 1.27** | −1.00 (ns) | −1.19* |
| DNA damage | |||||||
| Ankrd13a | Ankyrin repeat domain 13a | A_51_P190886 | NM_026718 | 1.03 (ns) | 1.13*** | −1.02 (ns) | −1.10* |
| Ankrd13a | Ankyrin repeat domain 13a | A_52_P665309 | NM_026718 | 1.26 (ns) | 1.09* | 1.03 (ns) | −1.09* |
| Oxr1 | Oxidation resistance 1 | A_51_P161893 | NM_130885 | 1.15 (ns) | 1.13* | −1.03 (ns) | −1.22*** |
| Sf3b2 | Splicing factor 3b, subunit 2 | A_51_P280158 | NM_030109 | 1.02 (ns) | 1.06* | −1.01 (ns) | −1.05* |
| Mitochonrial energy | |||||||
| Acsl4 | Acyl-CoA synthetase long-chain family member 4 | A_51_P268154 | NM_019477 | 1.03 | −1.10* | 1.07 (ns) | 1.09* |
| Atp5g3 | ATP synthase, H + transporting, mitochondrial F0 complex, subunit c (subunit 9), isoform 3 | A_51_P294849 | NM_175015 | 1.06 | 1.09* | −1.02 (ns) | −1.13* |
| Rhot1 | Ras homolog gene family, member T1 | A_52_P232663 | NM_021536 | −1.04 | 1.08* | 1.04 (ns) | 1.16** |
| DNA regulation | |||||||
| Dpys | Dihydropyrimidinase | A_51_P244950 | BC029718 | −1.06 (ns) | −1.34** | −1.05 (ns) | 1.60** |
| Rrm1 | Ribonucleotide reductase M1 | A_51_P502082 | NM_009103 | 1.05 (ns) | −1.19* | −1.04 (ns) | 1.14* |
| Apoptosis | |||||||
| Bcl2l11 | BCL2-like 11 (apoptosis facilitator) | A_52_P158527 | NM_207680 | 1.02 (ns) | 1.09* | −1.00 (ns) | −1.10* |
| Zc3h12a | Zinc finger CCCH type containing 12A | A_51_P297925 | NM_153159 | −1.06 (ns) | −1.10* | 1.01 (ns) | 1.13* |
| Transcription/translation | |||||||
| Cited4 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 | A_52_P426768 | NM_019563 | −1.23 (ns) | −1.21*** | 1.06 (ns) | 1.13* |
| Eif2s1 | Eukaryotic translation initiation factor 2, subunit 1 alpha | A_51_P448334 | NM_026114 | 1.01 (ns) | −1.15* | −1.07 (ns) | 1.27** |
| Snrpd3 | Small nuclear ribonucleoprotein D3 | A_52_P150588 | AK163643 | −1.07 (ns) | −1.09* | −1.04 (ns) | 1.13* |
| Other | |||||||
| Mlana | Melan-A | A_51_P499623 | AK020928 | 1.09 | 1.20 | −1.15 (ns) | −1.27* |
| Rex2 | Reduced expression 2 | A_52_P412109 | NM_009051 | −1.01 | −1.10** | 1.05 (ns) | 1.07* |
| Rnaset2b | Ribonuclease T2B | A_51_P310100 | AK029149 | −1.08 (ns) | −1.12* | 1.01 (ns) | 1.11* |
| Wdr33 | WD repeat domain 33 | A_52_P280899 | NM_028866 | 1.00 | 1.10** | −1.01 (ns) | −1.08* |
| Unannotated | |||||||
| 1700029I01Rik | RIKEN cDNA 1700029I01 gene | A_51_P109496 | NM_027285 | −1.05 (ns) | −1.26* | 1.07 (ns) | 1.19** |
| 4932432K03Rik | RIKEN cDNA 4932432K03 gene | A_52_P413907 | NM_144535 | −1.07 (ns) | −1.09* | −1.01 (ns) | 1.16* |
| AU020772 | Expressed sequence AU020772 | A_52_P564951 | NM_001017985 | −1.02 (ns) | 1.05* | 1.03 (ns) | −1.08*** |
| AU042671 | Expressed sequence AU042671 | A_51_P450764 | AK037367 | 1.04 (ns) | 1.12* | 1.01 (ns) | −1.13* |
| D14Ertd426e | DNA segment, Chr 14, ERATO Doi 426, expressed | A_52_P527268 | AK139611 | −1.09 (ns) | −1.07** | 1.08 (ns) | 1.13* |
| LOC435271 | Similar to schlafen 3 | A_52_P322927 | XM_487182 | −1.05 (ns) | −1.18*** | 1.01 (ns) | 1.19* |
| LOC631232 | Similar to zinc finger protein 665 | A_52_P17746 | XM_904987 | −1.01 (ns) | −1.12* | −1.00 (ns) | 1.11* |
| LOC631624 | Similar to reduced expression 2 | A_52_P501447 | XM_919663 | −1.03 (ns) | −1.10** | 1.04 (ns) | 1.08 |
| Zc3h12a | Zinc finger CCCH type containing 12A | A_51_P297925 | NM_153159 | −1.06 (ns) | −1.10* | 1.01(ns) | 1.13* |
| Glycerol-3-phosphate acyltransferase mRNA, complete cds | A_51_P471819 | M77003 | 1.08 (ns) | 1.16** | −1.04 (ns) | 1.13* | |
| Q3WEM2_9ACTO (Q3WEM2) Parallel beta-helix repeat precursor, partial (6%) | A_52_P214241 | TC1678456 | 1.01 (ns) | -1.11* | 1.01 (ns) | 1.10* | |
| 7 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:A730019A03 product:unclassifiable, full insert sequence | A_51_P482265 | AK163132 | −1.03 (ns) | −1.16 | 1.03 (ns) | 1.23** | |
| Adult male diencephalon cDNA, RIKEN full-length enriched library, clone:9330176N07 product:unclassifiable, full insert sequence | A_52_P448829 | AK034318 | 1.07 (ns) | 1.52* | −1.02 (ns) | −1.36* | |
| G protein-coupled receptor associated sorting protein 2 | A_51_P362373 | ENSMUST00000096320 | −1.13 (ns) | 1.18* | −1.05 (ns) | −1.19* | |
| BB736773 RIKEN full-length enriched, 6 days neonate spleen Mus musculus cDNA clone F430010B14 3′, mRNA sequence | A_52_P700030 | BB736773 | −1.19 (ns) | −1.23* | 1.02 (ns) | 1.34** | |
| K0288C02-5 N NIA Mouse Unfertilized Egg cDNA Library (Long) Mus musculus cDNA clone NIA:K0288C02 IMAGE:30053113 5′, mRNA sequence | A_52_P174014 | CA561520 | 1.01 (ns) | −1.18* | 1.01 (ns) | 1.18* | |
* Significant difference (p < 0.05) between BC and control diet fed mice
** Significant difference (p < 0.01) between BC and control diet fed mice
*** Significant difference (p < 0.001) between BC and control diet fed mice
ns no significant difference (p > 0.05) between BC and control diet fed mice
Table 2.
