Abstract
Mycoplasma pneumoniae, a pathogen causing human pneumonia, binds to solid surfaces at its membrane protrusion and glides by a unique mechanism. In this study, P1 adhesin, which functions as a “leg” in gliding, was isolated from mycoplasma culture and characterized. Using gel filtration, blue-native polyacrylamide gel electrophoresis (BN-PAGE), and chemical cross-linking, the isolated P1 adhesin was shown to form a complex with an accessory protein named P90. The complex included two molecules each of P1 adhesin and P90 (protein B), had a molecular mass of about 480 kDa, and was observed by electron microscopy to form 20-nm-diameter spheres. Partial digestion of isolated P1 adhesin by trypsin showed that the P1 adhesin molecule can be divided into three domains, consistent with the results from trypsin treatment of the cell surface. Sequence analysis of P1 adhesin and its orthologs showed that domain I is well conserved and that a transmembrane segment exists near the link between domains II and III.
Mycoplasmas are commensal, and occasionally parasitic, bacteria that lack a peptidoglycan layer and have small genomes (47). Mycoplasma pneumoniae, a cause of human “walking pneumonia,” forms a membrane protrusion at one pole and exhibits gliding motility in the direction of the protrusion (22, 32-35, 56). The maximum speed reaches 1 μm, one-half its cell length, per second (40, 51). This motility, combined with the ability to adhere to epithelial cells (20), is involved in the pathogenic process, enabling the cells to translocate from the tips of bronchial cilia to the host cell surface (24). Previous studies, including genome analyses, have shown that this motility is not related to other known mechanisms of bacterial movement, nor does it involve motor proteins known to be involved in eukaryotic cell motility (11, 17, 32, 39, 42). The gliding machinery is located at a membrane protrusion called the attachment organelle, which is composed of an internal rod-shaped cytoskeleton and nap-like surface protrusions (18, 20, 22, 32-35).
P1 adhesin, a 170-kDa protein found in mycoplasmas, binds to solid surfaces, such as host cells or glass (12, 51). In the genome of M. pneumoniae, P1 adhesin (MPN141) is encoded in the same operon with two other open reading frames (ORFs), MPN140 and MPN142 (25). The translation product of MPN142 is divided into two proteins by a putative endopeptidase activity (5), P40 (protein C; 45 kDa) and P90 (protein B; 83 kDa), both of which have been suggested to interact physically with P1 adhesin (26, 27, 52, 53). MPN140 encodes a putative phosphodiesterase whose role in cytadherence is not known. It has been suggested that P1 adhesin functions as a leg for gliding motility, repeatedly catching and releasing surface structure, because the monoclonal antibodies against the protein reduce gliding speed and the ability to bind to solid surfaces (51). P1 adhesin is also known as the immunodominant protein and exhibits sequence polymorphism among its clinical strains (19, 57-59, 65).
Knowing the structure and activity of P1 adhesin should be helpful in understanding the motility and antigenic variation of M. pneumoniae. In the present study, we isolated native P1 adhesin from M. pneumoniae cells and analyzed its structure.
MATERIALS AND METHODS
Isolation of the P1-P90 complex.
M. pneumoniae strain M129 (the type strain of group I) (29), whose genome has been sequenced (6, 10), was grown at 37°C in Aluotto medium (2, 38). The following procedures were done at 4°C unless otherwise noted. Cells from 1 liter of culture in the exponential phase were centrifuged at 14,000 × g for 10 min and washed twice with phosphate-buffered saline (PBS) consisting of 75 mM sodium phosphate (pH 7.3) and 68 mM NaCl. The cells were suspended to an optical density at 600 nm of 20 in PBS containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM β-mercaptoethanol, and 1 mM EDTA and then were mixed with CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} to 1% (vol/vol). After gentle shaking for 5 min, the suspension was centrifuged at 25,000 × g for 20 min. The pellet was dissolved in one-fifth of its original volume of PBS and then mixed with N-octyl-β-d-glucoside (octylglucoside) to 2% (vol/vol). After gentle shaking for 10 min at room temperature (RT), the suspension was centrifuged at 25,000 × g for 20 min at RT, as described previously (15). The supernatant was fractionated by salting out with ammonium sulfate at 45 to 55% saturation, and the insoluble fraction was recovered by centrifugation at 22,000 × g for 15 min. The pellet was dissolved and then dialyzed overnight against PBS-NT containing 75 mM sodium phosphate (pH 7.3), 400 mM NaCl, and 0.3% Triton X-100. The insoluble fraction was removed by centrifugation at 25,000 × g for 15 min. The soluble fraction was loaded onto a Hi Load 16/60 Superdex 200-pg set (GE Healthcare, Milwaukee, WI) on an AKTA purifier (GE Healthcare), eluted with PBS-NT at a flow rate of 1 ml/min, and fractionated into 1-ml aliquots. The fractions containing the P1-P90 complex were concentrated using Biomax-50 (Millipore, Bedford, MA) to 0.1 mg/ml. The homogeneity of the fraction was estimated by densitometry of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and stained with Coomassie brilliant blue (CBB), using a scanner (GT-9800F; Epson, Nagano, Japan) and analyzing software, Image J 1.41 (http://rsb.info.nih.gov/ij/). The focused protein bands were identified by peptide mass fingerprinting (PMF), as reported previously (41), by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), and by immunoblotting using 0.5 μg/ml monoclonal antibody (51).
Analyzing the construction of the P1-P90 complex.
