Abstract
A variety of molecular genetic approaches were used to study the effect of rabies virus (RV) infection on host gene expression in mouse brain. The down-regulation of gene expression was found to be a major effect of RV infection by using subtraction hybridization. However, a combination of techniques identified approximately 39 genes activated by infection. These included genes involved in regulation of cell metabolism, protein synthesis, synaptic activity, and cell growth and differentiation. Northern blot analysis to monitor temporal activation of several of these genes following infection revealed essentially two patterns of activation: (i) an early response with up-regulation beginning within 3 days after infection and correlating with transcription of RV nuclear protein; and (ii) a late response with enhanced expression occurring at days 6–7 after infection and associated with peak RV replication. The gene activation patterns and the known functions of their products suggest that a number of host genes may be involved in the replication and spread of RV in the brain.
The outcome of rabies virus (RV) infection is determined by the convergence of several different virus–host interactions, including those that contribute to virus replication and spread, pathogenic effects on cells, and antiviral responses. RV is particularly suitable for the study of virus–host interactions because of the small size of its genome and its high specificity for infection of neurons. Nevertheless, the mechanisms involved in the disease process are undoubtedly complex. For example, the virus, which uses the neuronal network to spread within the host, must initially replicate in neurons without causing significant functional impairment that would compromise the infection cycle (1). While there is some understanding of the roles of viral proteins in rabies pathogenesis, the contribution of host factors to RV transcription/replication and axonal/trans-synaptic spread remain unknown. Various technologies are now available to identify host genes that might be essential for the life cycle of RV, important in antiviral defense, or activated nonspecifically in RV infection (2–4). Because each technology is suited for the detection of different classes of genes and different levels of gene expression, we used a variety of assays, including differential display (DD), cDNA array hybridization, subtraction hybridization (SH), and restriction fragment differential display (RFDD), to assess gene expression in rabies-infected mouse brain. With these approaches, we have revealed the effects of rabies infection on select gene expression patterns in the brain. Based on known functional properties of several gene products as well as the timing of their up-regulation after infection, we discuss possible contributions of host genes to the pathogenesis of rabies.
Materials and Methods
Virus Infection of Mice.
Female, 6- to 8-week-old Swiss Webster mice were purchased from Taconic Farms. Mice were maintained under pathogen-free conditions and used to 10 weeks of age. Groups of five mice were infected intramuscularly in the masseter with 10 IM LD50 of the mouse-adapted CVS-N2c strain of RV (5). At different times after infection, mice were killed and their brains were removed and snap-frozen in liquid nitrogen. Analyses were performed on RNA prepared from individual mice, selected on the basis of RNA quality and the presence of rabies N mRNA.
RNA Isolation and DNase I Treatment.
Total cellular RNA was isolated from mouse brain using the Ultraspec RNA isolation system (Biotecx Laboratories, Houston) according to the manufacturer's instructions. Residual DNA contamination was removed by treatment of the RNA with RQ1 RNase-free DNase I (Promega). Samples were extracted with Ultraspec RNA reagent and precipitated with isopropanol. The RNA pellet was resuspended in 300 μl of diethyl pyrocarbonate-treated water.
DD Analysis.
Single-stranded cDNAs generated from control and rabies-infected brain mRNA preparations were used as templates for PCR reactions with random and anchor primers and radiolabeled nucleotides (6). The labeled PCR products of the different samples were electrophoresed in adjacent lanes of a polyacrylamide gel, and differentially expressed genes were identified by comparison of the intensity of comigrating bands. The genes involved were identified by sequencing and by searching in GenBank.
cDNA Array Hybridization.
Highly labeled single-strand cDNAs synthesized from normal and infected total mouse brain RNAs (day 6 after infection) were directly hybridized with the Atlas mouse 1.2 Array (CLONTECH) according to the manufacturer's recommendations. The hybridization spots were detected by phosphorimaging and autoradiography. Densities of hybridization signals were estimated using the Beta 3b version of the Scion image computer program (Scion Corporation, Frederick, MD) and normalized to the expression of housekeeping genes. Genes, up-regulated by rabies infection were reexamined by using Northern hybridization.
SH.
SH was performed using the PCR-Select cDNA Subtraction Kit (CLONTECH) with some modifications. Briefly, double-stranded cDNAs were synthesized by the SMART PCR cDNA Synthesis Kit (CLONTECH). The first and second rounds of SH were performed according to the manufacturer's recommendation with a 4-fold excess of driver cDNA. The resulting cDNA products were random-labeled and used as probes for hybridization with cDNA array membranes.
RFDD.