Classification of genes that were significantly regulated by BC in both female and male Bcmo1 +/+ mice
| Gene symbol | Gene name | Probe name | Systematic name | BC-induced fold change | |||
|---|---|---|---|---|---|---|---|
| Female | Male | ||||||
| Bcmo1 +/+ | Bcmo1 −/− | Bcmo1 +/+ | Bcmo1 −/− | ||||
| Immune response | |||||||
| Cd207 | CD 207 antigen | A_51_P110381 | NM_144943 | −1.40** | 1.16 (ns) | −1.16* | −1.21 (ns) |
| Cd209b | CD209b antigen | A_52_P267717 | NM_026972 | −1.29*** | −1.05 (ns) | −1.15* | 1.10 (ns) |
| Cd5 l | CD5 antigen-like | A_51_P205779 | NM_009690 | −2.02* | −1.93 (ns) | −1.87* | −1.15 (ns) |
| Csf2rb1 | Colony stimulating factor 2 receptor, beta 1, low-affinity (granulocyte–macrophage) | A_52_P664853 | NM_007780 | −1.14* | 1.03 (ns) | 1.08* | 1.01 (ns) |
| Ly6g5b | Lymphocyte antigen 6 complex, locus G5B | A_51_P369443 | NM_148939 | −1.09* | −1.08 (ns) | 1.06* | 1.03 (ns) |
| Xpnpep2 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound | A_51_P152585 | NM_133213 | 1.15* | 1.12 (ns) | 1.13* | 1.09* |
| Apoptosis | |||||||
| Dnase1l3 | Deoxyribonuclease 1-like 3 | A_52_P165657 | NM_007870 | −1.26* | −1.15 (ns) | −1.16* | −1.43* |
| Hsd11b1 | Hydroxysteroid 11-beta dehydrogenase 1 | A_51_P127297 | NM_008288 | 1.22* | −1.07 (ns) | 1.09* | −1.05 (ns) |
| Retinol metabolism | |||||||
| Lrat | Lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) | A_52_P669005 | NM_023624 | 1.58** | 1.34 (ns) | 1.57* | 1.49 (ns) |
| Rdh12 | Retinol dehydrogenase 12 | A_51_P141960 | NM_030017 | −1.07* | 1.01 (ns) | 1.05** | 1.02 (ns) |
| Mucus production | |||||||
| Muc4 | Mucin 4 | A_51_P392277 | AF218265 | 1.23* | −1.25 (ns) | 1.27* | 1.06 (ns) |
| Mitochondrial functioning | |||||||
| Sirt4 | Sirtuin 4 (silent mating type information regulation 2 homolog) 4 (S. cerevisiae) | A_51_P424641 | BC022653 | 1.06* | 1.02 (ns) | −1.06* | 1.02 (ns) |
| Trit1 | tRNA isopentenyltransferase 1 | A_51_P137317 | NM_025873 | 1.08* | −1.03 (ns) | −1.09* | 1.08 (ns) |
| Intracellular trafficking | |||||||
| Tom1 | Target of myb1 homolog (chicken) | A_52_P1480 | NM_011622 | −1.13* | 1.04 (ns) | 1.08* | 1.02 (ns) |
| Ion chanel | |||||||
| Tpcn2 | Two pore segment channel 2 | A_51_P517952 | NM_146206 | −1.23* | −1.02 (ns) | 1.10* | 1.19 (ns) |
| Protein modification | |||||||
| Ube1l2 | Ubiquitin-activating enzyme E1-like 2 | A_52_P671062 | NM_172712 | −1.13* | −1.03 (ns) | 1.18* | 1.11 (ns) |
| Unknown | |||||||
| Phgdhl1 | Phosphoglycerate dehydrogenase like 1 | A_52_P82229 | NM_026861 | −1.11* | −1.02 (ns) | 1.07* | 1.04 (ns) |
| Zmynd10 | Zinc finger, MYND domain containing 10 | A_51_P392967 | NM_053253 | 1.16* | −1.02 (ns) | −1.08* | −1.02 (ns) |
| AI508575 | A_52_P724015 | AI508575 | 1.12* | −1.04 (ns) | −1.10* | −1.06 (ns) | |
| AK080164 | A_52_P33067 | AK080164 | −1.18* | 1.12 (ns) | 1.14* | 1.02 (ns) | |
* Significant difference (p < 0.05) between BC and control diet fed mice
** Significant difference (p < 0.01) between BC and control diet fed mice
*** Significant difference (p < 0.001) between BC and control diet fed mice
ns no significant difference (p > 0.05) between BC and control diet fed mice
Fig. 3.
