Abstract
We report here the first demonstration of intra- and interspecies conjugative plasmid DNA transfer for Campylobacter fetus. Gene regions carried by a Campylobacter coli plasmid were identified that are sufficient for conjugative mobilization to Escherichia coli and C. fetus recipients. A broader functional range is predicted. Efficient DNA transfer involves the virB9 and virD4 genes of the type IV bacterial secretion system encoded by a pathogenicity island of C. fetus subsp. venerealis. Complementation of these phenotypes from expression constructions based on the promoter of the C. fetus surface antigen protein (sap) locus was temperature dependent, and a temperature regulation of the sap promoter was subsequently confirmed under laboratory conditions. Gene transfer was sensitive to surface or entry exclusion functions in potential recipient cells carrying IncPα plasmid RP4 implying functional relatedness to C. fetus proteins. The virB/virD4 locus is also known to be involved in bacterial invasion and killing of cultured human cells in vitro. Whether specifically secreted effector proteins contribute to host colonization and infection activities is currently unknown. Two putative effector proteins carrying an FIC domain conserved in a few bacterial type III and type IV secreted proteins of pathogens were analyzed for secretion by the C. fetus or heterologous conjugative systems. No evidence for interbacterial translocation of the Fic proteins was found.
Type IV secretion systems (T4SS) are membrane-associated transporter complexes used by Gram-negative and Gram-positive bacteria to deliver substrate molecules to a variety of target cells in a contact-dependent manner (for reviews, see references 3, 5, 12, 16, 21, and 32). Secreted macromolecules are proteins or protein-DNA complexes; thus, some T4SS support horizontal gene transfer. The mechanisms involved include conjugation, oncogenic T-DNA delivery to plant cells by the phytopathogen Agrobacterium tumefaciens, and DNA uptake and transformation, as well as DNA release into the extracellular milieu (15). Moreover, several mammalian pathogens utilize their T4 translocation machinery for toxin secretion and the targeted delivery of virulence factors into eukaryotic host cells during infection. Accordingly, T4SS contribute to pathogenesis in various ways, including increased genome plasticity, antibiotic resistance spread, enhanced surface colonization and biofilm formation, and the specific injection of virulence proteins. In the latter case, the translocated proteins affect basic cellular functions of the host, resulting in the induction of disease (9, 23, 56, 70).
Macromolecular secretion across bacterial and host cell membranes requires the assembly of a multisubunit, cell envelope-spanning machinery comprising a secretion channel and often a pilus or cell surface filament. In paradigm conjugation systems, these components are collectively referred to as the Mating pair formation (Mpf) proteins (58). Systems related to the F-plasmid share 8 of 10 highly conserved Mpf components common in T4SS (40). Conjugative P-like (e.g., IncP, -W, and -N) and I-type (e.g., R64 and ColIb-P9) systems require distinct sets of auxiliary genes. The T4SS associated with bacterial virulence have been genetically categorized into distinct classes, T4a and T4b, based on whether they share common ancestries with the A. tumefaciens VirB/D4 (T4a) or ColIb-P9 (T4b) archetypal systems (19).
T4a pathways are highly conserved and well studied. The Agrobacterium apparatus includes 12 envelope-spanning components (VirB1-B11) expressed from a single operon. In addition, an inner membrane anchored T4 coupling protein T4CP (VirD4) governs the selective uptake of secretion substrates into the VirB translocation channel (3). T4CPs are universally required for transferring protein-DNA complexes and are typically necessary for protein translocation and virulence in pathogen-associated systems (22, 41, 54, 59). Several virulence-associated T4SS appear to have retained the capacity to mobilize plasmids by conjugation, while gaining proficiency in specific protein translocation (10, 69). Our work with the human and animal pathogen Campylobacter fetus recently revealed a subspecies-specific pathogenicity island (PAI) encoding a functional bacterial T4a secretion apparatus (28). This pathogen belongs to the group of epsilonproteobacteria and is highly adapted to mucosal surfaces (31). The two subspecies, C. fetus subsp. fetus and C. fetus subsp. venerealis, show a clonal population structure (65). Despite that level of genetic relatedness the subspecies exhibit distinct host and tissue preferences. C. fetus subsp. venerealis is largely restricted to the bovine genital tract, causing epidemic abortion in these animals and substantial economic losses to the cattle industry (8). C. fetus subsp. fetus is an important agent in ovine abortion worldwide and can also induce severe disease in humans (7, 8, 61, 63). This subspecies is the predominant Campylobacter species isolated from human blood and is considered to be an emerging pathogen (7). We are interested in the contribution of the C. fetus subsp. venerealis chromosomal PAI and its resident T4SS to the host and tissue tropism of this subspecies. Mutational inactivation of the virD4 and virB9 components in virulent C. fetus subsp. venerealis isolates attenuated invasion and cell-killing phenotypes in cultured human cell lines (28). Nonetheless, progress in understanding the role this chromosomally encoded T4SS plays in host-pathogen interactions remains challenging; this is due to the difficulty of manipulating the organism genetically and also to the current absence of well-characterized in vitro models of tissue-specific infection (34).