All steps of RFDD except total RNA isolation were performed by PCG Scientific Corp. (Frederick, MD). Briefly, total RNA from normal brain and from infected mice (day 6 p.i.) were used for synthesis of double-stranded cDNAs, digested with TaqI restriction enzyme, and ligated with two different DNA adaptors. These were amplified by using a conventional primer and a set of 64 selection primers, which allow selection of different cDNA subpopulations in 64 PCR reactions. Amplified fragments were separated on a polyacrylamide gel, and fragments were excised from the gel and used for gene array slide preparation. Slides containing 2,688 different fragments were hybridized simultaneously with two fluorescently labeled cDNA probes obtained from brain RNAs of uninfected mice and mice infected 6 days previously, and the fluorescent signals were quantified by an automated scanner system.
Northern Blot Hybridization.
Total RNAs (20 μg) from brain tissue obtained from individual mice that were either uninfected or infected 3–7 days previously with CVS-N2c strain of RV virus were size-fractionated on 1.2% formaldehyde agarose gel and transferred to nylon membranes. Probes for Northern hybridization, consisting of cDNAs identified by the approaches detailed above, were labeled by using the Random-Primed DNA Labeling Kit (Boeringer Mannheim). After hybridization and washing, Northern blots were quantitated by PhosphoImager system (Molecular Dynamics), exposed to Fuji film, and rehybridized with a probe for glyceraldehyde-3-phospho-dehydrogenase (G3PDH) to monitor RNA loading.
Results
Identification of Genes Differentially Expressed in RV-Infected Mouse Brain.
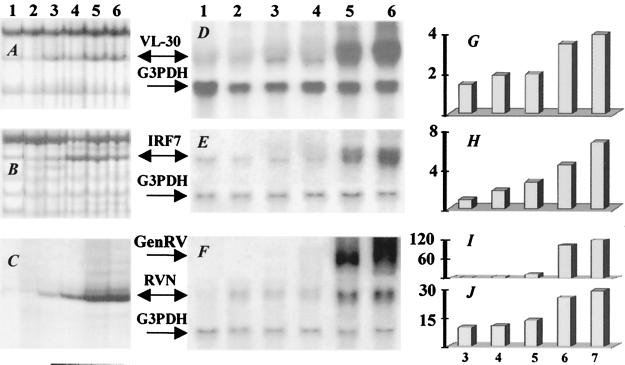
DD using random primer sets identified 29 differentially expressed gene fragments in brains of uninfected mice or mice at 3, 4, 5, 6, or 7 days after infection with CVS-N2c. Nine of these gene fragments represent previously identified genes (Table 1). The sequences of the remaining 20 fragments were identical to different, previously reported mouse ESTs (expressed sequence tags) that have not yet been associated with specific genes. Reverse slot hybridization analysis to confirm the differential expression of the amplified DNA fragments between uninfected and rabies-infected (day 6) mice revealed only three cDNA fragments, corresponding respectively to VL-30 retrotransposon, interferon regulatory factor 7 (IRF7), and RV nuclear (N) protein, that were strongly up-regulated (data not shown). The DD expression patterns of these genes are shown in Fig. 1 (A, B, and C). Northern blot analysis of the expression of the RV N gene in the brains of mice at different days after CVS-N2c infection demonstrated low levels of transcription beginning on day 3 and becoming elevated at days 6–7 (Fig. 1 C, F, and J) when negative-strand RV genomic RNA became abundant (Fig. 1 F and I). Similarly, enhanced expression of IRF7 and VL-30 was also a relatively late occurrence at days 6 and 7 p.i. (Fig. 1 D, E, G, and H).
Table 1.