Classification of genes that are regulated by BC (p < 0.05) in male as well as in female Bcmo1 −/− mice into different biological process categories
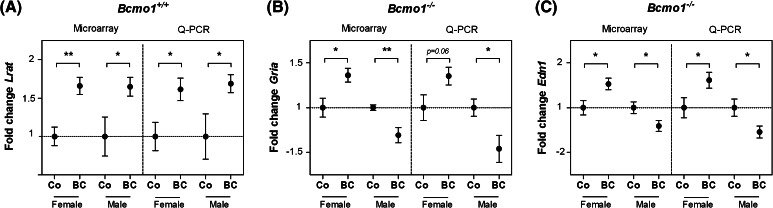
We used Q-PCR to validate the microarray experiment. For validation of the effects in Bcmo1 +/+ mice, lecitin retinol acyltransferase (phosphatidylcholine-retinol O-acyltransferase) (Lrat), an enzyme involved in downstream BC metabolism, was selected. For validation of effects induced by BC supplementation in Bcmo1 −/− mice, the genes glutamate receptor, ionotropic, AMPA1 (alpha 1) (Gria1) and endothelin 1 (Edn1) were chosen because of their relatively high expression level and relatively high fold changes. All three selected genes showed an identical expression using the microarray technique and Q-PCR (Fig. 4).
Fig. 4.
The fold change gene expression of Lrat in Bcmo1 +/+ mice (a), Gria in Bcmo1 −/− mice (b) and Edn1 in Bcmo1 −/− mice (c) in BC supplementated mice relative to the control diet fed mice in the microarray, validated by Q-PCR using the reference genes Stx5a and Rnf130. Data represent the average gene expression ± SEM compared to control diet fed mice. Using Student’s t-test statistics on log transformed data: *p < 0.05 and **p < 0.01
BC supplementation results in the overrepresentation of the steroid biosynthetic process in male Bcmo1−/− mice
Gene-ontology processes affected by BC supplementation in male and female Bcmo1 −/− mice were examined to further explain the opposite direction of gene expression induced by BC. This overrepresentation analysis was performed using ErmineJ overrepresentation analysis [23] on all genes and their corresponding p-value. We found a significant overrepresentation of the steroid biosynthetic process (GO:0006694) in male Bcmo1 −/− mice by BC supplementation (p = 4.6 × 10−6), but not in female Bcmo1 −/− mice (p = 0.17). Genes regulated by BC resulting in the overrepresentation of the steroid biosynthetic process in male Bcmo1 −/− mice are listed in Table 4 (genes presented until p = 0.1).
Table 4.
Genes regulated by BC in male Bcmo1 −/− mice resulting in a significant overrepresentation of the steroid biosynthetic process (GO: 0006694)a
| Probe namer | Source symbol | Gene name | BC-induced fold change gene expression Bcmo1 −/− mice | |
|---|---|---|---|---|
| Female | Male | |||
| A_52_P137371 | Hmgcr | 3-Hydroxy-3-methylglutaryl-Coenzyme A reductase | −1.08 (0.29) | 1.14 (0.01) |
| A_51_P413238 | Pbx1 | Pre B-cell leukemia transcription factor 1 | 1.02 (0.68) | −1.11 (0.02) |
| A_51_P309733 | Cyp11a1 | Cytochrome P450, family 11, subfamily a, polypeptide 1 | 1.06 (0.50) | 1.20 (0.02) |
| A_51_P169527 | Mvk | Mevalonate kinase | −1.05 (0.48) | 1.11 (0.03) |
| A_52_P165654 | Star | Steroidogenic acute regulatory protein | 1.09 (0.41) | 1.12 (0.03) |
| A_52_P384574 | Stard4 | StAR-related lipid transfer (START) domain containing 4 | 1.07 (0.54) | −1.27 (0.04) |
| A_52_P223224 | Dhrs8 | Dehydrogenase/reductase (SDR family) member 8 | −1.02 (0.63) | 1.11 (0.04) |
| A_51_P515446 | Cyp39a1 | Cytochrome P450, family 39, subfamily a, polypeptide 1 | 1.01 (0.83) | 1.08 (0.05) |