BS3 (bis[sulfosuccinimidyl] suberate) is a noncleavable and membrane-impermeable cross-linker containing an amine-reactive N-hydroxysulfosuccinimide (NHS) ester at each end of an 8-carbon spacer arm. The P1-P90 complex fraction in PBS-NT was treated with various concentrations of BS3 (Thermo Scientific, Waltham, MA). After incubation at RT for 15 min with gentle mixing, the reaction was quenched for 15 min by the addition of β-mercaptoethanol, and then the sample was subjected to SDS-PAGE. The resulting fragments were subjected to in-gel digestion and identified by PMF. Blue-native polyacrylamide gel electrophoresis (BN-PAGE) was done as described in the user manual of the Native PAGE Novex Bis-Tris Gel System (Invitrogen). Rotary-shadowing electron microscopy (EM) was done as previously described (1, 3).
Partial digestion of the P1-P90 complex under isolated and “on-cell” conditions.
For the on-cell conditions, mycoplasma cells were collected by centrifugation, washed twice with PBS containing 10 mM glucose (PBS-G), and resuspended in PBS-G at 1,000 times their concentration in the cell culture. The cells, or the isolated P1-P90 complexes, were treated at 25°C for 15 min with various concentrations of trypsin. The digestion was terminated with 10 mM PMSF, and the digestion products were analyzed by SDS-PAGE. The whole lysates of the trypsin-treated cells were subjected to immunoblotting, using 0.5 μg/ml monoclonal antibody (51). The protein bands of the products were identified by PMF and subjected to Edman degradation.
Sequence analysis.
Phylogenetic trees of the 16S rRNA sequences and amino acid sequences of P1 adhesin orthologs were constructed with the software GENETYX v. 7.0 (Genetyx, Tokyo, Japan). The multiple alignment of P1 adhesin orthologs was computed by the T-Coffee algorithm (http://tcoffee.vital-it.ch/cgi-bin/Tcoffee/tcoffee_cgi/index.cgi). The transmembrane (TM) segment was predicted by SMART 6 (http://smart.embl-heidelberg.de/smart/change_mode.pl).
RESULTS
Isolation of P1 adhesin and P90.
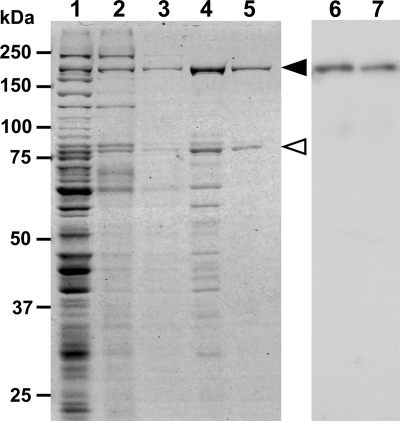
The P1 adhesin was identified as a protein band in SDS-PAGE by immunoblotting analyses using the monocolonal antibody raised against a recombinant protein fragment of P1 adhesin (51). Therefore, we can trace the protein during the cell fractionation process. In the present study, the P1 adhesin was isolated by a three-step biochemical procedure: (i) solubilization by two detergents, (ii) salting out, and (iii) gel filtration (Fig. 1). Briefly, the suspension of mycoplasma cells treated with 1% CHAPS was centrifuged, and the pellet was treated with 2% octylglucoside. Most P1 adhesin was found in the insoluble fraction of CHAPS (Fig. 1, lane 2) and in the soluble fraction of octylglucoside (Fig. 1, lane 3), as previously reported (15). Then, the soluble fraction of octylglucoside was subjected to the salting-out process. P1 adhesin, along with some other proteins, was found in the precipitate after treatment with a 45 to 55% ammonium sulfate solution (Fig. 1, lane 4). The precipitate was dissolved, dialyzed, and applied to a gel filtration column. SDS-PAGE showed that the P1 adhesin fraction from gel filtration contained only two proteins (Fig. 1, lane 5). Immunoblotting analysis showed that the anti-P1 adhesin antibody reacts with the larger protein band (Fig. 1, lane 7), and PMF also showed that the protein band is P1 adhesin encoded by MPN141 with an expect value of 3.8 × 10−8, where the detected peptides covered 26% of the amino acid sequence. The smaller band was shown by PMF to be P90, where the detected peptides covered 39% of the amino acid sequence (data not shown). P90 is known to be produced as the C-terminal fragment after cleavage between residues 454 and 455 of the protein encoded by MPN142 (5). Based on the intensities of the P1 and P90 bands on SDS-PAGE gels, the purity of the P1-P90 samples was estimated to be 97%, and the relative intensity of the two bands was 1 to 0.45. Subsequent experiments were performed in the presence of 0.3% Triton X-100 because the P1-P90 complex forms aggregates in the absence of detergent.
FIG. 1.
Protein profiles of fractions in the P1-P90 complex isolation procedure. The protein fractions were applied to SDS-10% PAGE and subjected to CBB staining (lanes 1 to 5) or immunoblotting using an anti-P1 adhesion monoclonal antibody (lanes 6 and 7). Lanes 1 and 6, whole-cell lysate; lane 2, CHAPS-insoluble fraction; lane 3, octylglucoside-soluble fraction; lane 4, precipitate of 45 to 55% saturated ammonium sulfate; lanes 5 and 7, P1-P90 complex fraction eluted during gel filtration. Molecular masses are indicated on the left. The protein bands of P1 adhesin and P90 are marked by solid and open triangles, respectively.
Molecular mass of the P1-P90 complex.