Host CNS genes induced by RV
| Gene/protein identification | GenBank accession no. | Method of identification | Function |
|---|---|---|---|
| IRF7 | U73037 | DD | Antiviral response |
| EBI-1 ligand chemokine | AF059208 | DD | Chemotactic factor for CD4 cells |
| Mouse MHC class I H2-D-b | M18523 | DD | Cytotoxic T-lymphocyte response |
| NADH-ubiquinone oxidoreductase | P03893 | DD | Respiratory chain protein |
| Genes encoding mitochondrial tRNA | MIMM01 | DD | The tRNAs for Phe, Val, and Leu |
| PSD-95 | D50621 | DD | Postsynaptic density protein |
| L26 | X80699 | DD | Ribosomal protein L26 |
| VL-30 retrotransposon (VL-30) | M21123 | DD | Virus-like retro-element |
| Replication-dependent histone H2A.1 | AA409282 | DD | DNA replication protein |
| Eyes absent homolog 2 | U71208 | cDNA array | Early eye development protein |
| Early growth response protein 3 | S40835 | cDNA array | Transcription factor |
| STAT-1 | U06924 | cDNA array | Transcription factor |
| Major prion protein precursor | M13685 | cDNA array | Scrapie-associated fibril protein |
| Programmed cell death 2 | U10903 | cDNA array | Cell death-associated protein |
| Trombomodulin | X14432 | cDNA array | Anticoagulant |
| VAMP1 | U61751 | cDNA array | Exo- and endocytosis |
| N-cadherin | M31131 | cDNA array | Cell adhesion |
| DNA-binding protein SMBP2 | L10075 | SH | DNA-binding protein |
| Semaphorin G | X97818 | SH | Axonal guidance |
| HSP-90 | S46109 | SH | Stress response |
| Glial cells missing gene homolog | U59876 | SH | Transcription factor |
| Insulin-like growth factor I receptor | U00182 | SH | Growth factor receptor |
| Sp4 zinc finger transcription factor | U62522 | SH | Transcription factor |
| Transcription termination factor 1 | X83974 | SH | DNA binding protein |
| Laminin receptor 1 | J02870 | SH | Receptor for laminin |
| Bcl-xL | L35049 | SH | Apoptosis-associated protein |
| GM-CSF receptor | M85078 | SH | Transmembrane receptor |
| Neuroleukin | M14220 | SH | Growth factor |
| Interleukin-5 receptor alpha subunit | D90205 | SH | Cytokine receptor |
| FHF-4 | P70379 | SH | Growth factor |
| Frizzled homolog 6 | U43319 | SH | Transmembrane receptor |
| Guanine nucleotide binding protein | Y00703 | SH | Signals transducer |
| Calmodulin | X61432 | SH | Calcium sensor protein |
| Apolipoprotein D (ApoD) | L39123 | RFDD | Ligand transporter |
| CDC10 | AJ223794 | RFDD | Postsynaptic density protein |
| Replacement histone H3.3 gene | X13605 | RFDD | DNA synthesis protein |
| GTP binding protein Gz | AF056972 | RFDD | Signal transduction |
| Metallothionein II | J03848 | RFDD | Metal inducible gene |
| L1 retrotransposon | M68842 | RFDD | Retro-element |
Figure 1.
Differential expression genes identified by DD. (Left) RNAs prepared from noninfected (lane 1) and infected mouse brain at days 3–7 p.i. (lanes 2, 3, 4, 5, and 6, respectively) were analyzed as indicated in Materials and Methods: DD patterns of VL-30 (A), IRF7 (B), and RV N protein (C). Confirmation of up-regulation of VL-30 (D) and IRF7 (E) mRNAs by Northern blot. Total RNA prepared as in D and E was assayed with RV N-protein probe (F). The hybridization showed different patterns of expression for RV N protein (RVN) and RV genomic RNAs (GenRV). The blots D, E, and F were reprobed with G3PDH as a loading control. (G–J) PhosphorImage quantification of D, E, and F Northern blots. The quantifications were normalized according to G3PDH-hybridized band intensities. Vertical y axis on graphs shows the expression levels of mRNAs relative to noninfected samples at 3, 4, 5, 6, and 7 days p.i. (x axis): G, VL-30; H, IRF7; I, RV genomic; J, RV N protein.
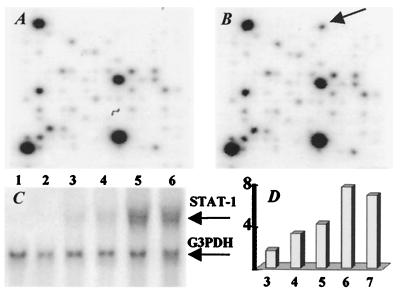
cDNA Array hybridization with 32P-labeled cDNA probes generated from total RNA of uninfected and 6-day p.i. brain revealed eight up-regulated genes at day 6 after RV infection (Table 1). Only STAT-1 was consistently up-regulated in Northern blot analysis (Fig. 2 A, B, and C). STAT-1 mRNA, which is constitutively expressed at low levels, was up-regulated on day 4 p.i., increasing to maximal levels by day 6 p.i. (Fig. 3D).
Figure 2.
Enhanced expression of STAT-1 in mouse brain after RV infection. CLONTECH Atlas Mouse 1.2 Expression Array filters were hybridized with 32P-labeled cDNA probes generated from normal brain RNA (A) and RNA from CVS-N2c infected brain at 6 days p.i. (B). The arrow in B indicates the position of STAT-1 on membrane that was up-regulated in this study. (C) STAT-1 mRNA expression levels were measured by Northern analysis in brain RNAs from uninfected (lane 1) or infected 3, 4, 5, 6, and 7 days previously (lanes 2–6). (D) Histogram shows the quantification of Northern blot signals. The expression ratios of STAT-1 relative to control sample (y axis) at 3, 4, 5, 6, and 7 days p.i. (x axis) indicate a higher expression level of STAT-1 gene at day 6 p.i.
