Abstract
The initiation of DNA replication requires the binding of the initiator protein, DnaA, to specific binding sites in the chromosomal origin of replication, oriC. DnaA also binds to many sites around the chromosome, outside oriC, and acts as a transcription factor at several of these. In low-G+C Gram-positive bacteria, the primosomal proteins DnaD and DnaB, in conjunction with loader ATPase DnaI, load the replicative helicase at oriC, and this depends on DnaA. DnaD and DnaB also are required to load the replicative helicase outside oriC during replication restart, independently of DnaA. Using chromatin immunoprecipitation, we found that DnaD and DnaB, but not the replicative helicase, are associated with many of the chromosomal regions bound by DnaA in Bacillus subtilis. This association was dependent on DnaA, and the order of recruitment was the same as that at oriC, but it was independent of a functional oriC and suggests that DnaD and DnaB do not require open complex formation for the stable association with DNA. These secondary binding regions for DnaA could be serving as a reservoir for excess DnaA, DnaD, and DnaB to help properly regulate replication initiation and perhaps are analogous to the proposed function of the datA locus in Escherichia coli. Alternatively, DnaD and DnaB might modulate the activity of DnaA at the secondary binding regions. All three of these proteins are widely conserved and likely have similar functions in a range of organisms.
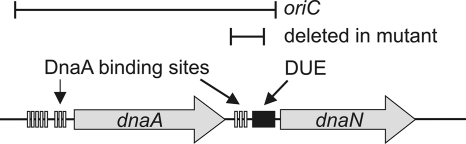
The replication initiation protein and transcription factor DnaA is an AAA+ ATPase that binds to many regions on the chromosome. The primary binding region is at the origin of chromosomal replication, oriC (at 0° on the B. subtilis chromosome), where there are many individual DnaA binding sites (Fig. 1). DnaA binds to oriC and causes local unwinding (melting) of the A+T-rich DNA unwinding element (DUE) (Fig. 1) and the subsequent assembly of the replicative helicase, followed by the assembly of the rest of the replication machinery (reviewed in references 27, 29, 49, and 56).
FIG. 1.
Map of the B. subtilis oriC region. The oriC regions contains the clusters of DnaA binding sites upstream and downstream from dnaA and the DNA unwinding element (DUE). The binding sites upstream of DnaA contribute to the autoregulation of DnaA and also are needed for oriC function. Part of the DUE and the DnaA binding sites downstream from dnaA are missing in the oriC-6 mutant (1).
The assembly of the replicative helicase is mediated by helicase loader proteins. In Escherichia coli, a single protein, the AAA+ ATPase DnaC, functions to load the helicase (reviewed in references 13 and 34). In contrast, in B. subtilis and other low-G+C Gram-positive bacteria, three different proteins, DnaD, DnaB, and the AAA+ ATPase DnaI, are needed to load the replicative helicase (DnaC in B. subtilis) during replication initiation at oriC and replication restart at stalled replication forks (5-7, 13, 24, 40, 54, 55, 63). There is a defined order of stable association of the replication initiation proteins with oriC. DnaA binds first, followed by DnaD and then DnaB, and finally the DnaI-mediated loading of helicase occurs (58). It is not known, however, if the association of DnaD and DnaB with oriC requires the melting of the DUE.
In addition to its primary role in replication initiation and binding to sites in the oriC region, DnaA also binds to many secondary sites around the chromosome, away from oriC. These secondary sites have been detected by chromatin immunoprecipitation (ChIP)-PCR, ChIP-chip, and analogous (ChAP-chip) approaches (3, 18, 25). Many of these secondary sites are in promoter regions, and DnaA functions as a transcription factor in several of these regions. DnaA modulates the transcription of many genes, including its own, likely under normal growth conditions and in response to replication stress and the inactivation of replication proteins (3, 9, 12, 16, 18, 22, 23, 29, 43). When replication stress is induced by DNA damage, the inhibition of replication elongation, or the inactivation of replication initiation protein DnaD, DnaB, or DnaI, DnaA becomes more active as a transcription factor, and the expression of target genes changes (3, 9, 17, 18, 22). DnaA activates the expression of some genes and represses the expression of others, and this apparently is dependent on the position of its binding sites relative to the RNA polymerase binding site (3, 18).
Because DnaD and DnaB are associated with and are recruited to oriC by DnaA, we wished to determine if they also were associated with other chromosomal regions that are bound by DnaA. We analyzed the association of DnaD and DnaB throughout the B. subtilis genome in response to replication stress. We found that DnaD and DnaB are associated with many (perhaps all) of the chromosomal regions bound by DnaA, and that there is the same dependence of association at these secondary DnaA binding regions as that at oriC: DnaA, then DnaD, then DnaB. In contrast to oriC, there was little or no detectable association of the replicative helicase (DnaC) at most of the secondary DnaA binding regions. Our findings indicate that the association of DnaD and DnaB with oriC likely does not depend on the melting of the DUE, and that there is a role for DnaD and DnaB outside oriC and independent of their function in replication restart.
We propose that in addition to their roles in the loading of the replicative helicase, DnaD and/or DnaB is modulating one or more aspects of DnaA function. In addition, it is possible that the secondary DnaA binding regions are acting to modulate oriC function by titrating excess DnaA, DnaD, and DnaB.
MATERIALS AND METHODS
B. subtilis strains and alleles.
All B. subtilis strains (Table 1) were isogenic with the laboratory wild type, JH642, and contain the trpC2 and pheA1 alleles, unless indicated otherwise. dnaA1, dnaB134, dnaD23, and dnaI2 (6, 7, 31, 48) are temperature-sensitive alleles that prevent replication initiation at the nonpermissive temperature. The transposon insertions Tn917ΩHU163, Tn917ΩHU151, and zhb83::Tn917 are linked to dnaA, dnaD, and the dnaB-dnaI operon, respectively. The oriC deletion mutation (Fig. 1) removes DnaA binding sites and part of the DUE between dnaA and dnaN (1, 28). The heterologous origin, oriN, and its initiator, repN, support replication in the absence of oriC and dnaA (20) and were integrated into spoIIIJ (1).
TABLE 1.
B. subtilis strains used
Media and growth conditions.