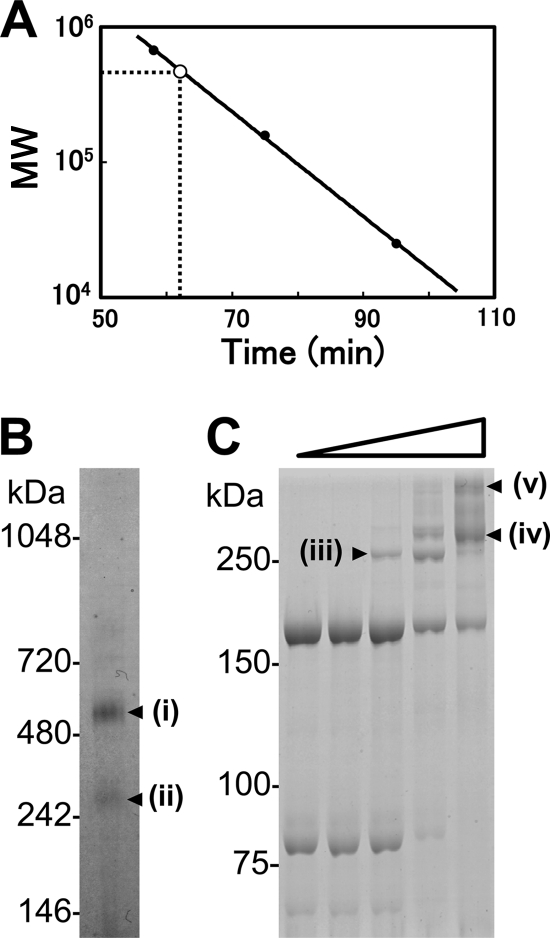
In the gel filtration step of isolation, P1-P90 eluted as a single peak with an estimated mass of 480 kDa, based on the retention time (Fig. 2 A). As the molecular masses of mature P1 adhesin and P90 were previously estimated as 170 and 84 kDa, respectively, based on the predicted amino acid sequences and Edman analyses (46), the above data suggest that P1 adhesin and P90 form an equimolar complex. To determine the molecular mass of the complex by another method, the isolated protein sample was also analyzed by BN-PAGE, a high-resolution native electrophoresis using Coomassie R-250 as a charge shift molecule. This technique allows accurate estimation of the sizes of native protein complexes, although the bands are generally broad compared to those of SDS-PAGE (50). Major and minor bands were detected at 531 and 274 kDa, designated i and ii, respectively, in Fig. 2B. Band i was analyzed by SDS-PAGE and PMF and was shown to be composed of P1 adhesin and P90. The ratio of P1 adhesin band intensities to those of P90 was estimated to be 1 to 0.45. Considering the known molecular weights of the two proteins, the molar ratio of P90 to P1 adhesin was calculated to be 1 to 1. Because the molecular mass of the complex of one P1 adhesin with one P90 is estimated to be 254 kDa, band i presumably contains two subunits of each protein and band ii contains one subunit of each protein.
FIG. 2.
Size and composition of isolated P1-P90 complex. (A) Size estimation by gel filtration. The retention time and apparent molecular mass during gel filtration are presented on the x and y axes, respectively. The solid circles show size standards: thyroglobulin, 670 kDa; aldolase, 158 kDa; and chymotrypsinogen, 25 kDa. The open circle shows the position of the P1-P90 complex. (B) Protein profiles of the P1-P90 complex analyzed by BN-PAGE. Molecular masses are indicated on the left. (C) Protein profiles of the P1-P90 complex cross-linked by various concentrations of BS3 as represented by the open triangle. The protein fraction was treated with 0, 0.002, 0.02, 0.2, and 2 mM BS3 and subjected to SDS-6.0% PAGE. Molecular masses are indicated on the left. The protein bands marked i to v were analyzed by PMF.
To analyze the composition of P1-P90 by another method, we treated the protein fraction with BS3, an amine-reactive 1.14-nm cross-linker, and subjected the treated complex to SDS-PAGE. The band intensities of P1 adhesin and P90 were reduced as the concentration of cross-linker increased. Simultaneously, new bands emerged at 261, 291, and 396 kDa, marked iii, iv, and v, respectively, in Fig. 2C. These bands were all shown to be composed of P1 adhesin and P90 by PMF (data not shown). The quantitative ratio of decrease in non-cross-linked P1 adhesin to that of non-cross-linked P90 was estimated to be 1.85, based on the band intensities of SDS-PAGE gels. Considering the molecular weights of the two proteins, the molar ratio of P1 adhesin to P90 was calculated as 0.92. This result also suggests that one P1 adhesin molecule binds one P90 molecule. Considering that the molecular mass of a complex of each monomer was estimated to be 254 kDa, bands iii and iv appear to contain one molecule of each, and band v contains two molecules of each. The difference in the positions of bands iii and iv may be caused by the difference in the cross-linking numbers.
EM images of the P1-P90 complex.
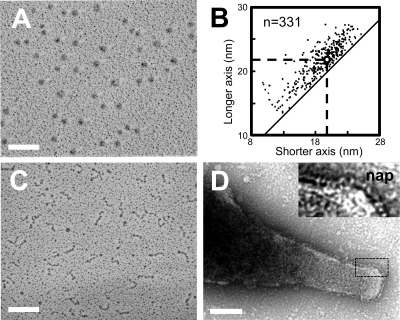
The protein samples of the P1-P90 complex were applied to rotary-shadowing EM (Fig. 3 A). Spherical particles were found in almost all fields of the EM grid, and the frequency of their appearance changed in correspondence with the concentration of the protein, suggesting that the particle is the P1-P90 complex. These images are totally different from the images of Gli349, the leg protein of the fastest-gliding mycoplasma, obtained by the same method (Fig. 3C) (1). We measured the dimensions of 331 P1-P90 complexes and found that the shorter and longer axes averaged 18.8 ± 3.1 and 21.8 ± 3.1 nm, respectively, with a mean difference of 2.7 ± 1.6 nm (Fig. 3B).
FIG. 3.