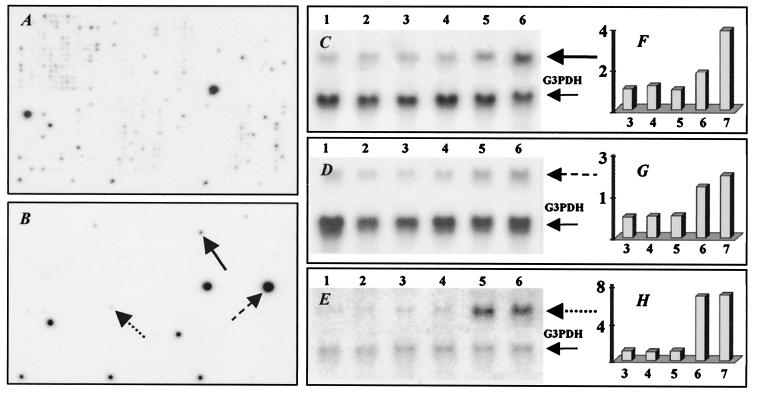
Figure 3.
Expression profiles for genes isolated from RV-infected mouse brain by SH. Hybridization of 32P-labeled cDNA probes from normal brain with 6 day infected brain cDNA subtracted (A) and from RV-infected brain at day 6 p.i. with normal brain cDNA subtracted (B) to Atlas cDNA mouse 1.2 Expression Array. Northern blot analysis of genes identified by SH (C–E). Total RNAs prepared from uninfected brain (lanes 1) or infected for 3 days (lane 2), 4 days (lane 3), 5 days (lane 4), 6 days (lane 5), and 7 days (lane 6) were hybridized with 32P-labeled probes for semaphorin G (solid line in C and B); laminin receptor 1 (dashed line in D and B); neuroleukin (dotted line in E and B). Expression values for each gene (F–H) were determined as described in the legend to Fig. 1.
Subtraction of mRNAs expressed in normal and rabies-infected mouse brain was performed in two directions. “Forward” subtraction of normal brain cDNA with excess cDNA from brain at day 6 p.i. was used to identify genes that are expressed in normal tissue but down-regulated by rabies infection. “Reverse” subtraction of cDNA from brain at day 6 p.i. with excess normal cDNA was used to identify genes up-regulated by infection. Hybridization of forward- and reverse-subtracted cDNA pools with two Atlas Mouse 1.2 Array membranes revealed more hybridization signals for the forward-subtracted probe compared with the reverse-subtracted probe (Fig. 3 A and B). Approximately 190 (16%) of the 1,176 genes represented on the array were expressed at detectable levels in the cDNA preparation from normal brain subtracted with a 4-fold excess of cDNA from the rabies-infected brain. This indicates that at least 190 genes are expressed at significantly higher levels in the normal as opposed to rabies-infected (day 6) mouse brain. However, only 16 (1.4%) of the genes tested were overexpressed in infected brain (Table 1). Subtraction efficiency was confirmed in both cases by depletion of abundant housekeeping genes. Northern blot analysis on brain mRNA samples from individual mice infected 3, 4, 5, 6, and 7 days previously indicated consistent up-regulation of semaphorin G, laminin receptor 1, neuroleukin (Fig. 3 C, D, and E, respectively), 90-kDa heat shock protein (HSP-90), and fibroblast growth factor homologous factors 4 (FHF-4) (data not shown). Expression levels semaphorin G and laminin receptor 1 were initially up-regulated in infected mouse brain at day 6 p.i. and more highly at day 7 p.i. (Fig. 3 C and F), whereas neuroleukin (Fig. 3 E and H) became strongly and abruptly elevated at day 6 p.i. and remained at high levels through day 7. Up-regulation of the remainder of the genes apparently stimulated in RV-infected brain could not be clearly confirmed by Northern analysis (data not shown).
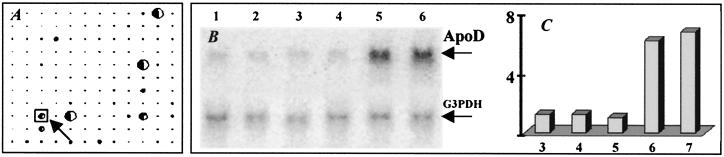
The comparison of gene expression in brain of uninfected mice and mice at day 6 p.i. using RFDD revealed about 2,688 differentially expressed gene fragments. Array analysis of fragments hybridized with fluorescent-labeled cDNA probes generated from both normal and infected brains revealed nine genes expressed at 2-fold or greater levels in RV-infected mice, six of which were associated with a known function (Table 1). Northern hybridization confirmed up-regulation of two genes (ApoD, Fig. 4 B and C, and CDC10, data not shown).
