Abstract
AI-2 is an autoinducer made by many bacteria. LsrB binds AI-2 in the periplasm, and Tsr is the l-serine chemoreceptor. We show that AI-2 strongly attracts Escherichia coli. Both LsrB and Tsr are necessary for sensing AI-2, but AI-2 uptake is not, suggesting that LsrB and Tsr interact directly in the periplasm.
Many functions in bacteria are regulated by population density, including formation of biofilms and production of virulence factors (5). Assessment of population density, known as quorum sensing, relies on the ability of cells to determine the concentrations of compounds known as autoinducers (AIs). As the cell density increases, an AI accumulates to a concentration that triggers a quorum-sensing response. Autoinducers activate some genes and repress others. Induced genes typically include those responsible for production of the autoinducer, resulting in a positive feedback loop. Cell densities required to accumulate enough AI for good induction are 108 per ml or higher.
AIs are of two basic types: species specific and general (22). Species-specific AI-1s are acyl homoserine lactones in Gram-negative bacteria and modified peptides in Gram-positive bacteria. Full induction of bioluminescence in the marine bacterium Vibrio harveyi, which colonizes dead organic matter, requires both a specific AI-1 inducer and a general autoinducer, called AI-2 (6). AI-2 is derived from spontaneous cyclization of 4,5-dihydroxy-2,3-pentanedione (DPD). DPD is made from S-ribosylhomocysteine by the enzyme LuxS (25). S-Ribosylhomocysteine is an intermediate in the breakdown of S-adenosylhomocysteine, the product remaining after methyl group donation by S-adenosylmethionine.
AI-2 is produced by a wide range of Gram-negative and Gram-positive bacteria and exists in multiple forms that are in equilibrium with each other (5). The form that is active in V. harveyi is (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate (S-THMF borate) (8), which binds to the periplasmic protein LuxP. In S. enterica serovar Typhimurium, a boron-free isomer of AI-2 [(2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetra-hydrofuran (R-THMF)] binds to the periplasmic LsrB protein (21). LsrB is the recognition component of an ABC transporter for AI-2. LsrACD are the membrane-bound components of the ABC transporter for AI-2. AI-2 is generated by the enzyme LuxS, and the YdgG (TqsA) protein has been implicated in AI-2 export from the cytoplasm (14).
AI-2 is a known chemoattractant for Escherichia coli (4, 10), but the receptor(s) involved in AI-2 sensing has not been identified. This work was initiated to characterize the proteins involved in AI-2 recognition by E. coli strain CV1, which is equivalent to the standard wild-type chemotaxis strain RP437. The strains used in this study are shown in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype | Resistancea | Source |
|---|---|---|---|
| Escherichia coli strains | |||
| CV1 | Chemotaxis wild type (same as RP437) | Str | 24 |
| TG1/pDS-Red Express | Wild type; dead-cell control | Amp | Stratagene |
| CV5 | CV1 Δtsr | Str | This studyb |
| CV12 | CV1 Δtar-tap trg::Tn10 | Str, Tet | This studyc |
| MJ101 | CV1 lsrBΩKanr | Str, Kan | This studyd |
| MJ102 | CV1 lsrCΩKanr | Str, Kan | This studye |
| BW25113 ΔlsrB | lsrBΩKanr | Kan | 3 |
| BW25113 ΔlsrC | lsrCΩKanr | Kan | 3 |
| Plasmids | |||
| pCM18 | GFP-expressing vector | Erm | 12 |
| pDS-RedExpress | RFP-expressing vector | Amp | Clontech |
| pCA24N-lsrB | pCA24N PT5-lac::lsrB; expresses E. coli lsrB from placZYA | Cm | 15 |
Str, streptomycin; Tet, tetracycline; Kan, kanamycin; Erm, erythromycin; Amp, ampicillin; Cm, chloramphenicol.
Made by introducing Δtsr9101 (7) into CV1 by phage P1 transduction (16), with selection for Thr+ and screening for Tsr−.
Made in two steps: Δtar-tap5201 (26) was introduced into CV1 by phage P1 transduction with selection for Eda+, followed by screening for Tar−, and then trg::Tn10 was introduced by phage P1 transduction followed by selection for Tetr on lysis broth (LB) (20) agar plates containing 10 μg/ml tetracycline, followed by screening for Trg−.