Molecular shape and dimensions of the P1-P90 complex. (A) Rotary-shadowed EM image. (B) Distribution of shorter and longer axes of the individual complex. The averages, shown by broken lines, are 18.8 ± 3.1 nm and 21.8 ± 3.1 nm for the shorter and longer axes, respectively. The diagonal solid line shows the plot of the same axes. (C) Rotary-shadowed EM image of Gli349, a leg protein of the fastest-gliding species, M. mobile (modified from reference 1). (D) Surface structure of M. pneumoniae. The area outlined by the dashed box is magnified in the inset to show the nap structures. Bars, 100 nm.
Domain structures of the P1-P90 complex suggested by partial digestion of isolated and in situ structures.
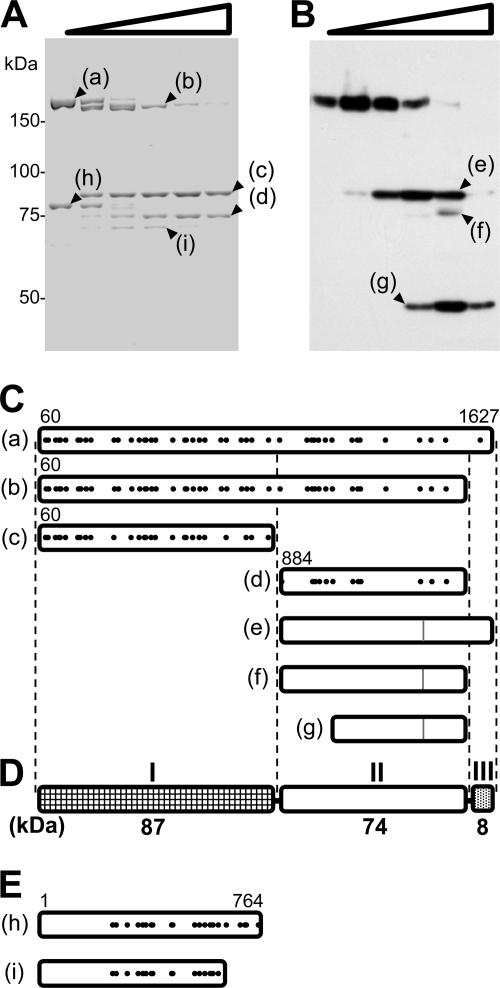
To determine the domain structure of the P1-P90 complex, the isolated complex was partially digested with trypsin and analyzed by SDS-PAGE. The original proteins and four major products, labeled a, b, c, d, h, and i, were detected at 169, 161, 87, 74, 82, and 70 kDa, respectively (Fig. 4 A). The results of Edman analysis showed that the N termini of the fragments started at amino acid residue 60 in a, b, and c and at residue 884 in d of the P1 adhesin ORF (Fig. 4C). Residue 883 was arginine, consistent with the enzymatic character of trypsin. Based on the residue positions of the N termini and the estimated lengths of the peptides, the products were assigned to the P1 adhesin sequence as presented in Fig. 4C. PMF showed that bands a, b, c, and d contained at least amino acid residues 66 to 1584, 66 to 1473, 66 to 865, and 993 to 1475 of the P1 adhesin ORF, respectively, supporting the above assignments. The detected peptides covered 27%, 28%, 28%, and 22% of the estimated region of the P1 adhesin ORF for a to d, respectively (Fig. 4C). These results suggest that the P1 adhesin molecule can be divided into three domains, designated I, II, and III (Fig. 4D). PMF of band h detected only the peptides corresponding to residues 258 to 762 of P90, suggesting that the peptides from the N-terminal 258 residues in P90 are difficult to ionize during MALDI-TOF. The PMF of i showed the same set of peptide signals as in h, except the peptides corresponding to amino acid residues 694 to 762. These observations suggest that the C-terminal 12 kDa is sensitive to proteases and was deleted in band i (Fig. 4E).
FIG. 4.
Partial digestion of the P1-P90 complex. (A) Protein profiles of the purified P1-P90 complex digested with various concentrations of trypsin. The protein fraction was treated with 0, 0.01, 0.03, 0.1, 0.3, and 1 mg/ml trypsin and subjected to SDS-8.75% PAGE. The resultant peptide fragments, marked a to i, were identified by PMF. (B) Immunoblot of M. pneumoniae cells treated with various concentrations of trypsin, using antibody against the C-terminal region of P1 adhesin for detection. The cells were treated with 0, 0.01, 0.03, 0.1, 0.3, and 1 mg/ml trypsin and subjected to SDS-PAGE and immunoblotting to detect the aa 1386 to 1394 sequence. The sizes of the resulting peptide fragments, marked e to g, were estimated to be 83, 77, and 48 kDa, respectively. The molecular masses are indicated on the left of panel A. (C) Mapping of products on the amino acid sequence of P1 adhesin. The 59 N-terminal residues predicted from the DNA sequence were processed in the mature form (46, 60). Protein bands a to d were identified by PMF. The positions of peptides detected in PMF are marked with solid dots. The N-terminal residues in bands a to d were determined by Edman analysis. The C terminus was estimated from the band size. The evident residue numbers of the ends are indicated. Peptide bands e to g were assigned from the band sizes, and the epitope positions are shown by gray lines. (D) Schematic of P1 adhesin, consisting of three domains, I, II, and III. The molecular mass is indicated below each domain. (E) Bands h and i mapped onto the amino acid sequence of P90, based on the results of PMF.
To analyze the domain structure of P1 adhesin on a cell, M. pneumoniae cells were treated with trypsin, and the whole-cell lysates were developed in SDS-PAGE and detected by a monoclonal antibody that recognizes residues 1386 to 1394 of P1 adhesin. The fragment signals marked e, f, and g were detected at apparent molecular masses of 83, 77, and 48 kDa, respectively (Fig. 4B). Signals e and f can be explained by the suggested domain structure as domains II and III and domain II, respectively. However, signal g does not correspond to any of the predicted domains. Another flexible region accessible to trypsin may exist around residue 1100, in the middle of domain II.