Cells were grown at 30°C in defined minimal medium (26) with 1% glucose, 0.1% glutamate and were supplemented with required amino acids. For the inactivation of the various replication initiation proteins by means of temperature-sensitive alleles, appropriate strains were shifted to the nonpermissive temperature (48°C for dnaB134 and dnaD23, 50°C for dnaI2, and 52°C for dnaA1) for 1 h. This allows ongoing rounds of replication to finish but prevents the initiation of a new round of replication from oriC. Replication elongation was arrested by adding 6-(p-hydroxyphenylazo)-uracil (HPUra) to a final concentration of 38 μg/ml to exponentially growing cells for 30 min. HPUra binds to the catalytic subunit (PolC) of DNA polymerase and blocks replication (4).
Chromatin immunoprecipitation (ChIP)-chip.
DNA microarrays contained almost all of the open reading frames of B. subtilis as well as 265 intergenic regions (3). They did not contain rRNA or tRNA genes and would not detect the recently described DnaA-independent association of DnaD, DnaB, and helicase with rRNA loci (42a). Details about the microarray have been deposited in NCBI's Gene Expression Omnibus (14) and are accessible through GEO platform accession number GPL10707.
Chromatin immunoprecipitation, DNA labeling, and subsequent hybridization to DNA microarrays were performed essentially as described previously (3). Briefly, cells were harvested from 50 ml of culture. After the pellets were washed with ice-cold phosphate-buffered saline (PBS), cells were lysed in 2.5 ml buffer A for 45 min at 37°C. Subsequently, 2.5 ml 2× IP buffer (0.1 M Tris-HCl, pH 7, 0.3 M NaCl, 10 mM EDTA, 0.02% [vol/vol] Triton X-100) was added and lysates were sonicated. Aliquots of 1 ml of each lysate were incubated with the appropriate antibodies as described previously (58). Results from the array hybridizations were analyzed using the Prep + 07 processing package (42), and data from three independent biological replicates were averaged and plotted using Microsoft Excel. Figures were further prepared in CorelDRAW Graphics Suite X4. The microarray data have been deposited in NCBI's Gene Expression Omnibus (14) and are accessible through GEO Series accession number GSE23686 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23686).
ChIP-qPCR.
Chromatin immunoprecipitation followed by quantitative real-time PCR (qPCR) was performed essentially as described previously (58), except that immunoprecipitations from cross-linked lysates were done either for 2 h at room temperature or overnight at 4°C. Results from the two conditions were indistinguishable (data not shown). Primers for amplifying the oriC region in the oriC deletion mutants were HM41 (5′-GGATTGATTTCACACAGCTTGTGT-3′) and HM42 (5′-CTTCCGGCACGTCCCTCCTT-3′), and yhaX was used as a control locus (18) using primers oWKS-145 (5′-CGAGCAAGGTGTCGCTTA-3′) and oWKS-146 (5′-GCAGGCGGTCATCATGTA-3′).
Purification of DnaA and DnaD. (i) DnaA.
An E. coli dnaA null mutant that overproduces wild-type (untagged) B. subtilis DnaA was constructed and kindly provided by A. Valbuzzi and W. F. Burkholder. pAV13 contains B. subtilis dnaA cloned between the NcoI and BglII sites of pQE60 (Qiagen). dnaA is expressed from a strong phage T5 promoter with lac operators. The plasmid is contained in E. coli strain MS3898 [ΔdnaA zia::pKN500(mini R1) asnB32 relA1 spoT1 thi-1 ilv192 mad1 recA1 λimm434 F−] (60) that contains pBB42, a derivative of pACYC184 that expresses lacIq.
DnaA was purified essentially as described previously (15). E. coli cells were grown in 4 liters of LB medium with isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) to induce the expression of dnaA. Cells were pelleted and resuspended and then lysed in 20 ml lysis buffer (25 mM HEPES, pH 7.6, 80 mM KCl, 1 mM EDTA, 20 mM spermidine, 2 mM dithiothreitol [DTT]) with 0.3 mg/ml lysozyme and a cocktail of protease inhibitors (Sigma P-8849) for 30 min on ice, followed by freezing and thawing. The cleared supernatant was precipitated with solid ammonium sulfate (0.35 mg/ml) for 30 min. The precipitate was centrifuged and resuspended in loading buffer containing 10 mM sodium phosphate, pH 7.6, 0.5 mM EDTA, 10 mM Mg acetate, 100 mM NaCl, 1 mM DTT, and 10% glycerol. This was passed through a PD-10 desalting column and then loaded onto a 5-ml HiTrap heparin column (GE Healthcare) that had been equilibrated with the same buffer. The column was washed with 50 ml of loading buffer, and DnaA was eluted with increasing NaCl concentrations, ranging from 200 to 900 mM in 100 mM increments. DnaA-containing fractions, as determined by SDS polyacrylamide gel electrophoresis, were concentrated and resuspended in storage buffer containing 45 mM HEPES, pH 7.6, 0.5 mM EDTA, 10 mM Mg acetate, 1 mM DTT, and 20% sucrose, with 100 mM potassium glutamate (KGlu). DnaA was loaded onto a Q column and eluted with storage buffer containing 1 M KGlu. DnaA purity was determined by SDS-PAGE, and fractions with >95% pure DnaA were pooled, concentrated, and frozen in storage buffer containing 500 mM KGlu.
(ii) DnaD.
B. subtilis dnaD was cloned between the BamHI and XhoI sites in pET21(+) (Novagen) to generate plasmid pCAL769. This plasmid was introduced into E. coli strain BL21-AI (Invitrogen) and produces DnaD with a C-terminal hexahistidine tag (DnaD-his6). Cells were grown in 500 ml of LB medium with 0.2% arabinose (to induce the expression of T7 RNA polymerase) and 1 mM IPTG (to induce the expression of the promoter driving dnaD-his6), harvested, and pelleted. DnaD-His6 was purified essentially as described previously (40). Cells were resuspended in 10 ml of HEN500 buffer (20 mm HEPES, pH 7.6, 0.1 mm EDTA, 500 mm NaCl, 10 mM imidazole) and broken by sonication. The cleared supernatant was incubated with 1 ml of nickel-nitrilotriacetic acid (Ni-NTA) beads for 1 h. The protein was eluted from the Ni-NTA beads by the addition of 500 mM imidazole. DnaD-His6 was diluted 5-fold with buffer Q50 (buffer Q is 50 mM Tris, 0.1 mM EDTA, 1 mM DTT, pH 8, supplemented with 50 mM NaCl) and applied to a Q column equilibrated with Q50 buffer. DnaD-His6 was eluted with increasing NaCl concentrations (from 200 to 800 mM in increments of 100 mM) in Q buffer. DnaD-containing fractions were diluted 5-fold and bound to a HiTrap heparin column equilibrated with buffer Q50. DnaD-His6 was eluted with increasing NaCl concentrations (between 200 and 500 mM, in increments of 100 mM) in Q buffer. The fractions containing DnaD visible on SDS-PAGE were dialyzed against DnaA storage buffer containing 500 mM KGlu.