Made by introducing lsrBΩKanr (3) into CV1 by phage P1 transduction, with selection for Kanr and confirmation of lsrB gene disruption by PCR.
Made by introducing lsrCΩKanr (3) into CV1 by phage P1 transduction, with selection for Kanr and confirmation of lsrC gene disruption by PCR.
The microplug (μPlug) assay (9), a modified plug-in-pond assay, provides a qualitative but highly visual representation of chemotaxis. Cells containing the green fluorescent protein (GFP)-encoding plasmid pCM18 were grown overnight at 32°C in tryptone broth (TB) (20) containing 150 μg/ml erythromycin. Cells were back-diluted to a turbidity of 0.05 at 600 nm in 25 ml of TB lacking erythromycin and then grown to mid-logarithmic phase (turbidity of 0.5 at 600 nm) at 32°C. Cells were examined by phase-contrast microscopy to ensure robust motility and normal run-tumble swimming behavior. The cells were harvested by low-speed centrifugation at 400 × g for 5 min, gently washed twice with chemotaxis buffer (CB; phosphate-buffered saline, 0.1 mM EDTA, 0.01 mM l-methionine, and 10 mM d,l-lactate) and then gently resuspended in 2 ml of CB. These cells were again examined under the microscope to ensure robust motility and normal run-tumble swimming behavior. Cells were mixed with an approximately equal number of TG1 cells expressing red fluorescent protein (RFP) from plasmid pDS-Red Express that had been killed by a 1-h treatment with 1 mg/ml kanamycin. The dead RFP-containing cells served as a control to visualize any mixing due to turbulence within the microfluidic chamber.
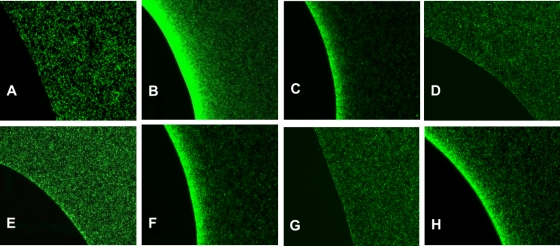
The responses of E. coli strain CV1 and its isogenic tsr and lsrB mutant derivatives are shown in Fig. 1. In the absence of AI-2 in the plug, CV1 cells distributed themselves randomly (Fig. 1A). However, when CV1 cells were exposed to plugs containing 200 μM l-serine or 200 μM AI-2, they exhibited strong attractant responses, shown by the accumulation of bacteria at the agarose plug-liquid interface (Fig. 1B and C). Strain CV5 (CV1 Δtsr), which lacks the l-serine receptor Tsr, did not respond to l-serine (Fig. 1D) and gave a severely attenuated response to AI-2 (Fig. 1E). Strain CV12 (CV1 Δtar-tap Δtrg), which has Tsr as its only functional receptor (other than Aer), responded to both l-serine and AI-2 (data not shown), although the accumulation was somewhat decreased relative to that of strain CV1. Thus, Tsr is both necessary and sufficient for good AI-2 chemotaxis in E. coli K-12, although there may be a small residual response in cells lacking Tsr.
FIG. 1.
Chemotactic responses to l-serine and AI-2, demonstrated by the results of μPlug assays. The μPlug assay was carried out as described in the text and as previously reported (10, 19); the distribution of dead, RFP-labeled cells is not shown, but it was always uniform except when the dead cells were displaced around the plug by accumulating GFP-labeled motile cells. (A) Distribution of wild-type (CV1) cells in the absence of any attractant in the plug. (B) Distribution of wild-type (CV1) cells with 200 μM l-serine in the plug. (C) Distribution of wild-type (CV1) cells with 200 μM AI-2 in the plug. (D) Distribution of Δtsr (CV5) cells with 200 μM l-serine in the plug. (E) Distribution of Δtsr (CV5) cells with 200 μM AI-2 in the plug. (F) Distribution of lsrBΩKanr (MJ101) cells with 200 μM l-serine in the plug. (G) Distribution of lsrBΩKanr (MJ101) cells with 200 μM AI-2 in the plug. (H) Distribution of lsrCΩKanr (MJ102) cells with 200 μM AI-2 in the plug.