Sequence analysis of P1 adhesin.
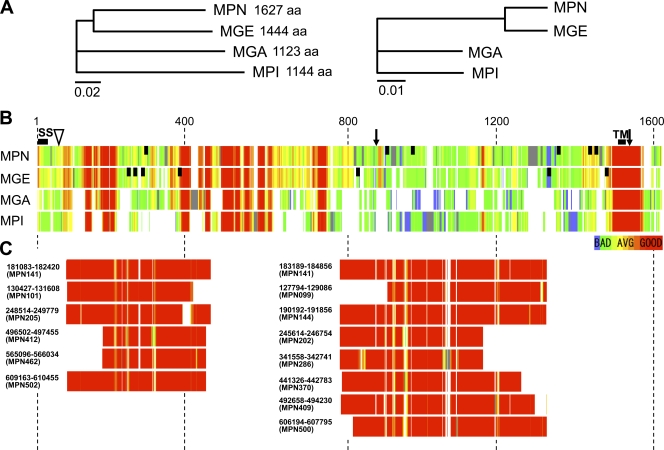
To date, the genomes of about 20 species of mollicutes have been sequenced. The orthologs of the P1 adhesin of Mycoplasma genitalium and Mycoplasma gallisepticum have been identified as MG_191 (MgPa) and MGA_0934 (GapA) (7, 44); that from Mycoplasma pirum has also been identified, although the genome has not yet been sequenced (61). The genome sequences of other mycoplasma species do not have orthologs of P1 adhesin. As the amino acid sequences of these orthologs have not been compared in detail, we analyzed them to obtain information about domain structures, recombination sites, and TM segments. First, we examined the relationship of the orthologs (Fig. 5 A, left). The sequence from M. genitalium is closely related to that of the P1 adhesin of M. pneumoniae, and those from M. gallisepticum and M. pirum are distantly related. This relationship is consistent with that of 16S rRNA, although the relative distances are slightly different (Fig. 5A, right). Next, we compared the amino acid sequences of P1 adhesin orthologs with the multiple-alignment algorithm T-Coffee. The 87-residue region starting at position 1509 in M. pneumoniae showed high similarity, represented by 84% identity with M. genitalium. The amino acid sequence is more conserved in domain I than in domain II, as shown by the respective 51% and 38% identities between M. pneumoniae and M. genitalium (Fig. 5B).
FIG. 5.
Sequence similarity of P1 adhesin homologs. (A) Phylogenetic tree of amino acid sequences of P1 adhesin orthologs (left) and 16S rRNA sequences (right) of M. pneumoniae (MPN) and its relatives, M. genitalium (MGE), M. gallisepticum (MGA), and M. pirum (MPI). The scale bars indicate the numbers of residue substitutions by position. (B) Multiple alignment of sequences of P1 adhesin orthologs performed by T-Coffee. The color indicates the sequence similarity score, as shown at the bottom right. The regions with no similarity to orthologs are colored gray. The N-terminal processing site is marked by a triangle between residues 59 and 60. The trypsin-sensitive sites are marked by arrows between residues 883 and 884 and ∼8 kDa from the C terminus. The predicted signal sequence (SS) and TM segment are labeled. The vertical black bars indicate the epitopes of adherence-inhibiting antibodies against P1 adhesin of M. pneumoniae and MgPa of M. genitalium (8, 43, 46). (C) Multiple alignment of amino acid sequences of P1 adhesin paralogs from M. pneumoniae performed with T-Coffee, showing the corresponding regions on the P1 adhesin sequence in panel B. The nucleotide numbers on the genome are indicated for the corresponding ORF. The paralogous sequences indicated on the left and right are classified as RepMP4 and RepMP2/3, respectively.
In the genome of M. pneumoniae, four groups of repeated DNA sequences (RepMP) have been found (65). Two of those groups, named RepMP2/3 and RepMP4, have been found to have high homology to the C-terminal and N-terminal parts of P1 adhesin, respectively, suggesting that the antigenic variation of P1 adhesin found in clinical isolates is generated by the genomic recombination between the P1 adhesin gene and RepMP paralogs (19, 57-59, 65). To discover the positional relationship between the possible recombination sites in the P1 adhesin gene and the domain structure of the protein, we performed multiple alignment of paralogs, including the P1 adhesin (Fig. 5C). We predicted the amino acid sequences of RepMP2/3 and RepMP4 paralogs, picked sequences longer than 300 residues, and compared them with the P1 adhesin sequence. The alignment showed that the amino acid sequences of RepMP2/3 and RepMP4 had high similarity to the corresponding region at residues 778 to 1333 and 76 to 464 of P1 adhesin, respectively. The amino acid (aa) 76 to 464 region of P1 that is homologous to RepMP4 corresponds to the N-terminal part of domain I, and the N terminus of the aa 778 to 1333 region, which is homologous to RepMP2/3, extends into domain I for about 106 residues.
We predicted the signal sequence and the TM segment with the SMART algorithm. For all 4 species, the N-terminal signal sequence was strongly suggested to be located at the positions corresponding to residues 1 to 27 in the sequence of M. pneumoniae P1 adhesin, consistent with the experimental data (Fig. 4) (46). A TM segment was also strongly suggested at the positions corresponding to residues 1523 to 1545, suggesting that the adhesin molecules likely penetrate the cell membrane in this region. Previously, two additional TM segments, located at residues 488 to 508 and 530 to 550, were predicted in P1 adhesin (43, 46), but they were not identified as TM segments in any of the 4 species analyzed in this work.
DISCUSSION
Effects of detergents on P1 adhesin.