Gel shift assays.
The DNA template for the gel shift assays was an end-labeled 135-bp fragment containing five DnaA binding sites from the dnaA promoter region. The fragment was generated by PCR using primers OCB67 (5′ AACTCTTGATTACTAATCCTACC 3′) and OCB68 (5′ ATATAGTAGATAAATAGCTTTTCG 3′) and B. subtilis chromosomal DNA as the template. The PCR product was purified with a PCR purification column (Qiagen) and end labeled with [γ-32]P-ATP using T4 polynucleotide kinase. The labeled DNA fragment then was separated from free ATP using a PCR purification column (Qiagen).
Conditions for the gel shift assays were performed essentially as described for DnaA (15). DnaA (50 nM) was incubated with 1 nM DNA in buffer containing 45 mM HEPES, pH 7.6, 50 mM KCl, 100 mM KGlu, 10 mM Mg acetate, 2.5 mM AMP-PNP, 0.5 mM EDTA, 1 mM DTT, 50 μg/ml bovine serum albumin (BSA), and 20% glycerol. Binding reactions were done with or without DnaD-His6 (300 nM) for 20 min at room temperature. The binding reactions were run on a 6% polyacrylamide gel (19:1 acrylamide/bisacrylamide) in 0.5× Tris-borate-EDTA (TBE) and 2.5% glycerol run in 0.5× TBE at approximately 12 V/cm for 3 h. Gels were imaged on a Typhoon scanner (GE Healthcare).
RESULTS
DnaD and DnaB are associated with chromosomal regions bound by DnaA.
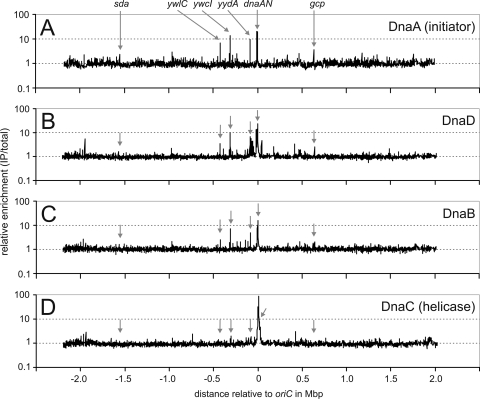
We performed chromatin immunoprecipitation (ChIP) assays using polyclonal antibodies against DnaA (Fig. 2A), DnaD (Fig. 2B), and DnaB (Fig. 2C), essentially as described previously (58). DNA from the immunoprecipitates was labeled and hybridized to DNA microarrays (ChIP-chip) containing probes for almost all open reading frames (but not rRNA and tRNA genes) and 265 intergenic regions (3). The association of DnaA with its secondary targets is higher during replication stress than during normal growth (3, 18), and these conditions were likely to increase the ability to detect other associated proteins. Wild-type B. subtilis cells were grown in defined minimal medium, and replication stress was induced by inhibiting the replicative polymerase PolC by the addition of HPUra for 30 min.
FIG. 2.
Genome-wide binding of DnaA, DnaB, DnaD, and the replicative helicase (DnaC). Strain JH642 was grown to mid-exponential phase and treated with HPUra for 30 min. DnaA (A), DnaD (B), DnaB (C), and helicase (D) were immunoprecipitated from cells after being cross-linked with formaldehyde. The enrichment of a given chromosomal region in the immunoprecipitates compared to total genomic DNA is plotted on the y axis relative to the chromosomal position on the x axis. The position of oriC is set as 0. Data represent averages from three independent biological replicates. These data are consistent with previous findings (3), although in the previous work cells were treated with HPUra for 60 min and the effects on the association of DnaA generally were greater than those detected here. Arrows indicate the location of characterized target sites of DnaA (3, 18, 25).
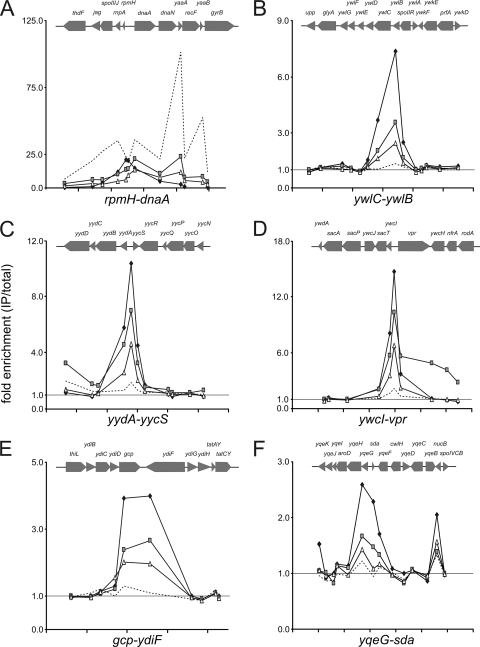
As expected, DnaA was detectably associated with oriC and several of its secondary targets under these conditions (Fig. 2A and 3 and Table 2). In addition to oriC, there were four predominant regions, along with yqeG-sda, detected in the ChIP-chip experiments, which is consistent with previous results (3, 11, 25). There was an approximately 2- to 4-fold increase in the association of DnaA with the secondary chromosomal regions, and a greater increase in association near oriC, during replication stress compared to that during exponential growth (Table 2). These results are consistent with previous findings (3). As reported previously (3, 18), these regions are characterized by a cluster of at least nine sequences that match the consensus DnaA binding site (allowing for one mismatch). These clustered sites are not known to have a conserved arrangement.
FIG. 3.
Detailed view of specific chromosomal regions bound by DnaA, DnaD, and DnaB. Data from Fig. 2 are replotted to show the association of DnaA (filled diamonds), DnaD (gray squares), DnaB (open triangles), and helicase (dotted lines) with specific chromosomal regions. Each y axis is a different scale. Tick marks represent 2-kb intervals. The gene organization is shown above each plot. Regions shown are rpmH-dnaA (oriC) (A), ywlC-ywlB (B), yydA-yycS (C), ywcI-vpr (D), gcp-ydiF (E), and yqeG-sda (F). These regions were chosen for the detailed view based on previous analyses of genes controlled by DnaA, in vivo analyses of binding, and descriptions of the potential DnaA binding sites (3, 9, 18, 25). Data represent averages from three independent biological replicates. Error bars are omitted for clarity (standard errors were within 10% of the means).