| A_52_P164570 | Hsd17b12 | Hydroxysteroid (17-beta) dehydrogenase 12 | 1.00 (0.95) | −1.15 (0.05) |
| A_52_P85765 | Stard6 | StAR-related lipid transfer (START) domain containing 6 | −1.05 (0.51) | 1.11 (0.05) |
| A_51_P420415 | Srd5a1 | Steroid 5 alpha-reductase 1 | −1.00 (0.98) | 1.12 (0.06) |
| A_51_P485791 | Cyp51 | Cytochrome P450, family 51 | −1.10 (0.14) | 1.14 (0.06) |
| A_52_P566605 | Hsd17b7 | Hydroxysteroid (17-beta) dehydrogenase 7 | −1.18 (0.03) | 1.09 (0.07) |
| A_52_P136138 | Fdft1 | Farnesyl diphosphate farnesyl transferase 1 | −1.08 (0.33) | 1.18 (0.07) |
| A_52_P388072 | Hmgcs1 | 3-Hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | −1.10 (0.22) | 1.16 (0.08) |
| A_52_P636752 | Cyp51 | Cytochrome P450, family 51 | −1.13 (0.19) | 1.15 (0.08) |
| A_51_P379798 | Fdps | Farnesyl diphosphate synthetase | −1.04 (0.65) | 1.14 (0.08) |
| A_51_P485946 | Fdft1 | Farnesyl diphosphate farnesyl transferase 1 | −1.15 (0.20) | 1.07 (0.10) |
aGenes presented: BC versus control in male Bcmo1−/− mice p < 0.1; between brackets the actual p-value
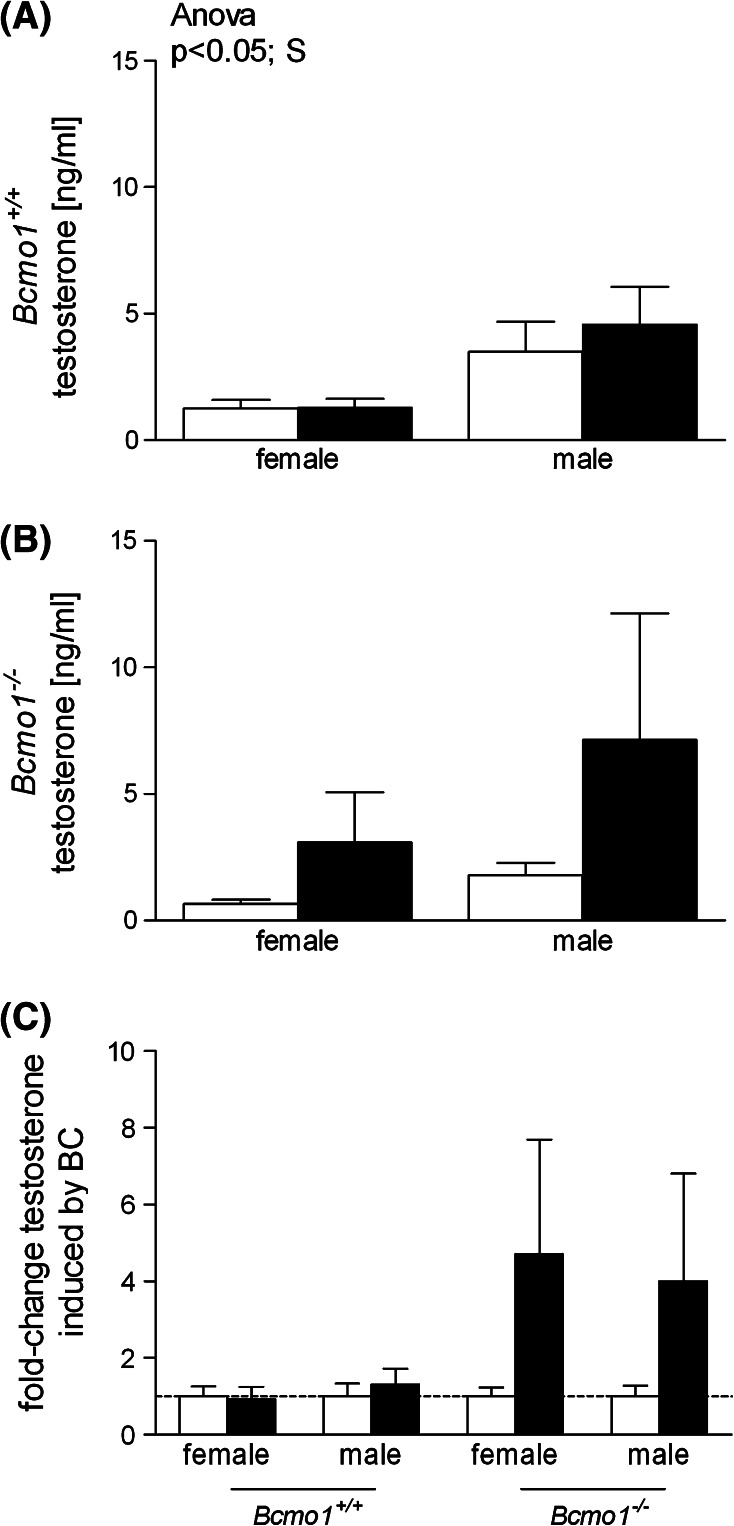
Effect of BC on serum testosterone levels
We determined serum concentrations of the sex hormone testosterone. Since only limited amounts of serum were available, other sex hormones requiring bigger serum amounts could not be determined. As expected, testosterone levels were significantly higher in Bcmo1 +/+ males than in Bcmo1 +/+ females (Fig. 5a). In Bcmo1 −/− mice, however, testosterone levels showed a high inter-individual variability (Fig. 5b). Due to this large variation, there was no statistically significant difference in testosterone levels between male and female Bcmo1 −/− mice. However, BC increased testosterone levels in both Bcmo1 −/− female and male mice with a fold change of 4.7 and 4.0, respectively (Fig. 5c), although, because of the high variation, differences did not reach statistical significance (ANOVA; effect of diet p = 0.12 and interaction effect of diet with Bcmo1 genotype p = 0.14).
Fig. 5.
Testosterone levels in serum of Bcmo1 +/+ mice (a), Bcmo1 −/− mice (b) and the corresponding fold change in testosterone (c) induced by BC supplementation (black bars) as compared to control mice (white bars). Data are expressed as mean ± SEM; significance was tested for diet (BC), gender (S) and genotype (G) using ANOVA and considered significant at p < 0.05. Significant effects using ANOVA are displayed on top of the figure
Discussion
We showed for the first time that BC supplementation to Bcmo1 −/− mice resulted in the regulation of gene expression in opposite directions in male and female mice. We hypothesize that BC alters systemic hormone production, which was supported by the overrepresentation of the steroid biosynthetic process (GO:0006694) in male Bcmo1 −/− mice upon consumption of a BC diet (p = 4.6 × 10−6). Moreover, testosterone concentrations were increased, but highly variable after BC supplementation in Bcmo1 −/− mice. The presence of an active Bcmo1 enzyme was of importance in the observed opposite direction of gene expression changes induced by BC supplementation in male and female mice, since these effects were not observed in Bcmo1 +/+ mice.