Figure 4.
Gene expression analysis using RFDD. Identification of differentially expressed genes in RV-infected mouse brain was performed as described in Materials and Methods. A is a pictorial representation shown as a GenePies chart with each spot corresponding to each gene. The expression level in each spot/gene is reflected by the size of the GenePies, i.e., spots with large intensities are shown as large pies, whereas spots with intensities close to the background level are very small. White and dark colors of each pie show the ratio of control to noncontrol expression levels. Up-regulation of ApoD at days 3, 4, 5, 6, and 7 p.i. (indicated by the arrow in A) was further assessed by Northern hybridization (B). Relative expression levels of ApoD mRNA (C) were determined as described in the legend to Fig. 1.
Confirmation of Host Gene Activation in Central Nervous System (CNS) of RV-Infected Mice.
Using DD, RFDD, cDNA array, and SH approaches, we identified no gene that was up-regulated in more than a single screening method. This finding might reflect the specificity of each technique for a select array of genes or a greater probability of detecting genes expressed at different levels. Nevertheless, each approach identified genes that were confirmed by Northern blot analysis to be up-regulated relative to the expression of the housekeeping gene G3PDH. However, confirmation of gene up-regulation may be limited to genes expressed at relatively high levels (at least 2-fold higher than those in control tissue). Northern analysis confirmed the up-regulation of 10 genes identified by the above techniques. Of the techniques used, SH yielded the highest number (five) of confirmed up-regulated genes (Table 2). Two basic patterns of gene activation during RV infection were discerned: STAT-1, VL-30, FHF-4, and IRF7 were all rapidly up-regulated at days 3 and 4 p.i., which correlated with the transcription of RV N protein, whereas expression of neuroleukin, laminin receptor 1, ApoD, semaphorin G, HSP-90, and CDC10 was enhanced late in infection (days 6 and 7), corresponding with replication of the RV genome.
Table 2.
Expression levels of host and RV mRNAs in mouse brain at different days after infection
| Gene name | Relative levels of gene expression*
|
||||
|---|---|---|---|---|---|
| Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | |
| STAT-1 | 1.67 | 3.22 | 4.16 | 7.65 | 6.85 |
| VL-30 | 1.45 | 2 | 2.31 | 3.48 | 4.11 |
| FHF-4 | 1.55 | 1.55 | 2.89 | 3.31 | 4.83 |
| IFRF7 | 0.99 | 1.81 | 2.75 | 4.43 | 6.68 |
| ApoD | 1.13 | 1.15 | 1.03 | 6.11 | 6.7 |
| SemG | 1.04 | 1.19 | 0.99 | 1.83 | 3.89 |
| NLK | 0.75 | 0.68 | 1.3 | 9.48 | 9.34 |
| Lam R1 | 0.74 | 0.69 | 0.79 | 1.8 | 2.26 |
| HSP-90 | 0.79 | 0.81 | 0.83 | 2.05 | 2.55 |
| CDC10 | 0.93 | 0.89 | 1.4 | 2.2 | 2.2 |
| RVN protein RNA | 9.64 | 10.64 | 13.31 | 25.16 | 29.04 |
| RV genomic RNA | 0.11 | 2.52 | 10.79 | 102.15 | 118.21 |
LamR1, laminin receptor 1; NLK, neuroleukin; SemG, semaphorin G.
Level of gene expression on the indicated day of infection, normalized to G3PDH, divided by normalized level of expression in uninfected animals.
Discussion
Clearly, the predominant effect of RV infection on host cells is the down-regulation of gene expression. SH analysis revealed that approximately 90% of the genes in normal brain detected on the arrays used were expressed at more than 4-fold lower levels in the day 6 infected mouse brain, whereas only 1.4% were up-regulated in the latter. In this regard, it should be noted that a large number of the genes on the array encode transcription factors and DNA binding proteins that are involved in normal cell maintenance functions may be unnecessary for RV replication. Because our studies were of total brain mRNA, this suggests that RV infection, which is restricted to neurons, inhibits the expression of such genes in the infected neuron as well as in other noninfected neuronal and non-neuronal CNS resident cells. Based on the levels of rabies genomic RNA detected in CNS tissue, we expect that the spread of CVS-N2c is limited for the first 5 days of infection but increases rapidly from day 6 until the death of the animal. We speculate that the inhibition of normal gene expression in uninfected CNS resident cells may make a significant contribution to the pathogenesis of rabies.