TABLE 2.
Enrichment of DnaA targets in immunoprecipitates
| Regiona | Fold increaseb (enrichment value, +HPUra/−HPUra) after IP |
|||
|---|---|---|---|---|
| DnaA | DnaD | DnaB | DnaC | |
| rpmH-dnaA (oriC) | 8.8 (21.8/2.5) | 8.4 (14.3/1.7) | 4.6 (7.0/1.5) | 17.6 (24.0/1.4) |
| ywlC-ywlB | 3.8 (7.4/2.0) | 3.1 (3.6/1.2) | 2.4 (2.4/1.0) | 1.2 (1.3/1.1) |
| yydA-yycS | 3.8 (10.4/2.8) | 4.3 (7.0/1.6) | 3.7 (4.6/1.2) | 1.3 (1.9/1.5) |
| ywcI-vpr | 3.5 (14.8/4.2) | 3.4 (10.3/3.1) | 4.1 (6.8/1.7) | 1.8 (2.1/1.2) |
| gcp-ydiF | 2.6 (4.0/1.5) | 2.5 (2.6/1.1) | 2.2 (2.0/0.9) | 1.1 (1.1/1.0) |
| yqeG-sda | 2.8 (2.3/0.8) | 1.5 (1.5/1.0) | 1.5 (1.1/0.7 | 1.1 (0.9/0.8) |
Data points used for each region are from rpmH(for oriC), ywlB(for ywlC/B), the intergenic region between yydA and yyc, the intergenic region between ywcI and vpr, ydiF(for gcp-ydiF), and sda(for yqeG-sda).
Fold increase in enrichment of the indicated locus in the specific immunoprecipitates, comparing replication arrest(+HPUra) to exponential growth(−HPUra). The actual enrichment values for each condition are shown in parentheses. Values are rounded to one decimal place and are averages of ChIP-chip results from three independent biological cultures, analyzed as described in Materials and Methods. Data for +HPUra are the same as those shown in Fig. 2 and 3.
We did similar ChIP-chip experiments with antibodies to the helicase loader proteins DnaD (Fig. 2B) and DnaB (Fig. 2C). We were not able to reliably immunoprecipitate DnaI under our experimental conditions, likely because the association of DnaI with DNA is transient (24, 58). We found that both DnaD and DnaB were associated with chromosomal regions outside oriC, and many of these regions appeared to correspond to the regions bound by DnaA (Fig. 2A and 3). As with DnaA, the association of DnaD and DnaB with these secondary (non-oriC) regions was greater during replication stress than during normal exponential growth (Table 2). From these experiments, we conclude that DnaD and DnaB are present at many, if not all, of the secondary (non-oriC) chromosomal regions that are bound by DnaA. This association might reflect recruitment by DnaA in a way analogous to that at oriC (58).
The replicative helicase (DnaC) is not associated with DnaD and DnaB at many of the chromosomal regions.
DnaD and DnaB function in recruiting the DnaI/helicase complex to oriC and places of replication restart (5-7, 24, 40, 54, 55, 58, 63). We monitored the association of the replicative helicase (DnaC) with chromosomal regions using ChIP-chip and antibodies to the replicative helicase (58). During normal exponential growth, there was some detectable association of helicase with oriC (Table 2). This association increased markedly after the inhibition of replication elongation caused by the addition of HPUra (Fig. 2D, 3A, and Table 2). Notably, the replicative helicase was not significantly associated with most of the identified secondary DnaA binding regions, even under conditions of replication stress (HPUra), when its association with oriC is high (Fig. 3B to E, Table 2). We cannot rule out the possibility that there is helicase at these regions, but it is below our limit of detection. However, the helicase signal at oriC after HPUra treatment is greater than that for the other three proteins (Fig. 2 and 3A), indicating that if helicase was recruited to these other regions, then we likely would detect it. We conclude that the association of DnaA, DnaD, and DnaB with these non-oriC regions is not sufficient to recruit significant amounts of the replicative helicase.
It is interesting that the replicative helicase was associated with several chromosomal regions outside oriC, in regions not known to be bound by DnaA (data not shown). Among these, the most notable was the association of helicase with the terminus region during exponential growth (∼14-fold enrichment compared to that of most other chromosomal regions), which is consistent with the function of the replication termination protein Rtp as an antagonist of helicase (30). This association was not detectable after treatment with HPUra (Fig. 2D). Helicase also was associated, to some extent, with other chromosomal regions (these data are available from NCBI's GEO database [14]). We do not yet know the significance of this association, but we suspect that it represents regions of replication fork stalling and/or PriA-dependent restart, perhaps analogous to that recently described at the rRNA operons (42a).
Hierarchical association of DnaA, DnaD, and DnaB.
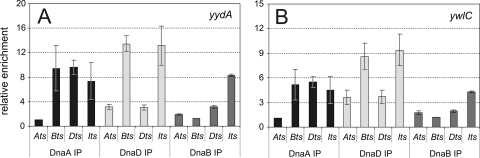
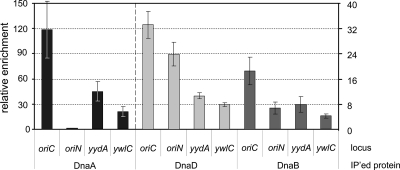
At oriC, there is a hierarchical dependence of the association of DnaA, DnaD, and DnaB, in that order, prior to the loading of the replicative helicase (58). We found that there is the same ordered dependence of the association of DnaA, DnaD, and DnaB at the secondary chromosomal regions, even though helicase is not loaded at these regions. We inactivated dnaA, dnaD, dnaB, or dnaI using temperature-sensitive alleles and determined the association of DnaA, DnaD, and DnaB with the secondary DnaA targets yydA (Fig. 4A) and ywlC (Fig. 4B) using ChIP and quantitative real-time PCR (ChIP-qPCR), essentially as described previously for the oriC region (58). The association of DnaA with these regions was independent of dnaD, dnaB, and dnaI (Fig. 4). That is, DnaA was associated with these regions in dnaD(Ts), dnaB(Ts), and dnaI(Ts) mutants at nonpermissive temperatures. This association at high temperature in the replication-temperature-sensitive mutants was greater than that following treatment with HPUra, which is consistent with previous findings (3). The association of DnaD was independent of DnaB and DnaI but was reduced in the dnaA(Ts) and dnaD(Ts) mutants. The association of DnaB was reduced in all mutants except dnaI(Ts) (Fig. 4). These results show that the helicase loader proteins DnaD and DnaB are recruited to the secondary targets of DnaA in the order DnaA, DnaD, then DnaB, the same ordered dependence as that at oriC (58).