Because AI-2 is known to bind to the periplasmic protein LsrB, we also looked at the responses of MJ101 (CV1 lsrBΩKanr) cells. These cells responded like strain CV1 to l-serine (Fig. 1F) but showed no accumulation around plugs containing 200 μM AI-2 (Fig. 1G). In contrast, MJ102 (CV1 lsrCΩKanr) cells, which should still produce LsrB but not be able to take up AI-2 into the cytoplasm, accumulated around plugs containing 200 μM l-serine (data not shown) or 200 μM AI-2 (Fig. 1H). The accumulation near AI-2 plugs was somewhat weaker than that of strain CV1, perhaps because of a polar effect of the lsrCΩKanr insertion on the downstream lsrB gene in the lsrACDBFGE operon (28). MJ101 cells containing plasmid pCA24N-PT5-lac::lsrB, which encodes wild-type E. coli LsrB, accumulated around AI-2-containing plugs about as well as MJ102 cells (data not shown) when 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce LsrB synthesis.
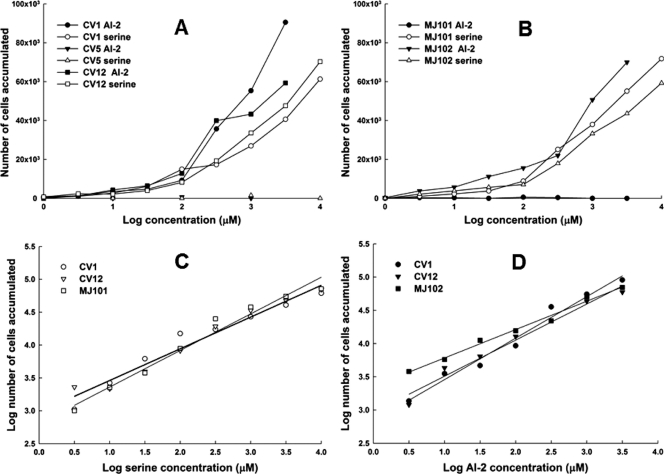
To quantify the response to AI-2 and to compare it to the response to l-serine, we performed capillary assays (1). As expected from the μPlug assay results, CV1 and CV12 cells accumulated in capillaries containing either l-serine or AI-2 (Fig. 2 A), with the CV1 strain giving a stronger response, whereas CV5 (Δtsr) cells did not accumulate in capillaries containing either compound. MJ101 cells (lsrBΩKanr) responded to l-serine but not to AI-2, whereas MJ102 cells (lsrCΩKanr) responded to both (Fig. 2B). Thus, the conclusions from the μPlug assay were confirmed.
FIG. 2.
Responses of cells to l-serine and AI-2 in the capillary assay. Assays were performed at 32°C, essentially as described by Adler (1). Cells were resuspended in CB to a cell density of about 5 × 107/ml. Assays were run twice, with three capillaries per run for each strain under each condition. Data shown are averages of six capillaries, and the standard deviations (not shown for purposes of clarity) ranged from 10 to 20%. The background accumulations in buffer-only capillaries were in the range of 500 to 1,000 cells. (A) Normalized values (with buffer-only control subtracted) of CV1 (wild-type) cells, CV5 (Δtsr) cells, and CV12 (Δtar-tap Δtrg) cells exposed to capillaries containing l-serine or AI-2. (B) Normalized values of MJ101 (lsrBΩKanr) cells and MJ102 (lsrCΩKanr) cells exposed to capillaries containing l-serine or AI-2. (C) Data for the responses of CV1, CV12, and MJ101 cells to l-serine, plotted on a log-log scale. The straight lines are linear regressions that can be extrapolated back to a threshold value. The extrapolated threshold concentrations, as predicted by Weber's law, are 1.7 × 10−12 M for strain CV12 and 3.5 × 10−12 M for strains CV1 and MJ101. The regression lines for CV12 and MJ101 were identical. (D) Data for the responses of CV1, CV12, and MJ102 cells to AI-2, plotted on a log-log scale. The linear regressions can be extrapolated back to a threshold value. The extrapolated threshold concentrations are 1.6 × 10−11 for strain CV1, 4.6 × 10−12 M for strain CV12, and 2.5 × 10−14 M for strain MJ102.