While it is somewhat premature, based on these initial results, to speculate about the contributions of the various up-regulated gene products to RV infection, certain known attributes of some of these proteins allow us to formulate testable hypotheses about their possible roles in the infection. As shown in Table 2, host gene responses evidently fall into two categories: (i) early activation beginning by day 3 and increasing throughout the infection in parallel with rabies N protein RNA transcription; and (ii) late activation at days 6 and 7 p.i. which is associated with an exponential increase in the replication of the RV genome. We consider that these temporal differences in host gene activation may be relevant to their functions in rabies infection. In our view, genes activated early in the infection are more likely to represent the innate host cell response to the infection as well as genes important for the replication of virus mRNAs and proteins. On the other hand, we expect that genes contributing to the cell-to-cell spread of the virus and neural dysfunction are more likely to be activated at the later stage when more virus particulars are being formed.
With respect to early events in virus infection, three transcription factors, IRF7, STAT-1, and VL-30, were found to be up-regulated by RV. IRF7 and STAT-1 are members of two different families of transcription factors that have been implicated in antiviral defense, cell growth, and immune regulation. Both are involved in the interferon response pathway (7, 8). On the other hand, VL-30 retro-elements represent a prototype of the “growth-regulated” class of early response nuclear transcription factors whose function, when activated, is to integrate extracellular second/third messenger signaling pathways into cellular responses (9). Retrotransposition of VL-30 elements can also cause insertional deregulation of cellular gene expression (10). Further studies are required to determine whether these transcription factors are induced in the infected neurons or in other CNS resident cells and whether their end results are to protect against RV infection or facilitate spread of the virus.
Gene products, which are neuron specific or have a selective effect on neuronal function, are most likely to be relevant to the replication and spread of the virus. Because neurons are essentially nondividing, we speculate that up-regulation of certain genes may be required to support RV replication, which, at high levels, is a relatively late event in the infection (see Table 2, RV genomic RNA). Growth factor genes are expected in this category and, indeed, two growth factor genes, encoding FHF-4 and neuroleukin, were confirmed by Northern hybridization to be up-regulated in the RV-infected CNS. FHF-4 is a member of the fibroblast growth factor homologous factors family that is predominantly expressed in the CNS (11). The functions of FHF-4 are not fully understood but are expected to include aspects of cell activation (12). Neuroleukin, a neurotropic growth factor that promotes motor neuron regeneration in vivo and the survival of peripheral and central neurons in vitro (13, 14), is strongly up-regulated late in the infection (days 6 and 7).
In addition to host cellular responses, which may support the production of RV RNA and proteins, the assembly of RV may also require host proteins. For example, chaperons may be necessary for proper folding of viral proteins. The stress protein HSP-90 is a highly conserved molecular chaperon that is constitutively expressed in the cytoplasm of many neuronal populations throughout the neocortex, hippocampus, striatum, and cerebellum of the brain (15, 16). HSP-90 possesses two chaperon sites located in the NH2 and COOH terminals and, through direct interactions, has important effects on the activity of a number of proteins (17). We speculate that this may also be the case for rabies proteins as HSP-90 expression is up-regulated during rabies infection, particularly at days 6 and 7 p.i., when extensive assembly of new RV particles is likely.
The virtually exclusive trans-synaptic spread of RV within neurons suggests that there may be specialized interactions between RV and proteins expressed at the synaptic junction. The expression of genes relevant to the spread of RV, but also necessary for synaptic function, may either be conserved or up-regulated, likely late during infection coincidental with high levels of virus production. A group of proteins expressed either in the synaptic region or neuromuscular junction of neurons, including CDC10, PSD-95, N-cadherin, the GTP binding protein Gz, and VAMP1 fall into this category. CDC10, N-cadherin, Gz, and VAMP1 are up-regulated late in rabies infection whereas, unlike the majority of other genes, the expression of PSD-95 is maintained throughout. CDC10, which is associated with the exocyst complex in the postsynaptic region, is thought to be involved in vesicle fusion at the plasma membrane (18, 19). VAMP1 (synaptobrevin), one of the key proteins in the endocytosis vesicle complex in the presynaptic membrane, is thought to have an important role in synaptic vesicle function (20, 21). Gz, which is predominantly expressed in the brain, retina, and adrenals (22), like other GTP binding proteins is likely to be involved in G-protein coupled signal transduction and regulation of membrane traffic (23). We hypothesize that CDC10 and VAMP1, which both have transmembrane domains, may physically interact with RV or its subunits to facilitate trans-synaptic transport of the virus while Gz may be involved in the regulation of this process.