In a previous study, denatured P1 adhesin was purified from M. pneumoniae cells for use as an antigen (15). In that procedure, a P1 adhesin-rich fraction was obtained by treatment with nonionic detergents, i.e., cells treated with 1% CHAPS were extracted with 2% octylglucoside. In the present study, we used this method to obtain the P1 adhesin-rich fraction. In the subsequent procedures, we used a milder detergent than octylglucoside, namely, Triton X-100 (Fig. 1). The sample obtained here seems not to be significantly affected in structure, because the pattern of trypsin digestion of the isolated P1 adhesin was consistent with the trypsin digestion of P1 adhesin on intact cells (Fig. 4).
Binding of P1 adhesin with P90.
We showed by gel filtration, BN-PAGE, and chemical cross-linking analysis that isolated P1 adhesin and P90 form a complex, suggesting that P1 adhesin physically interacts with P90 (Fig. 2). Previously, cross-linking studies showed that P90 is located at a position close to P1 adhesin on the cell (26, 27). Seto et al. showed that P90 localizes at a position on the cell close to that occupied by P1 adhesin when viewed by immunofluorescence microscopy (52, 53). These previous observations suggested that a physical interaction exists between P1 adhesin and P90, an interaction that is consistent with the present results. P40, encoded by the same operon as P1 adhesin, was also suggested to interact physically with P1 adhesin (26, 52); however, the present study did not find evidence for this interaction. P40 may require other factors, such as a lipid or another protein, to maintain its association with P1 adhesin or P90.
Structural model of the P1 adhesin-P90 complex.
We obtained the spherical outline of the P1 adhesin-P90 complex using rotary-shadowing EM, which gives high contrast but low image resolution. The complex should face in various directions relative to the imaging direction during EM. The alignment of the images cannot be reliably determined under the reduced resolution of rotary shadowing, but this assumption may explain the wide distribution of particle dimensions shown in Fig. 3.
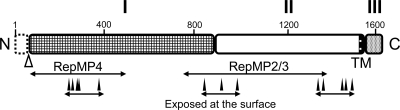
The global structure for P1 adhesin can be divided into domains I, II, and III, which are predicted to be linked by flexible hinges that can be digested preferentially by trypsin (Fig. 6). Domain I is highly conserved among all four known orthologs, which may suggest that it plays a role in binding to other proteins or to the substrate, because a specific function is often found in a conserved region. Some parts of domains I and II are suggested to be exposed to the outside of the complex, because they are recognized by adherence-inhibiting antibodies (Fig. 5 and 6) (8, 16, 43, 46, 51). Domain II contains the C-terminal TM segment. Domain III is a cytoplasmic domain. Considering the previous finding that P1 adhesin is diffusely distributed in a mutant lacking the cytoskeletal rod structure (52, 53), this domain may interact with other proteins in the cell. We suggest that one P1 adhesin molecule binds one P90 molecule and that these heterodimers then assemble into a complex that includes two molecules each of P1 adhesin and P90.
FIG. 6.
Schematic of P1 adhesin. The molecule is divided into domains I, II, and III, which are linked by predicted flexible hinges. Domain I is highly conserved. The TM segment is marked. The N-terminal 59 residues are removed during the maturation process at the position marked by an open triangle. The regions homologous to paralogs are indicated by lines marked RepMP4 and RepMP2/3. The binding sites of inhibitory antibodies, which should mark exposed regions, are indicated by filled triangles (8, 43, 46). Domain III is inside the cell and probably interacts with other proteins in the cell. Two molecules of P1 are predicted to fold into a globular complex with two molecules of P90.
Molecular shape of the P1-P90 complex compared to previous reports.
At the surface of the attachment organelle, a “nap” structure has been observed by EM and is suggested to be involved in the function of adhesin (12, 20, 21). This structure may be composed of P1 adhesin, because (i) the nap localized under EM at a position similar to that of P1 adhesin as seen with immunofluorescence microscopy (52, 53); (ii) P1 adhesin on a cell is sensitive to proteases, suggesting that the molecule projects from the cell surface (13); and (iii) the antibody against P1 adhesin inhibits P1 adhesin functions, suggesting that the molecule is exposed (12, 46, 51). Here, we show that the P1-P90 complex appears as a 17- to 20-nm sphere during rotary-shadowing EM (Fig. 3A). The outlines of the nap structure cannot be traced in negative-staining EM because many structures are in a cluster (Fig. 3D). The three-dimensional reconstitution of the nap images from the cryo-EM showed a knob with a length of 4 to 8 nm and a diameter of 8 nm (55). However, the current observations do not disprove that the nap structure is composed of P1 adhesin, because rotary-shadowing EM generally visualizes the object as larger than it is (41).
Previously, Adan-Kubo et al. isolated Gli349, the leg protein of Mycoplasma mobile, which is phylogenetically distant from M. pneumoniae (17), and showed that it is a 100-nm rod structure, reminiscent of the symbol for an eighth note in music (1, 28, 31, 36). The difference in the molecular shapes of P1 adhesin and Gli349 is consistent with the fact that these proteins do not share amino acid sequences (31). M. mobile glides as fast as 4.5 μm/s, five times faster than M. pneumoniae. The difference in gliding speeds between the two species may be caused by the difference in the morphologies of leg proteins (32, 37, 40, 62-64).
Variable region of P1 adhesin.
P1 adhesin is also known as the immunodeterminant protein and shows sequence variation between clinical strains, resulting in the structural change in immunodominant epitopes of the adhesin for evading the host immune system (19, 49, 57-59, 65). The variation of P1 adhesin is thought to be generated by intragenomic DNA recombination for the following reasons. (i) All P1 genotypes of M. pneumoniae found in clinical isolates can be explained by recombination between P1 adhesin and one of its paralogous DNA sequences on the genome, RepMP2/3 or RepMP4 (19, 57). (ii) In M. genitalium, which is closely related to M. pneumoniae, adherence-deficient mutants were obtained through recombination between MgPa and a paralogous sequence encoded in the neighboring downstream region (4, 14, 45). In the present study, we compared the amino acid sequence of P1 adhesin with its RepMP2/3 and RepMP4 paralogs by multiple alignment (Fig. 5C). The RepMP paralogs aligned primarily with domains I and II, but the boundaries do not precisely match those of the domains (Fig. 6), suggesting that the units of molecular structure and DNA recombination for antigenic variation can evolve in different ways.