This study aimed to identify macromolecular substrates translocated by the C. fetus T4SS. We demonstrate that C. fetus subsp. venerealis supports intra- and interspecies conjugative mobilization of plasmid DNA in a process requiring the T4SS. Functional overlap between C. fetus subsp. venerealis and IncP plasmid components was detected via plasmid RP4-induced fertility inhibition. Two putative secretion substrates encoded by the C. fetus subsp. venerealis PAI were investigated. The data provide evidence for conjugative DNA delivery but not interbacterial protein transfer.
MATERIALS AND METHODS
Sequence analysis and alignments.
The predicted protein sequences of the C. fetus subsp. venerealis virB/D4 locus (GenBank accession number EU443150) were analyzed by using BLAST-P (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Predicted proteins were compared over the whole amino acid sequence by using the CLUSTAL W function of MegAlign from Lasergene (DNAStar, Inc., Madison, WI) with default settings. Protein domains were identified by using the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/structure/index.shtml).
Bacterial strains.
Strains are listed in Table 1. Growth conditions for Campylobacter and Escherichia coli strains were as previously described (33). C. fetus subsp. venerealis vir gene mutants were described earlier (28).
TABLE 1.
Bacterial strains used in this study
| Strain | Descriptiona | Source or referenceb |
|---|---|---|
| C. fetus | ||
| subsp. fetus ATCC 27374 | Type strain; Nalr | ATCC |
| subsp. fetus F12 | Human isolate, septicemia; Cipr Nalr | 37 |
| subsp. fetus 82-40 | Human isolate, septicemia; GenBank NC_008599 | |
| subsp. venerealis ATCC 19438 | Type strain; Nalr | ATCC |
| subsp. venerealis 3-18 | Cfv ATCC 19438 virB9::PCc-aphA-3 | 28 |
| subsp. venerealis SK1 | Cfv ATCC 19438 virD4::PrpsJ-aphA-3 | 28 |
| subsp. venerealis JL1 | Cfv ATCC 19438 cdtB::PrpsJ-aphA-3 | 28 |
| subsp. venerealis 84-112 | Bovine isolate, genital secretion; Nalr | 50 |
| subsp. venerealis V81_SK1 | Cfv 84-112 virD4::PrpsJ-aphA-3 | 28 |
| E. coli | ||
| DH5α | endA1 recA1 gyrA96 thi-1 hsdR17 supE44 λ− relA1 deoR Δ(lacZYA-argF)U169 φ80dlacZΔM15 | 73 |
| MT102 | araD139 (ara-leu)7697 Δlac thi hsdR mcr rpsL | 4 |
| MS411 | ilvG rfb-50 thi | M. Schembri (DTU) |
| SAR18 | ara Δ(lac-pro) thi attB::bla-PA1/04/03-gfpmut3*-To | 51 |
| MS614 | Smr; ilvG rfb-50 thi rpsL | M. Schembri (DTU) |
| CHS26Cm::LKL | CSH26 galK::cat::loxP-Km-loxP | 39 |
| MT102Cm::LKL | MT102 galK::cat::loxP-Km-loxP | 39 |
| CHS26Cm::LTL | CSH26 galK::cat::loxP-TetRA-loxP | 39 |
| S17-1 λpir | Tpr Smr; recA thi pro hsdR−M+ RP4:2-Tc::Mu:Km Tn7 λpir | 18 |
Nalr, nalidixic acid resistance; Cipr, ciprofloxacin resistance; Smr, streptomycin resistance; Tpr, trimethoprim resistance.