By plotting the data from the capillary assay on a log-log plot, we could extrapolate a threshold concentration for both compounds in each strain (19). With l-serine, the extrapolated detection thresholds for strains CV1, CV12, and MJ101 were all in the range of 2 × 10−12 to 4 × 10−12 M (Fig. 2C). The result was quite different with AI-2, because strains CV1 and CV12 had extrapolated detection thresholds of ∼5 × 10−12 and 2 × 10−11 M, respectively, but the extrapolated detection threshold for the LsrC− strain MJ102 was at least 100-fold lower, ∼2 × 10−14 M (Fig. 2D). This is the expected result if the periplasmic AI-2 concentration in MJ102 cells is higher than in CV1 cells, because in the latter strain AI-2 is cleared from the periplasm by transport into the cell. The same phenomenon has been seen with cells containing maltose-binding protein in the absence of a functional maltose transport system (29).
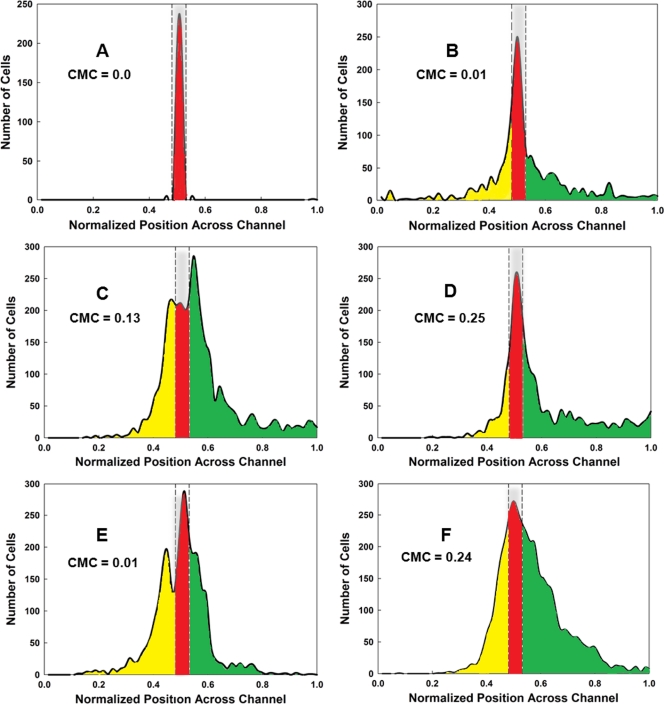
To test the relative sensitivity of cells to l-serine and AI-2 in an independent assay, we employed the μFlow device (9, 10). The gradients were created in two ways. In the first scenario, a linear gradient was generated across the 1,050-μm width of the microfluidic observation chamber by utilizing two input channels, delivering 0 and 200 μM chemoeffector. In the second scenario, a nonlinear gradient was generated by using five input channels to deliver 0, 0, 2, 20, and 200 μM chemoeffector. The concentration at the entry point for the cells was 100 μM in the linear gradient and 2 μM in the nonlinear gradient, and the cells were preequilibrated with these concentrations prior to their introduction into the observation chamber. The distribution profiles of fluorescently labeled CV1 cells in l-serine and AI-2 are shown in Fig. 3. For each set of conditions, the chemotaxis migration coefficients (CMC values) (10, 18) were calculated (Fig. 3). Basically, the CMC value represents the difference in the total number of cells migrating in the up-gradient direction minus the total number of cells migrating in the down-gradient direction, with the cell values weighted by the fractional distance they moved toward a boundary. Thus, cells migrating all the way to the right or left walls of the chamber received a weighting factor of 1 or −1, respectively, cells migrating halfway to the right or left walls received weighting factors of 0.5 or −0.5, and so forth.
FIG. 3.