BC supplementation to Bcmo1 −/− mice resulted in an opposite direction of gene regulation in males and females. A hypothesis to explain this is that BC supplementation induces alterations in sex hormone synthesis or metabolism in male and female mice. Interestingly, the biosynthetic pathway for BC production in plants partly overlaps with the steroid biosynthetic pathway in animals [25]. To support this hypothesis, we measured testosterone levels in serum, which were highly variable in BC-supplemented Bcmo1 −/− mice, but not in the other groups. Another important sex hormone that might have been changed after BC supplementation to Bcmo1 −/− is 17β-estradiol, but the amount of serum left did not allow us to measure this. Although we were not able to technically prove a change in sex hormones, a change in sex hormones would result in a gender-specific change since the lung contains all necessary elements for sex hormone action, such as enzymes involved in androgen and estrogen metabolism as well as estrogen and androgen receptors [26–28]. In line with this, the GO process for steroid biosynthesis was significantly regulated in male lung tissue, as can be seen in Table 4. Several enzymes involved in steroid conversion were shown to be significantly regulated in male Bcmo1 −/− mice. For example, testosterone can be converted in the lung into estrone and 17β-estradiol. 17β-Estradiol can bind to the estrogen receptor to induce estrogen-related responses [29]. Interestingly, the gene expression of 17-beta-hydroxysteroid dehydrogenase 12 (Hsd17b12), which converts estrone into estradiol, was significantly upregulated [30], and steroid 5 alpha-reductase 1 (Srd5a1), an important enzyme in the inactivation of testosterone [27], was downregulated (p = 0.055) in lungs of male Bcmo1 −/− mice upon BC supplementation. Moreover, 17-beta-hydroxysteroid dehydrogenase 7 (Hsd17b7), which oxidizes or reduces estradiol, and thus decreases the estrogenic potency, was significantly downregulated by BC in female Bcmo1 −/− mice. These data indicate that BC might have had an effect on sex hormone synthesis and had an effect on hormone conversion in male Bcmo1 −/− mice. Further research is needed to elucidate the exact mechanism underlying the observed opposite change in gene regulation induced by BC supplementation in male and female Bcmo1 −/− mice.
Although the number of studies that have assessed effects of BC on hormone production is limited, these studies actually support a relation between BC and steroid hormone alterations. BC supplementation to female dogs resulted in increased plasma progesterone concentrations [31], and cats supplemented with BC had increased estradiol concentrations [32]. In addition, it has been reported that downstream BC metabolites, such as vitamin A and the bioactive metabolite retinoic acid, also have an effect on steroidogenesis and that these effects are, at least in part, due to an increased expression of enzymes involved in steroidogenesis [33, 34]. It is not known whether previously described effects of downstream BC metabolites on steroidogenesis are regulated by binding of retinoic acid to the transcription factors retinoic acid receptor (RAR) or rexinoid X receptor (RXR).
Our results suggest that the BC-induced opposite direction of gene expression in the different genders of Bcmo1 −/− mice was due to an increase in BC concentration and not due to a change in BC metabolites such as retinoic acid, since this opposite direction of gene expression regulation was only observed in Bcmo1 −/− mice. Compared to Bcmo1 −/− mice, male and female Bcmo1 +/+ mice had higher concentrations of the BC metabolites; retinol and retinyl esters accumulated in lung tissue after BC supplementation. Furthermore, although still not fully understood, it is generally believed that retinoic acid concentrations are well regulated [35]. Nevertheless, it may be that the concentration of the bioactive metabolite retinoic acid, able to bind to RAR and RXR, is altered only in Bcmo1 −/− mice and not in Bcmo1 +/+ mice. We previously reported that the Bcmo1 −/− mice were more prone to a phenotypic vitamin A deficiency than Bcmo1 +/+ mice due to alterations in the expression of downstream BC-metabolizing enzymes important in the production of retinoic acid [17]. Supplemental BC was able to restore this phenotype. We similarly show in this study that downstream BC metabolism was altered in Bcmo1 −/− mice. Most striking was the upregulation of Lrat combined with the downregulation of Adh7 in Bcmo1 −/− mice. Lrat is involved in the storage of retinol by the formation of retinyl esters [36], while Adh7 is involved in the conversion of retinol into retinal, which is the precursor for retinoic acid [37]. This therefore implicates a more vitamin A-storing phenotype of Bcmo1 −/− mice compared to Bcmo1 +/+ mice. Moreover, more enzymes involved in downstream BC metabolism were altered, and altogether these data indicate that also a change in retinoic acid, possibly because of a less tight regulation of retinoic acid stores in Bcmo1 −/− mice compared to Bcmo1 +/+ mice, might explain the observed alterations in hormone production.
Another question is whether the processes that were regulated by BC supplementation in lung tissue of Bcmo1 −/− mice are known to respond to hormonal changes. The BC-regulated genes in lungs of Bcmo1 −/− mice in this study were mainly categorized into three biological categories: endothelial functioning, immune response and nervous response. A literature search revealed that these three categories are all affected by one gaseous mediator: nitric oxide (NO). NO is a radical and can be produced endogenously by the iron-containing enzyme nitric oxide synthase (Nos). Three distinct NOS isoforms have been characterized, neuronal NOS (nNOS or NOSI), inducible NOS (iNOS or NOSII) and endothelial NOS (eNOS or NOSIII), and all three NOS isoforms have been detected in the airway epithelium [38, 39]. The functions of NO in the airway include neurotransmission, vascular tone homeostasis and regulation of immune functioning [40]. In our microarray, only NOSIII had an expression twice above background, and there were no significant differences in gene expression after BC supplementation. It is known that estrogen and testosterone can have opposite effects on NO production resulting from NOS activity [41]. An effect of BC on sex hormone production may therefore result in a change in expression of genes that are mediated by NO in opposite directions in male and female lungs. Thus, when BC-induced changes in hormone production, this possibly results in differences in NO production, which might explain that particularly genes involved in immune response, nervous response and endothelial functioning were changed after BC supplementation to male and female Bcmo1 −/− mice.