ATCC, American Type Culture Collection; DTU, Technical University of Denmark.
Plasmids and PCR amplification.
Plasmids are listed in Table 2. Plasmid DNA was purified from E. coli and Campylobacter cells with a QIAprep spin miniprep kit (Qiagen, Hilden, Germany). Restriction endonucleases and DNA modifying enzymes, obtained from New England Biolabs, Inc. (Beverly, MA), and Fermentas GmbH (St. Leon-Rot, Germany) were used as recommended by the suppliers. Phusion High-Fidelity DNA polymerase (Finnzymes Oy, Espoo, Finland) was used to amplify DNA fragments for cloning. All other PCR amplifications utilized DyNAzyme II DNA polymerase (Finnzymes Oy) according to the manufacturer's specifications.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant featuresa | Source or reference |
|---|---|---|
| pRY111 | PCc-cat mobIncP oriVpIP1455 oriVpBR322 | 75 |
| pRYGG1 | PsapA in the pRY111 MCS | 33 |
| pRYSS1 | Fusion (Φ) (PsapA-aphA-3) in pRYGG1 | 33 |
| pRYVL1 | Φ(PsapA-aphA-3) in pRYGG1; BamHI and PstI linker inserted between PsapA and aphA-3 | 33 |
| pRYVL2 | Φ(PsapA-virD4-aphA-3) in pRYGG1 | 33 |
| pRYMJ2 | Φ(PsapA-Cfv-virB9-virB10-virB11-virD4) in pRYGG1 | 33 |
| pRYEL1 | Φ(PsapA-gfp) in pRYGG1 | 33 |
| pSK108-1 | 3,207 bp of pCFV108 in pBluescript KS II(−) | 33 |
| pRYSK2 | oriVpIP1455 replaced by oriVpCFV108 | 33 |
| pBlue-oriT | mobIncP in the pBluescript KS II(−) MCS | 33 |
| pRYSK3 | pCFV108 iterons; Φ(PsapA-aphA-3) in pBlue-oriT | 33 |
| pRYSK9 | mobApCfv108 in pRYSK3 | This study |
| pRYSK17 | nicpIP1455 in pRYSK3 | This study |
| pRYBM5 | Φ(PgatC-aphA-3); PsapA of pRYVL1 replaced by PgatC | This study |
| pRYSK12 | Φ(PgatC-virD4); PsapA of pRYVL2 replaced by PgatC | This study |
| CFP B | Cre fusion plasmid | 49 |
| pCreSK1 | Φ(Cre-fic1) in CFPB | This study |
| pCreSK2 | Φ(Cre-fic2) in CFPB | This study |
| pRYSK13 | Φ(Cre-fic1) in pRYSK3 | This study |
| pRYSK14 | Φ(Cre-fic2) in pRYSK3 | This study |
| pRYSK15 | Cre in pRYSK3 | This study |
| pCreTraI R1 | R1 traI encoding residues 3 to 1756 in CFP B | 39 |
| pCreTraI F | KpnI-SalI traI fragment from p99I+ in CFP B | 39 |
| pSU1501 | R388 trwC in pKK223-3 | 29 |
| pCreTrwC | R388 trwC encoding residues 2 to 966 in CFP B | This study |
| pJMTraD | Wild-type F traD in pBAD24 | 42 |
| pKD46 | Red recombinase expression plasmid: araBp gam-bet-exo oriR101 repA101(Ts) bla | 17 |
| RP4 | IncP conjugative plasmid | 48 |
| pSU2007 | IncW conjugative plasmid; Kmr | 44 |
| pAR119 | IncF conjugative plasmid; Kmr | 52 |
| pLG211 | IncI conjugative plasmid; Kmr | 14 |
| R1-16 | IncFII conjugative plasmid; Kmr fin- | 27 |
| pOX38 | IncFI, conjugative plasmid, KmR | 13 |
| pOX38traD411 | traD-null derivative of pOX38 | 43 |
Kmr, kanamycin resistance.