FIG. 4.
Hierarchy in the association of DnaA, DnaD, and DnaB with chromosomal regions. Temperature-sensitive mutants included dnaA(Ts) (WKS588), dnaD(Ts) (KPL73), and dnaB(Ts) (KPL69). Cells were grown in defined minimal medium at 30°C and shifted to the nonpermissive temperature (see Materials and Methods) for 1 h to inactive the mutant protein and allow ongoing rounds of replication to finish. The relative enrichment of yydA (A) and ywlC (B) after cross-linking and the immunoprecipitation of the indicated proteins, DnaA (black bars), DnaD (light gray bars), and DnaB (dark gray bars), was determined. Error bars represent standard errors of the means from three independent cultures.
ChIP experiments represent a population average of protein association. When multiple proteins are associated with a given region, they all could be there together, or different proteins could be associated separately in different subpopulations of cells. The association of DnaA was required for the association of DnaD, and DnaD was required for DnaB, indicating that all three proteins are present together. It is formally possible that DnaA is needed to recruit DnaD and DnaB and then DnaA is no longer present in the region, but we think this is unlikely.
Association of DnaA, DnaD, and DnaB with secondary chromosomal regions does not depend on replication from oriC.
We considered the possibility that a complex of DnaA, DnaD, and DnaB assembles at oriC and subsequently is recruited to the secondary sites in a replication-dependent manner, possibly through the postulated association of DnaA with the replisome (59). To test this, we measured the association of DnaA, DnaD, and DnaB with the secondary regions in a strain without a functional oriC. The chromosomal origin of replication of B. subtilis can be inactivated when a heterologous origin (oriN) is provided elsewhere in the chromosome (1, 20, 28). Replication from oriN requires a dedicated initiator protein, RepN, is dependent on DnaD, DnaB, and DnaC (the replicative helicase), but is independent of DnaA (20).
We used ChIP-qPCR to measure the association of DnaA, DnaD, and DnaB with the secondary regions yydA and ywlC, the mutant oriC region, and oriN in the absence of a functional oriC (Fig. 1). We found that DnaA, DnaD, and DnaB were associated with the secondary regions yydA and ywlC and the mutant oriC (Fig. 5). The amount of association was greater than that in wild-type (oriC+) cells, likely due to replication stress in cells initiating asynchronously from oriN. As expected, DnaD and DnaB also were associated with oriN, but DnaA was not (Fig. 5). Based on these results, we conclude that the association of DnaA, DnaD, and DnaB with the secondary target sites of DnaA does not depend on replication initiation from oriC or on the function of DnaA as a replication initiation protein. These results also indicate that the association of DnaD and DnaB likely does not require the melting of the DUE in oriC, as the oriC mutant is missing DUE sequences (Fig. 1). Since the dnaA promoter region is part of oriC (46, 53), this association is a reflection of DnaA binding to its own promoter region and part of oriC. It is possible that there is some melting nearby, but if so, this is insufficient to function as an origin of replication.
FIG. 5.
Association of DnaA, DnaD, and DnaB at chromosomal loci is independent of replication from oriC. The oriC mutant strain MMB170 (oriN ΔoriC-6) was grown in defined minimal medium at 30°C, and the relative enrichment of the oriC region, oriN, yydA, and ywlC was determined by quantitative real-time PCR after cross-linking and the immunoprecipitation of DnaA (black bars), DnaD (light gray bars), and DnaB (dark gray bars). Note the different scales for the DnaA IP compared to those for DnaD and DnaB. Error bars represent standard errors of the means (n = 3).
DnaA-dependent association of DnaD with DNA in vitro.
The results presented above indicate that DnaD is recruited to DNA by DnaA in vivo, and that DnaB is not need for this recruitment. However, it is not clear if other cellular proteins are required. To test this, we decided to monitor the association of purified DnaD and DnaA with DNA in vitro. We used a gel mobility shift assay to detect the association of each protein with a DNA fragment from the oriC region, upstream of dnaA, that contains five DnaA binding sites but not the DUE (Fig. 1). The addition of DnaD had no detectable effect on the mobility of this fragment in a polyacrylamide gel (Fig. 6, lanes 1, 2, and 6). In contrast, the addition of DnaA (with AMP-PNP in the reaction) caused a decrease in mobility, indicating the binding of DnaA to the fragment (Fig. 6, lanes 3 and 5). The addition of DnaD along with DnaA caused a supershift (Fig. 6, lane 4). Similar results were obtained with ADP, ATP, or no nucleotide in place of AMP-PNP (C. Y. Bonilla and A. D. Grossman, unpublished results). These in vitro results are consistent with the DnaA-dependent association of DnaD with DNA in vivo and indicate that DnaA and DNA are sufficient for this association, although we cannot rule out the participation of other proteins in vivo.
FIG. 6.
DnaA-dependent association of DnaD with DNA. Gel shift assay of a 135-bp PCR product from the dnaA promoter containing five DnaA binding sites was incubated either alone (lanes 1 and 6) or with DnaD (lane 2), DnaA (lanes 3 and 5), or both DnaA and DnaD (lane 4) and separated on a 6% polyacrylamide gel. Binding reaction mixtures contained DnaA at 50 nM, DnaD at 300 nM, and DNA at 1 nM final concentrations. We have not yet analyzed the association of DnaB with these proteins in vitro.
DISCUSSION
We found that the replication initiation and restart proteins DnaD and DnaB are associated with several chromosomal regions, not just oriC, and that this association depends on the replication initiation protein and transcription factor DnaA. DnaA binds to specific sites located in many chromosomal regions, some of which are in transcriptional regulatory regions and affect gene expression (3, 9, 18, 22, 25, 62). Several of the regions bound by DnaA are readily detected by ChIP-chip (3) and the analogous ChAP-chip (11, 25). Virtually all of these regions were associated with DnaD and DnaB (Fig. 2, 3, and Table 2), and this association was dependent on DnaA. The binding of DnaA to these regions increases during replication stress, because DnaA is more active and/or there is more DnaA per chromosome (3, 18). Similarly, the association of DnaD and DnaB with these regions also increased during replication stress (Table 2). We propose that the association of DnaA, DnaD, and DnaB at various chromosomal regions is indicative of a function for these proteins outside oriC and independent of the function of DnaD and DnaB in replication restart. Below we summarize the functions of DnaD and DnaB in replication initiation and restart and discuss possible functions related to their association with DnaA and its binding sites around the chromosome.