A last point that we would like to address is whether changes in hormone production or metabolism induced by BC could possibly explain an increase in lung cancer risk as observed in the CARET and ATBC studies. If this is the case, than hormones are a determining factor in lung carcinogenesis, resulting in gender-related differences in lung cancer risk. Indeed, females are thought to have a higher risk for the development of smoke-induced lung cancer than males [42, 43], and lung cancer is more frequent in never-smoking females than in never-smoking males [44]. It has been hypothesized that especially estrogens might be an important contributor to the differences in lung cancer risk between the different genders. For example, estrogen replacement therapy increases lung cancer risk, and early menopause, which results in decreased estrogen levels, which is associated with a reduced risk for lung cancer in women [45]. Therefore, BC-induced changes in hormone production could be an explanation for part of the increases in lung cancer risk in the CARET and ATBC studies.
In summary, we used Bcmo1 −/− mice to investigate BC-induced alterations in gene expression in lung. We unexpectedly found that BC supplementation resulted in an opposite direction of gene expression in male and female Bcmo1 −/− mice. We hypothesize that BC supplementation to Bcmo1 −/− mice resulted in changes in hormone production or metabolism, which would explain our opposite effects on lung gene expression. To fully understand and explain these effects, further research is needed specifically focusing on reproductive organs as potential target organs for BC and on quantitative measurements of sex hormones in response to BC supplementation in males and in females.
Acknowledgments
Yvonne van Helden was supported by a grant from NUTRIM/VLAG. We are grateful to Johannes von Lintig and M. Luisa Bonet for their contributions to the animal experiment. We thank Evelien Kramer, Dini Venema and Annelies Bunschoten for their technical assistance. We also thank DSM Nutraceuticals for the use of Bcmo1 −/− mice and the BC beadlets. None of the authors declare any commercial interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Ziegler RG. Vegetables, fruits, and carotenoids and the risk of cancer. Am J Clin Nutr. 1991;53:251S–259S. doi: 10.1093/ajcn/53.1.251S. [DOI] [PubMed] [Google Scholar]
- 2.van Poppel G. Epidemiological evidence for beta-carotene in prevention of cancer and cardiovascular disease. Eur J Clin Nutr. 1996;50(Suppl 3):S57–S61. [PubMed] [Google Scholar]
- 3.Ziegler RG. A review of epidemiologic evidence that carotenoids reduce the risk of cancer. J Nutr. 1989;119:116–122. doi: 10.1093/jn/119.1.116. [DOI] [PubMed] [Google Scholar]
- 4.Strobel M, Tinz J, Biesalski HK. The importance of beta-carotene as a source of vitamin A with special regard to pregnant and breastfeeding women. Eur J Nutr. 2007;46(Suppl 1):I1–I20. doi: 10.1007/s00394-007-1001-z. [DOI] [PubMed] [Google Scholar]
- 5.Albanes D, Heinonen OP, Taylor PR, Virtamo J, Edwards BK, Rautalahti M, Hartman AM, Palmgren J, Freedman LS, Haapakoski J, Barrett MJ, Pietinen P, Malila N, Tala E, Liippo K, Salomaa ER, Tangrea JA, Teppo L, Askin FB, Taskinen E, Erozan Y, Greenwald P, Huttunen JK. Alpha-tocopherol and beta-carotene supplements and lung cancer incidence in the alpha-tocopherol, beta-carotene cancer prevention study: effects of base-line characteristics and study compliance. J Natl Cancer Inst. 1996;88:1560–1570. doi: 10.1093/jnci/88.21.1560. [DOI] [PubMed] [Google Scholar]
- 6.Omenn GS, Goodman GE, Thornquist MD, Balmes J, Cullen MR, Glass A, Keogh JP, Meyskens FL, Jr, Valanis B, Williams JH, Jr, Barnhart S, Cherniack MG, Brodkin CA, Hammar S. Risk factors for lung cancer and for intervention effects in CARET, the Beta-Carotene and Retinol Efficacy Trial. J Natl Cancer Inst. 1996;88:1550–1559. doi: 10.1093/jnci/88.21.1550. [DOI] [PubMed] [Google Scholar]
- 7.Goodman GE, Thornquist MD, Balmes J, Cullen MR, Meyskens FL, Jr, Omenn GS, Valanis B, Williams JH., Jr The Beta-Carotene and Retinol Efficacy Trial: incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping beta-carotene and retinol supplements. J Natl Cancer Inst. 2004;96:1743–1750. doi: 10.1093/jnci/djh320. [DOI] [PubMed] [Google Scholar]
- 8.Cook NR, Le IM, Manson JE, Buring JE, Hennekens CH. Effects of beta-carotene supplementation on cancer incidence by baseline characteristics in the Physicians’ Health Study (United States) Cancer Causes Control. 2000;11:617–626. doi: 10.1023/A:1008995430664. [DOI] [PubMed] [Google Scholar]