Construction of plasmids.
Numbered oligonucleotide primers are described in Table 3. A 1.6-kb mobA fragment pSK108-1 was excised with SmaI and NdeI and then blunted using T4 DNA polymerase (New England Biolabs) before ligation with the SmaI site of pRYSK3 to create pRYSK9. A 220-bp NotI/PstI fragment containing the gatC promoter was amplified from C. fetus subsp. venerealis chromosomal DNA with the primers 5 and 6. This regulatory region was then used to replace the sapA promoter of pRYVL1. A kanamycin resistance phenotype in C. fetus subsp. venerealis confirmed promoter activity for the resulting construction, pRYBM5. To generate the complementation vector pRYSK12, the gatC promoter fragment was amplified from pRYBM5 with primers 5 and 7, digested with NotI/BamHI, and inserted into a NotI/BamHI-digested pRYVL2. A 420-bp fragment containing the pIP1455 iteron sequence and a putative nic site was amplified from pRYSS1 by using the primer pair 8 and 9 and inserted into the SmaI site of pRYSK3 to generate pRYSK17. To fuse the Cre recombinase with the FIC domain-containing proteins, Fic1 and Fic2, the fic genes were amplified from chromosomal C. fetus subsp. venerealis DNA using the primer pairs 12/13 and 14/15, respectively. The products were inserted in the KpnI/SalI-cut plasmid CFP B to generate C-terminal fusions with the Cre coding sequence, resulting in pCreSK1 and pCreSK2, respectively. The 1,028-bp cre gene, the 1,860-bp cre-fic1 fusion, and the 1,944-bp cre-fic2 fusion were amplified from either CFP B or pCreSK1 or pCreSK2 with primers 16 and 17. PCR products were cut and ligated with SmaI-digested pRYSK3 to generate pRYSK15, pRYSK13, and pRYSK14, respectively. For Cre-TrwC, the trwC gene was amplified from plasmid pSU1501 using the primers 18/19 and then ligated with a KpnI/SalI-cut CFP B vector.
TABLE 3.
Oligonucleotides used in this study
| No. | Oligonucleotidea | Sequenceb (5′-3′) | Description and/or accession no.c |
|---|---|---|---|
| 1 | aphA-3_f* | GAGGATCCGCTAAAATGAGAATATCACCG | aphA-3 from codon 2 (nt 607 to 626); M26832† |
| 2 | aphA-3_r* | GAGGATCCCTTTTTAGACATCTAAATCTAGG | aphA-3 (nt 1423 to 1401); M26832† |
| 3 | cat_BamHI_f* | TTGGATCCCAATTCACAAAGATTGATATA | cat from codon 2 (nt 312 to 332); M35190† |
| 4 | cat_BamHI_r* | GGGATCCTATTTATTCAGCAAGTCTTG | cat (nt 931 to 912); M35190† |
| 5 | 1146_NotI_f | TTGCGGCCGCAATAGTATCCTTAACATAAAATTTT | Upstream of gatC; (nt 113712 to 113745); NC_008599† |
| 6 | 1146_PstI_r | ATTCTGCAGAGAATACAACTCCATTTTTGATT | Upstream of gatC; (nt 1136947 to 1136925); NC_008599† |
| 7 | gatC_BamHI_r | TTGGATCCAGAATACAACTCCATTTTTGATT | Upstream of gatC; (nt 1136947 to 1136925); NC_008599† |
| 8 | nic_SmaI_f | TATCCCGGGTTAGGTATAATCTCACAAAACG | pIP1455 |
| 9 | nic_SmaI_r | TAACCCGGGTAGCACGGAAGACGGACAAA | pIP1455 |
| 10 | galK_f | TCCATCAGCGTGACTACCAT | E. coli K-12 galK (nt 787966-787985); NC_000913† |