Two other genes found to be up-regulated late in the RV-infected CNS, semaphorin G and laminin receptor 1, may be involved in establishing new connections between neurons, which facilitate spread of the virus. Semaphorin G, a member of the membrane-bound semaphorins that is expressed exclusively in brain, is sharply up-regulated from day 6 to day 7 p.i. when RV replication is maximal. Semaphorin G contains seven COOH-terminal thrombospondin repeats, a motif known to promote neurite outgrowth (24). Laminin receptor 1, which is up-regulated during the same period of infection, potentially has a related function. Laminin, a component of basal laminae, evidently mediates growth and spread of nerve fibers through binding to laminin receptor 1 (25). The activation of these genes in the RV-infected brain suggests that the establishment of new connections between neurons may be an element of the spread of RV in the CNS. Other genes relevant to CNS damage and repair, and possibly this process, are up-regulated in non-neuronal cells in the RV-infected brain. One example of this, ApoD, is thought to play a role in neuronal degeneration and regeneration in the CNS and the peripheral nervous system as it is up-regulated in several neurodegenerative disorders and in traumatic brain injury (26, 27).
We expect that the replication and spread of RV depends on a sequence of host gene functions that parallel the replication of RV proteins and, subsequently, production of RV. Early in the infection (days 3 and 4), several genes are up-regulated, which may be relevant to neuronal activation (Fig. 5). Without further experimentation, we cannot speculate as to whether these support or interfere with the transcription and translation of viral genes. However, we believe that several host genes either up-regulated late in the infection or expressed throughout are important to RV assembly and spread. We theorize that HSP-90 may be involved in the assembly of RV, whereas CDC10 and associated postsynaptic density proteins contribute to the trans-synaptic spread of the virus. We further propose that semaphorin G and laminin receptor 1 as well as, possibly, ApoD may contribute to the establishment of new neural connections to facilitate the spread of RV.
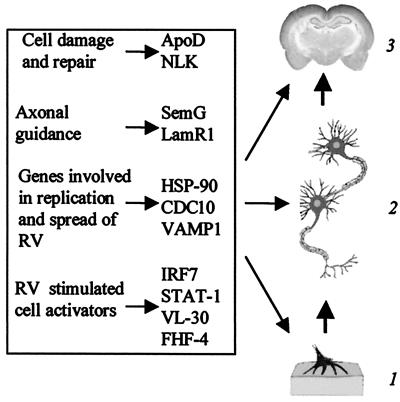
Figure 5.
Schematic representation of possible contributions of mouse genes to RV infection: 1, neuromuscular junction; 2, motor neurons; 3, brain.
Clinical rabies has an almost invariably fatal outcome generally attributed to respiratory failure. Nevertheless, there is most often little or no histopathological evidence of neural destruction in animals dying of rabies (28), and functional changes in RV-infected neurons in vitro are minimal (29). We speculate that death from rabies may be the result of what is essentially a short circuit of normal neural pathways caused by the formation of new associations between neurons as a consequence of genes activated by the infection. To test this and the other hypotheses presented in this study, further experiments in gene knockout mice as well as in cultures of primary neurons are required. It is conceivable that such studies may lead to novel rabies postexposure therapeutic strategies that may extend the period after infection before a lethal outcome becomes inevitable.
Acknowledgments
This study was supported by Public Health Service Grants AI-41544 and AI-09706.
Abbreviations
- RV
rabies virus
- p.i.