- 9.Hennekens CH, Buring JE, Manson JE, Stampfer M, Rosner B, Cook NR, Belanger C, LaMotte F, Gaziano JM, Ridker PM, Willett W, Peto R. Lack of effect of long-term supplementation with beta carotene on the incidence of malignant neoplasms and cardiovascular disease. N Engl J Med. 1996;334:1145–1149. doi: 10.1056/NEJM199605023341801. [DOI] [PubMed] [Google Scholar]
- 10.von Lintig J, Wyss A. Molecular analysis of vitamin A formation: cloning and characterization of beta-carotene 15, 15’-dioxygenases. Arch Biochem Biophys. 2001;385:47–52. doi: 10.1006/abbi.2000.2096. [DOI] [PubMed] [Google Scholar]
- 11.Hickenbottom SJ, Follett JR, Lin Y, Dueker SR, Burri BJ, Neidlinger TR, Clifford AJ. Variability in conversion of beta-carotene to vitamin A in men as measured by using a double-tracer study design. Am J Clin Nutr. 2002;75:900–907. doi: 10.1093/ajcn/75.5.900. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y, Dueker SR, Burri BJ, Neidlinger TR, Clifford AJ. Variability of the conversion of beta-carotene to vitamin A in women measured by using a double-tracer study design. Am J Clin Nutr. 2000;71:1545–1554. doi: 10.1093/ajcn/71.6.1545. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Yin S, Zhao X, Russell RM, Tang G. beta-Carotene-vitamin A equivalence in Chinese adults assessed by an isotope dilution technique. Br J Nutr. 2004;91:121–131. doi: 10.1079/BJN20031030. [DOI] [PubMed] [Google Scholar]
- 14.Leung WC, Hessel S, Meplan C, Flint J, Oberhauser V, Tourniaire F, Hesketh JE, von Lintig J, Lietz G. Two common single nucleotide polymorphisms in the gene encoding beta-carotene 15, 15′-monoxygenase alter beta-carotene metabolism in female volunteers. Faseb J. 2009;23:1041–1053. doi: 10.1096/fj.08-121962. [DOI] [PubMed] [Google Scholar]
- 15.Lindqvist A, Sharvill J, Sharvill DE, Andersson S. Loss-of-function mutation in carotenoid 15, 15′-monooxygenase identified in a patient with hypercarotenemia and hypovitaminosis A. J Nutr. 2007;137:2346–2350. doi: 10.1093/jn/137.11.2346. [DOI] [PubMed] [Google Scholar]
- 16.Hessel S, Eichinger A, Isken A, Amengual J, Hunzelmann S, Hoeller U, Elste V, Hunziker W, Goralczyk R, Oberhauser V, von Lintig J, Wyss A. CMO1 deficiency abolishes vitamin A production from beta-carotene and alters lipid metabolism in mice. J Biol Chem. 2007;282:33553–33561. doi: 10.1074/jbc.M706763200. [DOI] [PubMed] [Google Scholar]
- 17.van Helden G, Heil SG, van Schooten FJ, Kramer E, Hessel S, Amengual J, Ribot J, Teerds K, Wyss A, Lietz G, Bonet ML, von Lintig J, Godschalk RW, Keijer J. Knockout of the Bcmo1 gene results in an inflammatory response in female lung, which is suppressed by dietary beta-carotene. Cell Mol Life Sci. 2010;67:2039–2056. doi: 10.1007/s00018-010-0341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Helden YG, Godschalk RW, Heil SG, Bunschoten A, Hessel S, Amengual J, Bonet ML, von Lintig J, van Schooten FJ, Keijer J. Downregulation of Fzd6 and Cthrc1 and upregulation of olfactory receptors and protocadherins by dietary beta-carotene in lungs of Bcmo1-/- mice. Carcinogenesis (in press). doi: 10.1093/carcin/bgq083 [DOI] [PubMed]
- 19.Rodenburg W, Keijer J, Kramer E, Vink C, van der Meer R, Bovee-Oudenhoven IM. Impaired barrier function by dietary fructo-oligosaccharides (FOS) in rats is accompanied by increased colonic mitochondrial gene expression. BMC Genomics. 2008;9:144. doi: 10.1186/1471-2164-9-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Schothorst EM, Pagmantidis V, de Boer VC, Hesketh J, Keijer J. Assessment of reducing RNA input for Agilent oligo microarrays. Anal Biochem. 2007;363:315–317. doi: 10.1016/j.ab.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 22.Pellis L, Franssen-van Hal NL, Burema J, Keijer J. The intraclass correlation coefficient applied for evaluation of data correction, labeling methods, and rectal biopsy sampling in DNA microarray experiments. Physiol Genomics. 2003;16:99–106. doi: 10.1152/physiolgenomics.00111.2003. [DOI] [PubMed] [Google Scholar]
- 23.Lee HK, Braynen W, Keshav K, Pavlidis P. ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6:269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rijntjes E, Wientjes AT, Swarts HJ, de Rooij DG, Teerds KJ. Dietary-induced hyperthyroidism marginally affects neonatal testicular development. J Androl. 2008;29:643–653. doi: 10.2164/jandrol.108.005108. [DOI] [PubMed] [Google Scholar]
- 25.Gawienowski AM. Integration of the metabolic pathways of steroids, carotenoids, and retinoids. Crit Rev Biochem Mol Biol. 1999;34:405–410. doi: 10.1080/10409239991209372. [DOI] [PubMed] [Google Scholar]