| 11 | galK_r | TAGCCGTCGTGCCTTACT | E. coli K-12 galK (nt 789594-789577) |
| 12 | Fic1_KpnI_f | GCTGGTACCGATGGCGGTGTAAATTTAGGC | fic1 (nt 31745 to 31765) |
| 13 | Fic1_SalI_r | CTAGTCGACTTATCTCTCCTTTTCCTTTGAAT | fic1 (nt 32578 to 32556) |
| 14 | Fic2_KpnI_f | GCTGGTACCCAAGAACAATATACGGAAATCAA | fic2 (nt 32581 to 32603) |
| 15 | Fic2_SalI_r | CTAGTCGACTTATCTTTCCTTTTCTTTTGATTTT | fic2 (nt 33498 to 33474) |
| 16 | Cre_SmaI_f | TAACCCGGGATGTCCAATTTACTGACCGTA | Cre fusion plasmid CFP B (nt 245 to 265) |
| 17 | Cre_SmaI_r | TTACCCGGGTGAAGGCTCTCAAGGGCAT | Cre fusion plasmid CFP B (nt 1295 to 1313) |
| 18 | TrwC_SFw1 | CATGTAGGTACCCTCAGTCACATGGTATTGAC | trwC from codon 2 (nt 1542 to 1561); X63150.2† |
| 19 | TrwC_SRev1 | GCAATCGTCGACTTACCTTCCGGCCTCCATGCCG | trwC (nt 4439 to 4418); X63150.2† |
| 20 | Trans17 | TGATCCTGGTTTTGGAGTGA | Transposase A (nt 21254 to 21273) |
| 21 | Trans8 | GTATGAGCTTTATCCTTTGTTTC | Transposase B (nt 21992 to 21970) |
| 22 | Trans14 | GGGGCATCAACTCTTAGAGG | Transposase B (nt 22528 to 22547) |
| 23 | Trans18 | CATTGAGAATTTCGGCACCT | ORF3 (nt 23060 to 23041)‡ |
| 24 | Trans4 | ATCACTCAGCAGCAAGAGCG | ORF3 (nt 22739 to 22758) |
| 25 | TopTraE1 | TTGCACGTGCGAGTTCAGGC | Topoisomerase (nt 23294 to 23275) |
| 26 | TraE 7c | GCAAGAGCGGAAAGCTTCTGCC | Topoisomerase (nt 25347 to 25368) |
| 27 | TraE 21 | GTTGATAGCTGCTTTGCG | Chromosome segregation (nt 25844 to 25827) |
| 28 | TraE 8d | GGAGCAAAACAAAGAACCAAC | Chromosome segregation (nt 26410 to 26430) |
| 29 | TraE 9 | CAGCATTTAAGGCAGAAC | Lytic transglycosylase (nt 26860 to 26843) |
| 30 | TraE 10 | GCAAGTTAATTCAATACACG | Lytic transgycosylase (nt 27030 to 27049) |
| 31 | TraE 29 | GGCTATCGACGTCAAAGCC | dnaG (nt 27330 to 27312) |
| 32 | TraE 14 | GAAAGAACAGCTCAATGG | dnaG (nt 28322 to 28339) |
| 33 | TraE 13 | CGGCGCGAGTGGGAAAG | ORF8 (nt 28616 to 28600)‡ |
| 34 | TraE 32 | CGCGCCGCATATATTAGAA | ORF8 (nt 28610 to 28628)‡ |
| 35 | TraE 33 | CGACCTTTGTCGGTTGATTT | ORF9 (nt 28940 to 28921)‡ |
| 36 | TraE 20 | CCAGTGGCACAATCCGC | ORF9 (nt 28864 to 28880)‡ |
| 37 | TraE 34 | GCAGCCTTTCATTTGGGTAA | ORF10 (nt 29279 to 29260)‡ |
| 38 | TraE 28 | GCCAAAATTTAGCCAAATGGTAGC | ORF10 (nt 29731 to 29754)‡ |
| 39 | TraE 35 | CCCAACTCGCAGTTTTTCAT | ORF11 (nt 30142 to 30123)‡ |
| 40 | TraE 36 | GGTTGTGGCGAAGAAGCTAA | ORF11 (nt 29985 to 30004)‡ |
| 41 | TraE 24R | GAGTGCCAGCTTTATGGC | parA (nt 30353 to 30336) |
| 42 | TraE 24 | GCCATAAAGCTGGCACTC | parA (nt 30336 to 30353) |
| 43 | TraE 23 | GCGGTGTCTTTTGCAAAGC | Nucleotidyltransferase (nt 31112 to 31094) |
| 44 | NucTra1 | CCAAAACTGCAAGAAGACGG | Nucleotidyltransferase (nt 31048 to 31067) |
| 45 | Fic1-2 | TATCGTCGTCATTTTTGGCA | fic1 (nt 32195 to 32176) |
| 46 | Fic1-1 | GGCTCATCATAGCACAGCAA | fic1 (nt 31822 to 31841) |
| 47 | Fic2-2 | TTTCAAGCCCTTGTGGAAAG | fic2 (nt 32917 to 32902) |
| 48 | Fic2-5 | ATGGGAGCGATCAAACAACC | fic2 (nt 33445 to 33461) |