Binding activity of P1 adhesin.
P1 adhesin is essential for adhesion to animal cells and binding during gliding (12, 51). The binding target of adhesin has been suggested to be sialic acid, present at the tips of polysaccharides on animal cell surfaces (9, 23, 30, 48). Therefore, P1 adhesin may bind sialic acid. However, analysis by surface plasmon resonance could not detect any ability of the isolated P1-P90 complex to bind fetuin, a representative sialic acid-modified protein (data not shown). Other factors, including additional proteins and the presence of the cell membrane, may be necessary for the binding activity, because some mutants that lack cytoskeletal proteins located at the attachment organelle (HMW1, HMW2, and P30), encoded by ORFs MPN447, MPN310, and MPN453, are defective in cytadhesion, although P1 adhesin localizes at the cell surface in these mutants (22, 52, 53).
Sequence analyses of full-length P1 adhesin did not suggest any known protein functions. Generally, the specific activities of a protein are associated with conserved regions. Thus, in the present study, we analyzed three conserved regions of the P1 adhesin sequence, residues 131 to 206, 393 to 757, and 1509 to 1595, by PSI-BLAST, but no significant similarities to other proteins with known functions were found.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research on the Priority Areas “Applied Genomics” (to T.K.) and “Structures of Biological Macromolecular Assemblies” and “System Cell Engineering by Multiscale Manipulation” (to M.M.); by a Grant-in-Aid for Scientific Research (A) (to M.M.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and by a grant from the Institution for Fermentation Osaka (to M.M.).
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Adan-Kubo, J., A. Uenoyama, T. Arata, and M. Miyata. 2006. Morphology of isolated Gli349, a leg protein responsible for glass binding of Mycoplasma mobile gliding revealed by rotary-shadowing electron microscopy. J. Bacteriol. 188:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 3.Arata, T. 1998. Electron microscopic observation of monomeric actin attached to a myosin head. J. Struct. Biol. 123:8-16. [DOI] [PubMed] [Google Scholar]
- 4.Burgos, R., et al. 2006. Mycoplasma genitalium P140 and P110 cytadhesins are reciprocally stabilized and required for cell adhesion and terminal-organelle development. J. Bacteriol. 188:8627-8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catrein, I., R. Herrmann, A. Bosserhoff, and T. Ruppert. 2005. Experimental proof for a signal peptidase I like activity in Mycoplasma pneumoniae, but absence of a gene encoding a conserved bacterial type I SPase. FEBS J. 272:2892-2900. [DOI] [PubMed] [Google Scholar]
- 6.Dandekar, T., et al. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser, C. M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 8.Gerstenecker, B., and E. Jacobs. 1990. Topological mapping of the P1-adhesin of Mycoplasma pneumoniae with adherence-inhibiting monoclonal antibodies. J. Gen. Microbiol. 136:471-476. [DOI] [PubMed] [Google Scholar]
- 9.Glasgow, L. R., and R. L. Hill. 1980. Interaction of Mycoplasma gallisepticum with sialyl glycoproteins. Infect. Immun. 30:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Himmelreich, R., et al. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiratsuka, Y., M. Miyata, T. Tada, and T. Q. P. Uyeda. 2006. A microrotary motor powered by bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:13618-13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, P. C., et al. 1982. Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science 216:313-315. [DOI] [PubMed] [Google Scholar]
- 13.Hu, P. C., A. M. Collier, and J. B. Baseman. 1977. Surface parasitism by Mycoplasma pneumoniae of respiratory epithelium. J. Exp. Med. 145:1328-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iverson-Cabral, S. L., S. G. Astete, C. R. Cohen, E. P. Rocha, and P. A. Totten. 2006. Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences. Infect. Immun. 74:3715-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs, E., K. Fuchte, and W. Bredt. 1988. Isolation of the adherence protein of Mycoplasma pneumoniae by fractionated solubilization and size exclusion chromatography. Biol. Chem. Hoppe Seyler 369:1295-1299. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, E., R. Rock, and L. Dalehite. 1990. A B cell-, T cell-linked epitope located on the adhesin of Mycoplasma pneumoniae. Infect. Immun. 58:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaffe, J. D., et al. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenri, T., et al. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenri, T., et al. 1999. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun. 67:4557-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, DC.