Assays of chemotactic behavior in the μFlow device. Cells were grown and prepared as for the μPlug assay, as described previously (9). The CMC was calculated according to the methods described in the text and elsewhere (10, 18). (A) Typical distribution of RFP-labeled dead cells, shown in red. The distribution of cells from one run is shown; it is typical for that found for RFP-labeled dead cells in all runs. The area occupied by dead cells is delineated by the gray bar enclosed in dashed lines. (B) Typical distribution of CV1 (wild-type) GFP-labeled cells in the absence of a chemoeffector gradient. The distribution of cells moving in the up-gradient direction beyond the “dead” zone is highlighted in green, and the distribution of cells moving in the down-gradient direction is highlighted in yellow. GFP-labeled cells remaining in the region occupied by dead cells (highlighted in red) were not included in the calculations of CMC values. (C) Typical distribution of CV1 cells in a 0-to-200 μM nonlinear gradient of l-serine. (D) Typical distribution of CV1 cells in a 0-to-200 μM nonlinear gradient of AI-2. (E) Typical distribution of CV1 cells in a 0-to-200 μM linear gradient of l-serine. (F) Typical distribution of CV1 cells in a 0-to-200 μM linear gradient of l-serine. All assays were run a minimum of three times. The CMC values obtained are indicated on each graph.
CV1 cells responded to nonlinear and linear gradients of AI-2 (Fig. 3D and F) with similar CMC values (0.24 and 0.25), but they responded significantly only to the nonlinear gradient of l-serine, in which the gradient is very steep at the point at which the cells enter the chamber (compare Fig. 3C and E). Even in the nonlinear gradient, the CMC value for l-serine was only 0.13, about 50% that of the CMC value for AI-2. These results are consistent with the idea that chemotaxis to AI-2 is more sensitive at higher chemoeffector concentrations than is chemotaxis to l-serine.
We also tested the MJ102/pCA24N-PT5-lac::lsrB strain in a nonlinear AI-2 gradient in the μFlow device. It gave a CMC value of 0.18, which was lower than the value for CV1 cells in nonlinear AI-2 gradients but higher than that for CV1 cells in nonlinear gradients of l-serine. At present, we have no way of measuring periplasmic levels of LsrB, but the somewhat-attenuated response of MJ102/pCA24N-PT5-lac::lsrB cells relative to wild-type cells could reflect nonphysiological levels of LsrB in the complemented strain.
Preliminary evidence from μPlug assays (M. Hegde, unpublished data) suggests that the pathogenic S. Typhimurium strain 14028 also responds to AI-2 as an attractant and uses Tsr and LsrB to mediate that response. It may well be that many motile bacteria that produce the Tsr and LsrB proteins share the ability to perform chemotaxis to AI-2.
The reason why LsrB is essential for chemotaxis to AI-2 is not known. The role of LsrB in chemotaxis does not seem to be for transport of AI-2 into the cytoplasm, because strain MJ102, which carries the lsrCΩKanr mutation, is defective for AI-2 transport (27) but is still able to carry out AI-2 chemotaxis. By analogy with other binding protein-dependent chemoreceptor systems, such as those for maltose-binding protein (MBP) and Tar (29), it may be that AI-2-bound LsrB assumes a conformation that enables it to interact with Tsr directly in the periplasm. Similar interactions have been postulated for the involvement of galactose-binding protein (13) and ribose-binding protein (2) with Trg in taxis to these two sugars and for dipeptide-binding protein with Tap (17). Tsr is the only one of the four canonical chemoreceptors of E. coli not known to interact with a substrate-binding protein. It may be on the verge of losing that unique status.
The typical paradigm for indirectly binding chemoreceptors is that the ligands have a micromolar KD (equilibrium dissociation constant) for the periplasmic binding protein, and the ligand-bound binding protein has a low affinity for its chemoreceptor partner, estimated at about 200 μM for MBP and Tar (29). The KD for AI-2 binding to LsrB has been reported as ∼160 μM (30), which is at least 100-fold higher than the KD of MBP for maltose. We do not know the levels of LsrB in the periplasm of cells grown under various conditions, but they may be significantly lower than for other binding proteins, which typically far outnumber their cognate membrane-bound transport systems and chemoreceptors. Thus, the binding affinities of AI-2 for LsrB and of AI-2-bound LsrB for Tsr could be very different than for other known indirectly binding chemoreceptors.