| 49 | Cpp17-6 | ACCGATCAAGAAGCAGTCGT | cpp17 (nt 34120 to 34101) |
| 50 | NucTra2 | GCCGTATCGCATAGATCGAC | nt 31259 to 31240 |
| 51 | Cpp17-2 | CGATCTCGATGATCTACGCA | nt 34442 to 34423 |
| 52 | Km_screen_rev | GATCTTTAAATGGAGTGT | aphA-3 (nt 1105 to 1088); M26832† |
| 53 | glnA_RT_fwd | GCGAGTGGAATGATGGTAAAG | glnA (nt 1053483 to 1053504); NC_008599† |
| 54 | glnA_RT_rev | CTCTAACGCTTTTCTCTCCTG | glnA (nt 1054086 to 1054066); NC_008599† |
Plasmid mobilization from C. fetus to E. coli.
E. coli (2 × 107 CFU) from an overnight broth culture was mixed with 2 × 109 Campylobacter cells harvested from Columbia blood agar (CBA) plates after 24 h growth. The suspension was spotted onto a nitrocellulose filter (25 mm, 0.22 μm; Millipore), which was placed on an antibiotic-free CBA plate and incubated in a microaerobic atmosphere at 37 or 32°C for 18 to 24 h. C. fetus grows poorly at temperatures below 25°C and above 39°C, and DNA mobilization was not detected at these temperatures. In some experiments, the filters were initially soaked in buffer containing 20 mM Tris-HCl and 1 mM MgCl2 (pH 8) with or without 300 μg of DNase I (Roche, Mannheim, Germany). After incubation, cells were removed from the filter by vortexing in 1 ml of phosphate-buffered saline (PBS). Serial dilutions of the bacterial suspension were plated on LB agar containing 25 μg of kanamycin and/or 25 μg of chloramphenicol/ml to select for transconjugant E. coli cells. Transconjugant colonies were subcultured, and the plasmid content was verified either by DNA isolation, followed by restriction mapping, or by PCR analysis. Viable cell counts were determined for donor and recipient strains by serial dilutions and subsequent plating.
Plasmid mobilization to Campylobacter.
Conjugational DNA delivery from C. fetus to Campylobacter was performed as described for E. coli recipients. When C. fetus subsp. fetus served as a recipient, bacteria were mixed in a ratio of 10:1 (2 × 109 donor and 2 × 108 recipient cells, respectively, in suspension). The cell suspension was spotted onto a nitrocellulose filter and incubated under microaerophilic conditions at 37 or 32°C for 24 h either in the presence or absence of DNase I. Cells were removed from the filter by vortexing in 1 ml of PBS, and the suspension was spread over five CBA plates. Selection for transconjugant C. fetus subsp. fetus F12 combined ciprofloxacin (32 μg/ml) with selection for the newly acquired plasmid. Agar plates were incubated under microaerophilic conditions at 37°C for 5 days. In experiments with C. fetus subsp. venerealis JL1 recipients, equivalent cell numbers (2 × 109 CFU) were combined. Selection for transconjugants combined kanamycin for the chromosomally encoded aphA-3 gene and selection for the acquired plasmid. CBA plates were incubated under microaerophilic conditions at 37°C for up to 12 days.