Function of DnaD and DnaB at oriC.
dnaD and dnaB are needed for replication initiation at oriC. They were first identified in screens for temperature-sensitive mutations that block replication initiation but not replication elongation (31). Their characterized role at oriC is to help load the replicative helicase (DnaC). First, DnaA binds to sequences in oriC. This binding is needed for the association of DnaD, which in turn is necessary for the association of DnaB (58). These events are necessary for the loading of the replicative helicase by the AAA+ loader protein DnaI. There might be additional roles for DnaD and DnaB in chromosome architecture and DNA remodeling (66, 67).
One of the key steps in replication initiation is open complex formation; that is, the melting of the DNA unwinding element (DUE) in oriC and stabilization of the melted region. The DNA-remodeling activities of DnaD and DnaB might be important for these processes. In vitro, DnaA, but not DnaD, is needed for open complex formation (2, 35). The association of DnaD (and DnaB) with oriC requires DnaA function (58), and DnaD and DnaB can bind both single- and double-stranded DNA (40, 61, 66, 67). These findings are compatible with the possibility that the stable association of DnaD with oriC occurs after, and may even depend on, DnaA-mediated open complex formation. However, three lines of evidence from our results indicate that the association of DnaD with the oriC region does not depend on origin function or melting. First, in an oriC mutant largely missing the DUE and incapable of supporting replication initiation (Fig. 1), there still was the association of DnaA, DnaD, and DnaB with this region (Fig. 5). Second, DnaD and DnaB associate with several other chromosomal regions in a DnaA-dependent manner. These regions do not function as origins and likely do not undergo melting. Some are in promoter regions and appear to modulate gene expression, and others are separate from transcriptional regulatory regions (3, 9, 18, 25). Third, there is a DnaA-dependent association of DnaD with DNA on a linear DNA fragment. Since a supercoiled template is needed for DnaA-mediated melting (2, 35), the linear fragment should not have a melted region. The simplest explanation for our findings is that the stable recruitment of DnaD and DnaB to oriC requires DnaA and occurs independently of the melting of the DUE. We propose that the ability of DnaD to stimulate DNA looping and duplex melting (68) contributes to open complex formation at oriC in vivo, as has been postulated for DnaB (19). In this model, DnaD might be required for the melting or stabilization of the DUE in vivo (47), even though it is not required in vitro.
Function of DnaD and DnaB in replication restart.
In addition to their role in loading the replicative helicase at oriC during replication initiation, DnaD and DnaB (and DnaI) also are needed for loading helicase during replication restart (5-7, 24, 40, 54, 63). This occurs at various places around the chromosome where replication forks might collapse due to DNA lesions or other obstacles on the template. In contrast to replication initiation at oriC, replication restart does not require DnaA. Rather, it requires the primosomal protein PriA, which associates with Ssb at replication forks and functions to recruit the helicase loading machinery to regions of fork collapse (6, 7, 36, 40, 54).
It seems unlikely that the DnaA-dependent association of DnaD and DnaB with chromosomal regions is a reflection of replication restart at these regions. There is no known role for DnaA in replication restart, and we detected little or no helicase at these chromosomal regions. In contrast, there is a strong and detectable association of the replicative helicase with oriC during replication initiation in vivo (55, 58). Recent work indicates that highly transcribed rRNA genes are hotspots for replication fork stalling and restart (42a). The stalling and restart likely are due to conflicts between the replisome and RNA polymerase and result in the increased association of the replicative helicase and the restart proteins DnaD and DnaB with these regions. This association does not depend on DnaA (42a), making it mechanistically different from the association described here.
Possible functions for the DnaA-dependent association of DnaD and DnaB with chromosomal regions.
We considered several possible functions of DnaA, DnaD, and DnaB away from oriC. The function of DnaD and DnaB at the DnaA binding regions might be related to their DNA remodeling activities (10, 57, 61, 66-68). For example, DnaA could recruit DnaD and DnaB to the DnaA binding regions around the chromosome, and these regions could serve as hubs for an aspect of chromosome organization. This model is highly speculative, and we currently favor two other possibilities. DnaD and DnaB might function to modulate the activity of DnaA or affect the transcription of the genes regulated by DnaA. For example, DnaD and/or DnaB might be involved in inhibiting the activity of DnaA after the transcriptional response to replication stress (3, 17, 18). Alternatively, the secondary binding regions might serve as reservoirs for DnaA, DnaD, and DnaB to titrate excess protein away from oriC to help modulate origin activity. This could be analogous to the proposed function of the datA locus of E. coli (33, 44). These possibilities are not mutually exclusive. It also is possible that the association of DnaD and DnaB with DnaA at chromosomal regions outside oriC serves no physiological function and is simply a reflection of the interactions between these proteins. We do not favor this possibility.
Most of what we know about the regulation of the activity of DnaA comes from work with E. coli (reviewed in reference 29). However, many of the proteins and mechanisms so well studied in E. coli, e.g., Hda and RIDA, and the sequestration protein SeqA (29, 32, and references therein), do not exist outside the proteobacteria, and they certainly do not exist in B. subtilis (32, 65). YabA is one of the better characterized regulators of replication initiation and DnaA in B. subtilis (11, 17, 21, 51, 52, 59). YabA does not have a homolog in E. coli. Like Hda, YabA interacts with DnaA and DnaN and is a negative regulator of replication initiation (51, 52). However, it is not required for the transcriptional response to replication stress and does not appear to significantly affect the expression of genes controlled by DnaA (17).
Based on their interactions in vivo and in vitro, DnaD and/or DnaB could modulate the activity of DnaA. The binding of DnaA to several of its targets increases during replication stress (3), likely at different times during the replication cycle (62). DnaD and DnaB are not needed for the binding of DnaA or for the increase in binding during replication stress (3, 18). However, DnaD and DnaB might be involved in the recovery from replication stress, perhaps by antagonizing the activity of DnaA. We have not yet detected an effect of DnaD or DnaB on the activity of DnaA in vitro. In addition, experiments to test this in vivo are complicated by the fact that DnaD and DnaB are essential proteins needed for replication initiation, and mutations in these actually induce a replication stress response that causes the increased binding of DnaA to DNA and DnaA-mediated changes in gene expression (3, 18, 37).