AI-2, unlike other known attractants for E. coli, does not serve as food for bacteria (28) but is an intraspecific and interspecific signal of cell density. Therefore, chemotaxis to AI-2 may not have the rather narrow dose-response range that is characteristic of most indirectly binding chemoattractants (23). For nutrients, migration to concentrations higher than those needed for the maximum rate of uptake has no selective value. Given that AI-2 is produced by many different species of biofilm-forming bacteria (5), it may be that chemotaxis to AI-2 serves to recruit free-swimming, planktonic bacteria to biofilms (11). If so, there is no obvious reason why the response to AI-2 should saturate; the selective pressure may be to swim as close as possible to a source of AI-2. We are currently characterizing the molecular mechanism of AI-2 chemotaxis in order to understand how it has evolved to match the ecological context in which AI-2 chemotaxis occurs.
Acknowledgments
This work was funded in part by awards from the NSF to A.J. (CBET 0846453), the NIH (R01 GM089999) to T.K.W. and A.J., and the Bartoszek Fund for Basic Biological Science to M.D.M.
We are grateful for the Keio strains provided by the Genome Analysis Project and the National BioResource Project (Japan): E. coli. We thank Rasika Harshey for providing S. Typhimurium strain 14028 and its Δtsr derivative and Bonnie Bassler for providing S. Typhimurium strains MET 235 and MET259. Lily Bartoszek proofread the manuscript prior to submission.
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. [DOI] [PubMed] [Google Scholar]
- 2.Aksamit, R. R., and D. E. Koshland, Jr. 1974. Identification of the ribose binding protein as the receptor for ribose chemotaxis in Salmonella typhimurium. Biochemistry 13:4473-4478. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bansal, T., P. Jesudhasan, S. Pillai, T. K. Wood, and A. Jayaraman. 2008. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl. Microbiol. Biotechnol. 78:811-819. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L. 2002. Small talk: cell-to-cell communication in bacteria. Cell 109:421-424. [DOI] [PubMed] [Google Scholar]
- 6.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 7.Callahan, A. M., and J. S. Parkinson. 1985. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: cheD mutations affect the structure and function of the Tsr transducer. J. Bacteriol. 161:96-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X., et al. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 9.Englert, D. L., A. Jayaraman, and M. D. Manson. 2009. Microfluidic techniques for the analysis of bacterial chemotaxis. Methods Mol. Biol. 571:1-23. [DOI] [PubMed] [Google Scholar]
- 10.Englert, D. L., M. D. Manson, and A. Jayaraman. 2009. Flow-based microfluidic device for quantifying bacterial chemotaxis in stable, competing gradients. Appl. Environ. Microbiol. 75:4557-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez Barrios, A. F., et al. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 13.Hazelbauer, G. L., and J. Adler. 1971. Role of the galactose binding protein in chemotaxis of Escherichia coli toward galactose. Nat. New Biol. 230:101-104. [DOI] [PubMed] [Google Scholar]
- 14.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitagawa, M., et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291-299. [DOI] [PubMed] [Google Scholar]
- 16.Maeda, T., V. Sanchez-Torres, and T. K. Wood. 2008. Metabolic engineering to enhance bacterial hydrogen production. Microb. Biotechnol. 1:30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manson, M. D., V. Blank, G. Brade, and C. F. Higgins. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321:253-256. [DOI] [PubMed] [Google Scholar]
- 18.Mao, H., P. S. Cremer, and M. D. Manson. 2003. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 100:5449-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mesibov, R., G. W. Ordal, and J. Adler. 1973. The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over this range. Weber law and related phenomena. J. Gen. Physiol. 62:203-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 21.Miller, S. T., et al. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 22.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann, S., C. H. Hansen, N. S. Wingreen, and V. Sourjik. 2010. Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis. EMBO J. 29:3484-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 26.Slocum, M. K., and J. S. Parkinson. 1983. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J. Bacteriol. 155:565-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 28.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Y., et al. 1999. Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc. Natl. Acad. Sci. U. S. A. 96:939-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu, J., and D. Pei. 2008. A LuxP-based fluorescent sensor for bacterial autoinducer II. ACS Chem. Biol. 3:110-119. [DOI] [PubMed] [Google Scholar]