Cre recombinase assay for translocation (CRAfT).
To generate E. coli MT102Cm::LKL, a cat::loxP-Km-loxP cassette was inserted into galK of E. coli MT102. The primer pair 10/11 was used to amplify the cat::loxP-Km-loxP cassette from E. coli CSH26Cm::LKL. The temperature-sensitive pKD46 was introduced into MT102, and the cat::loxP-Km-loxP cassette was integrated via homologous recombination as described previously (17).
Campylobacter strains were thawed 72 h prior to the assay and subcultured every 24 h on kanamycin-containing CBA plates. The assay for protein translocation was performed as described for DNA transfer from Campylobacter to E. coli. CSH26Cm::LKL or MT102Cm::LKL served as E. coli recipients. Recombinant colonies resulting from Cre-mediated reconstitution of cat expression were selected on LB agar containing 20 μg of chloramphenicol/ml.
To screen for Cre-Fic translocation by a heterologous T4SS E. coli MS614 donor strains harbored plasmid R1-16, pOX38 or pSU2007. E. coli S17-1 λpir provided the P-type transfer system. The E. coli indicator strain was replaced with CSH26Cm::LTL. This strain was created with the lambda RED system as described above using targeting DNA made with primers 12 and 13 and template pAR183. All donor strains additionally contained a CFP B-based plasmid expressing the fusion protein. Drug-free LB was inoculated to an optical density at 600 nm of 0.005 with an overnight culture of the donor strain and incubated at 37°C for 60 min. A 10-fold excess of recipient strain E. coli CSH26Cm::LTL was added, and the mixture was incubated for 2.5 h at 37°C in liquid (pOX38 or R1-16) or after spotting onto a nitrocellulose filter (25 mm, 0.45 μm; Millipore) placed on antibiotic-free LB agar (S17-1 λpir or pSU2007). Mating was stopped by vortexing the suspension or the filter in 1 ml of LB medium for 1 min and immediately chilling the sample on ice. Transconjugants were selected on LB agar containing kanamycin (25 μg/ml) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 50 μg/ml). Recombinant colonies resulting from Cre-mediated reconstitution of cat expression were selected on LB agar containing 10 μg of chloramphenicol/ml. Donors were selected on streptomycin (100 μg/ml). Frequencies of conjugation and protein translocation were determined as transconjugants or recombinants per donor, respectively.
Prior to all CRAfT assays, every test protein fusion was assessed for stability and Cre recombinase activity in vivo by transformation of the indicator strains E. coli CSH26Cm:: LKL, CSH26Cm::LTL or E. coli MT102Cm::LKL with each expression vector. Transformants expressing the test fusion proteins, Cre alone, or the control fusions with known T4S substrates all exhibited an equivalent recombination frequency.
Isolation of RNA and preparation of cDNA.
Total RNA was isolated from C. fetus subsp. venerealis strain ATCC 19438 with or without plasmid pRYSS1 after 24 or 48 h of culture by using a RNeasy Protect bacteria kit (Qiagen) according to the manufacturer's instruction. Contaminating DNA was removed by incubation with 1.5 U of DNase I per μg of total RNA as recommended by the manufacturer (Fermentas). cDNA was prepared from 0.3 to 0.5 μg of RNA using specific primers (15 pmol) for first-strand synthesis and Moloney murine leukemia virus reverse transcriptase (RT; Fermentas) in a total volume of 20 μl. Negative controls were incubated without RT. Reaction products were treated with RNase H (New England Biolabs) at 37°C for 20 min without heat deactivation.
Analysis of sapA promoter temperature regulation.
RNA was isolated from C. fetus subsp. venerealis ATCC 19438(pRYSS1) grown at 32, 37, or 39°C. For second-strand synthesis, PCR primers were positioned to amplify a 500-kb fragment of aphA-3 from pRYSS1 and a 600-kb fragment of glnA (Table 3, primers 1 and 52 to 54). PCR fragments were amplified from 2 μl of the RT product as a template using Phusion High-Fidelity DNA polymerase (Finnzymes). Plasmid DNA (for aphA-3) and genomic DNA (for glnA) were used as positive control PCR templates.