- 21.Kirchhoff, H., R. Rosengarten, W. Lotz, M. Fischer, and D. Lopatta. 1984. Flask-shaped mycoplasmas: properties and pathogenicity for man and animals. Israel J. Med. Sci. 20:848-853. [PubMed] [Google Scholar]
- 22.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 23.Krivan, H. C., L. D. Olson, M. F. Barile, V. Ginsburg, and D. D. Roberts. 1989. Adhesion of Mycoplasma pneumoniae to sulfated glycolipids and inhibition by dextran sulfate. J. Biol. Chem. 264:9283-9288. [PubMed] [Google Scholar]
- 24.Krunkosky, T. M., J. L. Jordan, E. Chambers, and D. C. Krause. 2007. Mycoplasma pneumoniae host-pathogen studies in an air-liquid culture of differentiated human airway epithelial cells. Microb. Pathog. 42:98-103. [DOI] [PubMed] [Google Scholar]
- 25.Layh-Schmitt, G., and R. Herrmann. 1992. Localization and biochemical characterization of the ORF6 gene product of the Mycoplasma pneumoniae P1 operon. Infect. Immun. 60:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Layh-Schmitt, G., and R. Herrmann. 1994. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect. Immun. 62:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Layh-Schmitt, G., A. Podtelejnikov, and M. Mann. 2000. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology 146:741-747. [DOI] [PubMed] [Google Scholar]
- 28.Lesoil, C., et al. 2010. Molecular shape and binding force of Mycoplasma mobile's leg protein Gli349 revealed by an AFM study. Biochem. Biophys. Res. Commun. 391:1312-1317. [DOI] [PubMed] [Google Scholar]
- 29.Lipman, R. P., W. A. Clyde, Jr., and F. W. Denny. 1969. Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J. Bacteriol. 100:1037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loomes, L. M., et al. 1984. Erythrocyte receptors for Mycoplasma pneumoniae are sialylated oligosaccharides of Ii antigen type. Nature 307:560-563. [DOI] [PubMed] [Google Scholar]
- 31.Metsugi, S., et al. 2005. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics 1:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyata, M. 2008. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 16:6-12. [DOI] [PubMed] [Google Scholar]
- 33.Miyata, M. 2007. Molecular mechanism of mycoplasma gliding—a novel cell motility system, p. 137-175. In P. Lenz (ed.), Cell motility. Springer, New York, NY.
- 34.Miyata, M. 2010. Unique centipede mechanism of Mycoplasma gliding. Annu. Rev. Microbiol. 64:519-537. [DOI] [PubMed] [Google Scholar]
- 35.Miyata, M., and H. Ogaki. 2006. Cytoskeleton of Mollicutes. J. Mol. Microbiol. Biotechnol. 11:256-264. [DOI] [PubMed] [Google Scholar]
- 36.Miyata, M., and J. Petersen. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 186:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyata, M., et al. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 39.Nakane, D., and M. Miyata. 2007. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc. Natl. Acad. Sci. U. S. A. 104:19518-19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakane, D., and M. Miyata. 2009. Cytoskeletal asymmetrical-dumbbell structure of a gliding mycoplasma, Mycoplasma gallisepticum, revealed by negative-staining electron microscopy. J. Bacteriol. 191:3256-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nonaka, T., J. Adan-Kubo, and M. Miyata. 2010. Triskelion structure of Gli521 protein involved in gliding mechanism of Mycoplasma mobile. J. Bacteriol. 192:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohtani, N., and M. Miyata. 2007. Identification of a novel nucleoside triphosphatase from Mycoplasma mobile: a prime candidate for motor of gliding motility. Biochem. J. 403:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Opitz, O., and E. Jacobs. 1992. Adherence epitopes of Mycoplasma genitalium adhesin. J. Gen. Microbiol. 138:1785-1790. [DOI] [PubMed] [Google Scholar]
- 44.Papazisi, L., et al. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 149:2307-2316. [DOI] [PubMed] [Google Scholar]
- 45.Peterson, S. N., et al. 1995. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc. Natl. Acad. Sci. U. S. A. 92:11829-11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesion. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 47.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts, D. D., L. D. Olson, M. F. Barile, V. Ginsburg, and H. C. Krivan. 1989. Sialic acid-dependent adhesion of Mycoplasma pneumoniae to purified glycoproteins. J. Biol. Chem. 264:9289-9293. [PubMed] [Google Scholar]
- 49.Ruland, K., R. Wenzel, and R. Herrmann. 1990. Analysis of three different repeated DNA elements present in the P1 operon of Mycoplasma pneumoniae: size, number and distribution on the genome. Nucleic Acids Res. 18:6311-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 51.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seto, S., and M. Miyata. 2003. The attachment organelle formation represented by localization of cytadherence protein and formation of electron-dense core in the wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seybert, A., R. Herrmann, and A. S. Frangakis. 2006. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J. Struct. Biol. 156:342-354. [DOI] [PubMed] [Google Scholar]
- 56.Shimizu, T., and M. Miyata. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44:431-434. [DOI] [PubMed] [Google Scholar]
- 57.Spuesens, E. B., et al. 2009. Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in Mycoplasma pneumoniae. Microbiology 155:2182-2196. [DOI] [PubMed] [Google Scholar]
- 58.Su, C. J., A. Chavoya, S. F. Dallo, and J. B. Baseman. 1990. Sequence divergency of the cytadhesin gene of Mycoplasma pneumoniae. Infect. Immun. 58:2669-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su, C. J., S. F. Dallo, A. Chavoya, and J. B. Baseman. 1993. Possible origin of sequence divergence in the P1 cytadhesin gene of Mycoplasma pneumoniae. Infect. Immun. 61:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su, C. J., V. V. Tryon, and J. B. Baseman. 1987. Cloning and sequence analysis of cytadhesin P1 gene from Mycoplasma pneumoniae. Infect. Immun. 55:3023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tham, T. N., et al. 1994. Molecular characterization of the P1-like adhesin gene from Mycoplasma pirum. J. Bacteriol. 176:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uenoyama, A., and M. Miyata. 2005. Gliding ghosts of Mycoplasma mobile. Proc. Natl. Acad. Sci. U. S. A. 102:12754-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uenoyama, A., and M. Miyata. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 187:5578-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uenoyama, A., S. Seto, D. Nakane, and M. Miyata. 2009. Regions on Gli349 and Gli521 protein molecules directly involved in movements of Mycoplasma mobile gliding machinery suggested by inhibitory antibodies and mutants. J. Bacteriol. 191:1982-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wenzel, R., and R. Herrmann. 1988. Repetitive DNA sequences in Mycoplasma pneumoniae. Nucleic Acids Res. 16:8337-8350. [DOI] [PMC free article] [PubMed] [Google Scholar]