- 26.Mollerup S, Jorgensen K, Berge G, Haugen A. Expression of estrogen receptors alpha and beta in human lung tissue and cell lines. Lung Cancer. 2002;37:153–159. doi: 10.1016/S0169-5002(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 27.Provost PR, Blomquist CH, Drolet R, Flamand N, Tremblay Y. Androgen inactivation in human lung fibroblasts: variations in levels of 17 beta-hydroxysteroid dehydrogenase type 2 and 5 alpha-reductase activity compatible with androgen inactivation. J Clin Endocrinol Metab. 2002;87:3883–3892. doi: 10.1210/jc.87.8.3883. [DOI] [PubMed] [Google Scholar]
- 28.Wilson CM, McPhaul MJ. A and B forms of the androgen receptor are expressed in a variety of human tissues. Mol Cell Endocrinol. 1996;120:51–57. doi: 10.1016/0303-7207(96)03819-1. [DOI] [PubMed] [Google Scholar]
- 29.Gasperino J, Rom WN. Gender and lung cancer. Clin Lung Cancer. 2004;5:353–359. doi: 10.3816/CLC.2004.n.013. [DOI] [PubMed] [Google Scholar]
- 30.Luu-The V, Tremblay P, Labrie F. Characterization of type 12 17beta-hydroxysteroid dehydrogenase, an isoform of type 3 17beta-hydroxysteroid dehydrogenase responsible for estradiol formation in women. Mol Endocrinol. 2006;20:437–443. doi: 10.1210/me.2005-0058. [DOI] [PubMed] [Google Scholar]
- 31.Weng BC, Chew BP, Wong TS, Park JS, Kim HW, Lepine AJ. Beta-carotene uptake and changes in ovarian steroids and uterine proteins during the estrous cycle in the canine. J Anim Sci. 2000;78:1284–1290. doi: 10.2527/2000.7851284x. [DOI] [PubMed] [Google Scholar]
- 32.Chew BP, Weng BB, Kim HW, Wong TS, Park JS, Lepine AJ. Uptake of beta-carotene by ovarian and uterine tissues and effects on steroidogenesis during the estrous cycle in cats. Am J Vet Res. 2001;62:1063–1067. doi: 10.2460/ajvr.2001.62.1063. [DOI] [PubMed] [Google Scholar]
- 33.Livera G, Pairault C, Lambrot R, Lelievre-Pegorier M, Saez JM, Habert R, Rouiller-Fabre V. Retinoid-sensitive steps in steroidogenesis in fetal and neonatal rat testes: in vitro and in vivo studies. Biol Reprod. 2004;70:1814–1821. doi: 10.1095/biolreprod.103.021451. [DOI] [PubMed] [Google Scholar]
- 34.Livera G, Rouiller-Fabre V, Pairault C, Levacher C, Habert R. Regulation and perturbation of testicular functions by vitamin A. Reproduction. 2002;124:173–180. doi: 10.1530/rep.0.1240173. [DOI] [PubMed] [Google Scholar]
- 35.Thatcher JE, Isoherranen N. The role of CYP26 enzymes in retinoic acid clearance. Expert Opin Drug Metab Toxicol. 2009;5:875–886. doi: 10.1517/17425250903032681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang ZN, Davis GJ, Hurley TD, Stone CL, Li TK, Bosron WF. Catalytic efficiency of human alcohol dehydrogenases for retinol oxidation and retinal reduction. Alcohol Clin Exp Res. 1994;18:587–591. doi: 10.1111/j.1530-0277.1994.tb00914.x. [DOI] [PubMed] [Google Scholar]
- 37.Ross AC. Retinoid production and catabolism: role of diet in regulating retinol esterification and retinoic Acid oxidation. J Nutr. 2003;133:291S–296S. doi: 10.1093/jn/133.1.291S. [DOI] [PubMed] [Google Scholar]
- 38.Asano K, Chee CB, Gaston B, Lilly CM, Gerard C, Drazen JM, Stamler JS. Constitutive and inducible nitric oxide synthase gene expression, regulation, and activity in human lung epithelial cells. Proc Natl Acad Sci USA. 1994;91:10089–10093. doi: 10.1073/pnas.91.21.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giaid A, Saleh D. Reduced expression of endothelial nitric oxide synthase in the lungs of patients with pulmonary hypertension. N Engl J Med. 1995;333:214–221. doi: 10.1056/NEJM199507273330403. [DOI] [PubMed] [Google Scholar]
- 40.Ricciardolo FL, Sterk PJ, Gaston B, Folkerts G. Nitric oxide in health and disease of the respiratory system. Physiol Rev. 2004;84:731–765. doi: 10.1152/physrev.00034.2003. [DOI] [PubMed] [Google Scholar]
- 41.Singh R, Pervin S, Shryne J, Gorski R, Chaudhuri G. Castration increases and androgens decrease nitric oxide synthase activity in the brain: physiologic implications. Proc Natl Acad Sci USA. 2000;97:3672–3677. doi: 10.1073/pnas.050583297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park SK, Cho LY, Yang JJ, Park B, Chang SH, Lee KS, Kim H, Yoo KY, Lee CT. Lung cancer risk and cigarette smoking, lung tuberculosis according to histologic type and gender in a population based case-control study. Lung Cancer. 2009;68:20–26. doi: 10.1016/j.lungcan.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 43.Risch HA, Howe GR, Jain M, Burch JD, Holowaty EJ, Miller AB. Are female smokers at higher risk for lung cancer than male smokers? A case-control analysis by histologic type. Am J Epidemiol. 1993;138:281–293. doi: 10.1093/oxfordjournals.aje.a116857. [DOI] [PubMed] [Google Scholar]
- 44.Siegfried JM. Women and lung cancer: does oestrogen play a role? Lancet Oncol. 2001;2:506–513. doi: 10.1016/S1470-2045(01)00457-0. [DOI] [PubMed] [Google Scholar]
- 45.Taioli E, Wynder EL. Re: Endocrine factors and adenocarcinoma of the lung in women. J Natl Cancer Inst. 1994;86:869–870. doi: 10.1093/jnci/86.11.869. [DOI] [PubMed] [Google Scholar]