postinfection
- N protein
nuclear protein
- CNS
central nervous system
- DD
differential display
- SH
subtraction hybridization
- RFDD
restriction fragment differential display
- G3PDH, glyceraldehyde-3-phospho-dehydrogenase
IRF7, interferon regulatory factor 7
- HSP-90
90-kDa heat shock protein
- FHF-4
fibroblast growth factor homologous factors 4
References
- 1.Kelly R M, Strick P L. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/s0165-0270(00)00296-x. [DOI] [PubMed] [Google Scholar]
- 2.Raux H, Flam A, Blondel D. J Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castellanos J E, Martinez M, Acosta O, Hurtado H. Brain Res. 2000;871:120–126. doi: 10.1016/s0006-8993(00)02408-2. [DOI] [PubMed] [Google Scholar]
- 4.Irwin D J, Wunner W H, Ertl H C, Jackson A C. Clin Infect Dis. 2000;30:4–12. [Google Scholar]
- 5.Morimoto K, Hooper D C, Spitsin S, Koprowski H, Dietzschold B. J Virol. 1999;73:510–518. doi: 10.1128/jvi.73.1.510-518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 7.Marie I, Smith E, Prakash A, Levy D E. Mol Cell Biol. 2000;20:8803–8814. doi: 10.1128/mcb.20.23.8803-8814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen V T, Benveniste E N. J Biol Chem. 2000;275:23674–23684. doi: 10.1074/jbc.M002482200. [DOI] [PubMed] [Google Scholar]
- 9.Carter A T, Norton J D, Avery R J. Biochim Biophys Acta. 1988;951:130–138. doi: 10.1016/0167-4781(88)90033-4. [DOI] [PubMed] [Google Scholar]
- 10.French N S, Norton J D. Biochim Biophys Acta. 1997;1352:33–47. doi: 10.1016/s0167-4781(97)00009-2. [DOI] [PubMed] [Google Scholar]
- 11.Munoz-Sanjuan I, Fallon J F, Nathans J. Mech Dev. 2000;95:101–112. doi: 10.1016/s0925-4773(00)00336-1. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto S, Mikami T, Ohbayashi N, Ohta M, Itoh N. Biochim Biophys Acta. 1998;1398:38–41. doi: 10.1016/s0167-4781(98)00050-5. [DOI] [PubMed] [Google Scholar]
- 13.Haga A, Niinaka Y, Raz A. Biochim Biophys Acta. 2000;1480:235–244. doi: 10.1016/s0167-4838(00)00075-3. [DOI] [PubMed] [Google Scholar]
- 14.Leclerc N, Vallee A, Nabi I R. J Neurosci Res. 2000;60:602–612. doi: 10.1002/(SICI)1097-4547(20000601)60:5<602::AID-JNR5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 15.Ishimoto T, Kamei A, Koyanagi S, Nishide N, Uyeda A, Kasai M, Taguchi T. Biochem Biophys Res Commun. 1998;253:283–287. doi: 10.1006/bbrc.1998.9701. [DOI] [PubMed] [Google Scholar]
- 16.Gass P, Schroder H, Prior P, Kiessling M. Neurosci Lett. 1994;182:188–192. doi: 10.1016/0304-3940(94)90794-3. [DOI] [PubMed] [Google Scholar]
- 17.Itoh H, Ogura M, Komatsuda A, Wakui H, Miura A B, Tashima Y. Biochem J. 1999;343:697–703. [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu S C, Hazuka C D, Roth R, Foletti D L, Heuser J, Scheller R H. Neuron. 1998;20:1111–1122. doi: 10.1016/s0896-6273(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 19.Walikonis R S, Jensen O N, Mann M, Provance D W, Jr, Mercer J A, Kennedy M B. J Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berglund L, Hoffmann H J, Dahl R, Petersen T E. Biochem Biophys Res Commun. 1999;264:777–780. doi: 10.1006/bbrc.1999.1588. [DOI] [PubMed] [Google Scholar]
- 21.Kim P K, Hollerbach C, Trimble W S, Leber B, Andrews D W. J Biol Chem. 1999;274:36876–36882. doi: 10.1074/jbc.274.52.36876. [DOI] [PubMed] [Google Scholar]
- 22.Zigman J M, Westermark G T, LaMendola J, Steiner D F. Endocrinology. 1994;135:31–37. doi: 10.1210/endo.135.1.8013366. [DOI] [PubMed] [Google Scholar]
- 23.Bomsel M, Mostov K. Mol Biol Cell. 1992;3:1317–1328. doi: 10.1091/mbc.3.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams R H, Betz H, Puschel A W. Mech Dev. 1996;57:33–45. doi: 10.1016/0925-4773(96)00525-4. [DOI] [PubMed] [Google Scholar]
- 25.Massia S P, Rao S S, Hubbell J A. J Biol Chem. 1993;268:8053–8059. [PubMed] [Google Scholar]
- 26.Franz G, Reindl M, Patel S C, Beer R, Unterrichter I, Berger T, Schmutzhard E, Poewe W, Kampfl A. J Neurochem. 1999;73:1615–1625. doi: 10.1046/j.1471-4159.1999.0731615.x. [DOI] [PubMed] [Google Scholar]
- 27.Kalman J, McConathy W, Araoz C, Kasa P, Lacko A G. Neurol Res. 2000;22:330–336. doi: 10.1080/01616412.2000.11740678. [DOI] [PubMed] [Google Scholar]
- 28.Plotkin S A. Clin Infect Dis. 2000;30:4–12. doi: 10.1086/313632. [DOI] [PubMed] [Google Scholar]
- 29.Ohashi H, Komori S. Eur J Pharmacol. 2000;404:79–88. doi: 10.1016/s0014-2999(00)00621-x. [DOI] [PubMed] [Google Scholar]