Maintaining the proper amount of DnaA is critical for proper replication control (29, 32). In E. coli, a site called datA is thought to serve to titrate excess DnaA away from oriC to help maintain proper replication control (33, 44, 45). Although regions that function similarly have not been defined in other organisms, one or more of the DnaA binding regions in B. subtilis could serve a similar function. Furthermore, the overproduction of DnaD is toxic (41). The association of DnaA, DnaD, and DnaB with several chromosomal regions outside oriC might represent a conserved strategy to help maintain proper replication control by titrating replication initiation proteins away from oriC.
DnaD and DnaB homologues are found in low-G+C Gram-positive bacteria, and, where characterized, they are required for replication initiation (8, 38, 39), and their functions in replication are almost certainly the same as those in B. subtilis. We suspect that DnaD and DnaB also are associated with DnaA outside the oriC region in other organisms, and that their function outside oriC in these organisms is similar to that in B. subtilis. E. coli and its relatives do not have homologues of B. subtilis DnaD and DnaB. However, in E. coli, the AAA+ helicase loader protein (E. coli DnaC) interacts directly with DnaA (50), indicating that the E. coli helicase loader also might associate with DnaA at chromosomal regions outside oriC. Thus, the functions of these DnaA-associated primosomal proteins outside oriC might be similar in a wide range of organisms.
Supplementary Material
Acknowledgments
We thank G. Wright for providing HPUra; A. Valbuzzi and W. F. Burkholder for advice on purifying B. subtilis DnaA and providing the DnaA overexpression strain; C. Lee for providing the DnaD-His6 expression plasmid; T. A. Baker, J. E. Grimwade, J. D. Wang, and A. C. Leonard for discussions; P. Soultanas for communicating results prior to publication and comments on the manuscript; and members of the Grossman laboratory for discussions.
This work was supported in part by a Rubicon fellowship from the Netherlands Organization for Scientific Research to W.K.S., NIH postdoctoral fellowship F32GM093408 to H.M., and NIH grant GM41934 to A.D.G.
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Berkmen, M. B., and A. D. Grossman. 2007. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol. Microbiol. 63:150-165. [DOI] [PubMed] [Google Scholar]
- 2.Bramhill, D., and A. Kornberg. 1988. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52:743-755. [DOI] [PubMed] [Google Scholar]
- 3.Breier, A. M., and A. D. Grossman. 2009. Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J. Bacteriol. 191:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, N. C. 1970. 6-(p-hydroxyphenylazo)-uracil: a selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 67:1454-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruand, C., S. D. Ehrlich, and L. Janniere. 1995. Primosome assembly site in Bacillus subtilis. EMBO J. 14:2642-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruand, C., M. Farache, S. McGovern, S. D. Ehrlich, and P. Polard. 2001. DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol. Microbiol. 42:245-255. [DOI] [PubMed] [Google Scholar]
- 7.Bruand, C., et al. 2005. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol. Microbiol. 55:1138-1150. [DOI] [PubMed] [Google Scholar]
- 8.Bruck, I., and M. O'Donnell. 2000. The DNA replication machine of a gram-positive organism. J. Biol. Chem. 275:28971-28983. [DOI] [PubMed] [Google Scholar]
- 9.Burkholder, W. F., I. Kurtser, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269-279. [DOI] [PubMed] [Google Scholar]
- 10.Carneiro, M. J., et al. 2006. The DNA-remodelling activity of DnaD is the sum of oligomerization and DNA-binding activities on separate domains. Mol. Microbiol. 60:917-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, E., N. Ogasawara, and S. Ishikawa. 2008. The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtilis. Genes Genet. Syst. 83:111-125. [DOI] [PubMed] [Google Scholar]
- 12.Collier, J., S. R. Murray, and L. Shapiro. 2006. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 25:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey, M. J., and M. O'Donnell. 2003. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13:R594-R596. [DOI] [PubMed] [Google Scholar]
- 14.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuoka, T., S. Moriya, H. Yoshikawa, and N. Ogasawara. 1990. Purification and characterization of an initiation protein for chromosomal replication, DnaA, in Bacillus subtilis. J. Biochem. 107:732-739. [DOI] [PubMed] [Google Scholar]
- 16.Gon, S., et al. 2006. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 25:1137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goranov, A. I., A. M. Breier, H. Merrikh, and A. D. Grossman. 2009. YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress. Mol. Microbiol. 74:454-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goranov, A. I., L. Katz, A. M. Breier, C. B. Burge, and A. D. Grossman. 2005. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. U. S. A. 102:12932-12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grainger, W. H., C. Machon, D. J. Scott, and P. Soultanas. 2010. DnaB proteolysis in vivo regulates oligomerization and its localization at oriC in Bacillus subtilis. Nucleic Acids Res. 38:2851-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan, A. K., et al. 1997. Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J. Bacteriol. 179:2494-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi, M., Y. Ogura, E. J. Harry, N. Ogasawara, and S. Moriya. 2005. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol. Lett. 247:73-79. [DOI] [PubMed] [Google Scholar]
- 22.Hoover, S. E., W. Xu, W. Xiao, and W. F. Burkholder. 2010. Changes in DnaA-dependent gene expression contribute to the transcriptional and developmental response of Bacillus subtilis to manganese limitation in Luria-Bertani medium. J. Bacteriol. 192:3915-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hottes, A. K., L. Shapiro, and H. H. McAdams. 2005. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 58:1340-1353. [DOI] [PubMed] [Google Scholar]
- 24.Ioannou, C., P. M. Schaeffer, N. E. Dixon, and P. Soultanas. 2006. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 34:5247-5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa, S., et al. 2007. Distribution of stable DnaA-binding sites on the Bacillus Subtilis genome detected using a modified ChIP-chip method. DNA Res. 14:155-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaacks, K. J., J. Healy, R. Losick, and A. D. Grossman. 1989. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 171:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, A., and M. O'Donnell. 2005. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74:283-315. [DOI] [PubMed] [Google Scholar]
- 28.Kadoya, R., A. K. Hassan, Y. Kasahara, N. Ogasawara, and S. Moriya. 2002. Two separate DNA sequences within oriC participate in accurate chromosome segregation in Bacillus subtilis. Mol. Microbiol. 45:73-87. [DOI] [PubMed] [Google Scholar]
- 29.Kaguni, J. M. 2006. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 60:351-375. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan, D. L., and D. Bastia. 2009. Mechanisms of polar arrest of a replication fork. Mol. Microbiol. 72:279-285. [DOI] [PubMed] [Google Scholar]
- 31.Karamata, D., and J. D. Gross. 1970. Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol. Gen. Genet. 108:277-287. [DOI] [PubMed] [Google Scholar]
- 32.Katayama, T., S. Ozaki, K. Keyamura, and K. Fujimitsu. 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 8:163-170. [DOI] [PubMed] [Google Scholar]
- 33.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. W. H. Freeman and Co., New York, NY.
- 35.Krause, M., B. Ruckert, R. Lurz, and W. Messer. 1997. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol. 274:365-380. [DOI] [PubMed] [Google Scholar]
- 36.Lecointe, F., et al. 2007. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 26:4239-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemon, K. P., I. Kurtser, J. Wu, and A. D. Grossman. 2000. Control of initiation of sporulation by replication initiation genes in Bacillus subtilis. J. Bacteriol. 182:2989-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, Y., et al. 2004. Identification of temperature-sensitive dnaD mutants of Staphylococcus aureus that are defective in chromosomal DNA replication. Mol. Genet. Genomics 271:447-457. [DOI] [PubMed] [Google Scholar]
- 39.Li, Y., et al. 2007. DnaB and DnaI temperature-sensitive mutants of Staphylococcus aureus: evidence for involvement of DnaB and DnaI in synchrony regulation of chromosome replication. Microbiology 153:3370-3379. [DOI] [PubMed] [Google Scholar]
- 40.Marsin, S., S. McGovern, S. D. Ehrlich, C. Bruand, and P. Polard. 2001. Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem. 276:45818-45825. [DOI] [PubMed] [Google Scholar]
- 41.Marston, F. Y., et al. 2010. When simple sequence comparison fails: the cryptic case of the shared domains of the bacterial replication initiation proteins DnaB and DnaD. Nucleic Acids Res. 38:6930-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin-Requena, V., A. Munoz-Merida, M. G. Claros, and O. Trelles. 2009. PreP+07: improvements of a user friendly tool to preprocess and analyse microarray data. BMC Bioinformatics 10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Merrikh, H., C. Machon, W. H. Grainger, A. D. Grossman, and P. Soultanas. Co-directional replication-transcription conflicts lead to replication restart. Nature, in press. [DOI] [PMC free article] [PubMed]
- 43.Messer, W., and C. Weigel. 1997. DnaA initiator-also a transcription factor. Mol. Microbiol. 24:1-6. [DOI] [PubMed] [Google Scholar]
- 44.Morigen, A. L. Olesen, and K. Skarstad. 2003. Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol. Microbiol. 50:349-362. [DOI] [PubMed] [Google Scholar]
- 45.Morigen, F. Molina, and K. Skarstad. 2005. Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J. Bacteriol. 187:3913-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moriya, S., T. Atlung, F. G. Hansen, H. Yoshikawa, and N. Ogasawara. 1992. Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol. Microbiol. 6:309-315. [DOI] [PubMed] [Google Scholar]
- 47.Moriya, S., Y. Imai, A. K. Hassan, and N. Ogasawara. 1999. Regulation of initiation of Bacillus subtilis chromosome replication. Plasmid 41:17-29. [DOI] [PubMed] [Google Scholar]
- 48.Moriya, S., K. Kato, H. Yoshikawa, and N. Ogasawara. 1990. Isolation of a dnaA mutant of Bacillus subtilis defective in initiation of replication: amount of DnaA protein determines cells' initiation potential. EMBO J. 9:2905-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mott, M. L., and J. M. Berger. 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5:343-354. [DOI] [PubMed] [Google Scholar]
- 50.Mott, M. L., J. P. Erzberger, M. M. Coons, and J. M. Berger. 2008. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 135:623-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noirot-Gros, M. F., et al. 2002. An expanded view of bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 99:8342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noirot-Gros, M. F., et al. 2006. Functional dissection of YabA, a negative regulator of DNA replication initiation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 103:2368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogura, Y., Y. Imai, N. Ogasawara, and S. Moriya. 2001. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 183:3833-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polard, P., et al. 2002. Restart of DNA replication in Gram-positive bacteria: functional characterisation of the Bacillus subtilis PriA initiator. Nucleic Acids Res. 30:1593-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rokop, M. E., J. M. Auchtung, and A. D. Grossman. 2004. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol. Microbiol. 52:1757-1767. [DOI] [PubMed] [Google Scholar]
- 56.Schaeffer, P. M., M. J. Headlam, and N. E. Dixon. 2005. Protein-protein interactions in the eubacterial replisome. IUBMB Life 57:5-12. [DOI] [PubMed] [Google Scholar]
- 57.Schneider, S., W. Zhang, P. Soultanas, and M. Paoli. 2008. Structure of the N-terminal oligomerization domain of DnaD reveals a unique tetramerization motif and provides insights into scaffold formation. J. Mol. Biol. 376:1237-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smits, W. K., A. I. Goranov, and A. D. Grossman. 2010. Ordered association of helicase loader proteins with the Bacillus subtilis origin of replication in vivo. Mol. Microbiol. 75:452-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soufo, C. D., et al. 2008. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev. Cell 15:935-941. [DOI] [PubMed] [Google Scholar]
- 60.Sutton, M. D., and J. M. Kaguni. 1995. Novel alleles of the Escherichia coli dnaA gene are defective in replication of pSC101 but not of oriC. J. Bacteriol. 177:6657-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner, I. J., D. J. Scott, S. Allen, C. J. Roberts, and P. Soultanas. 2004. The Bacillus subtilis DnaD protein: a putative link between DNA remodeling and initiation of DNA replication. FEBS Lett. 577:460-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veening, J. W., H. Murray, and J. Errington. 2009. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 23:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Velten, M., et al. 2003. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell 11:1009-1020. [DOI] [PubMed] [Google Scholar]
- 64.Wang, J. D., G. M. Sanders, and A. D. Grossman. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128:865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zakrzewska-Czerwińsk, J., D. Jakimowicz, A. Zawilak-Pawlik, and W. Messer. 2007. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 31:378-387. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, W., S. Allen, C. J. Roberts, and P. Soultanas. 2006. The Bacillus subtilis primosomal protein DnaD untwists supercoiled DNA. J. Bacteriol. 188:5487-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, W., et al. 2005. The Bacillus subtilis DnaD and DnaB proteins exhibit different DNA remodelling activities. J. Mol. Biol. 351:66-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, W., et al. 2008. Single-molecule atomic force spectroscopy reveals that DnaD forms scaffolds and enhances duplex melting. J. Mol. Biol. 377:706-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.