Operon analysis.
RNA was isolated from C. fetus subsp. venerealis ATCC 19438 and cDNA prepared as described above. For second strand synthesis, PCR primers were positioned to amplify fragments spanning at least two open reading frames (ORFs) (Table 3, primers 20 to 51). PCR fragments were amplified from 2 μl (10%) of the RNase H-treated RT product as a template using Phusion High-Fidelity DNA polymerase (Finnzymes). Genomic DNA from C. fetus subsp. venerealis reference strain ATCC 19438 was used as a positive control PCR template.
RESULTS
C. fetus virB/virD4 gene homology and structure predict nucleoprotein and effector protein translocation.
CLUSTAL W comparisons were performed for the mating pore formation (Mpf) proteins and the putative T4CP (VirD4) of the C. fetus subsp. venerealis T4SS to ortholog proteins of public database entries (Table 4). Reference systems were selected to obtain broad distribution within the major branches of the phylogenetic trees of the ATPases VirB4 and VirB11 created by Fernandez-Lopez et al. (20) and Frank et al. (24). Our analysis focused on well-characterized systems of known function that additionally encode a T4CP and the most closely related T4SS of Campylobacter. Protein identities were generally low. BLAST search revealed that the Mpf proteins of the chromosomally encoded T4SS of C. hominis exhibited the highest overall homology to the C. fetus subsp. venerealis components, followed by plasmid pCC31 of C. coli and the pVT745 plasmid of Actinobacillus actinomycetemcomitans. The function of the C. hominis system is currently unknown. pCC31 and pVT745 are conjugative plasmids. Comparison to pCC31 revealed identities greater than 50% for VirB8, VirB9, VirB11, and VirD4. The C. hominis Mpf proteins display the highest overall identity, while the C. fetus subsp. venerealis VirD4 overlaps most closely with CmgD4 of pCC31 (59%).
TABLE 4.
Comparative identities of C. fetus subsp. venerealis Vir proteins and T4SS orthologs
| Organism or systema | Protein identity (%)b in: |
Locationc | Accession no.d | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VirB3/4 | VirB5 | VirB6 | VirB8 | VirB9 | VirB10 | VirB11 | VirD4 | Sum | |||
| C. fetus subsp. venerealis (vir)* | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 800 | C | EU553150 |
| C. hominis | 56 | 22 | 29 | 56 | 59 | 45 | 62 | 54 | 382 | C | NC_009714 |
| C. coli pCC31* | 46 | 22 | 25 | 56 | 52 | 39 | 54 | 59 | 352 | P | NC_006134 |
| A. actinomycetemcomitans pVT745* | 37 | 22 | 18 | 31 | 42 | 32 | 42 | 43 | 266 | P | NC_002579 |
| Broad-host-range plasmid pIP02T* | 32 | 14 | 16 | 31 | 33 | 28 | 38 | 42 | 223 | P | NC_003213 |
| X. axonopodis* | 31 | - | 17 | 25 | 22 | 23 | 31 | 36 | 184 | C | NC_003919 |
| B. henselae (vir) | 28 | 11 | 15 | 27 | 20 | 24 | 31 | 15 | 171 | C | BX897699 |
| L. pneumophila (lvhB)* | 21 | 7 | 17 | 18 | 24 | 20 | 25 | 20 | 152 | C | AE017354 |
| H. pylori (cag orHP) | 24 | - | - | 15 | 27 | 21 | 37 | 23 | 147 | C | NC_000915 |
| A. tumefaciens pTi* | 26 | 1 | 13 | 17 | 22 | 20 | 27 | 20 | 146 | P | NC_003065 |
| R. prowazekii | 30 | - | - | 14 | 21 | 18 | 29 | 24 | 135 | C | NC_000963 |
| Broad-host-range plasmid RP4* | 17 | 2 | 13 | - | - | 18 | 24 | 19 | 93 | P | NC_001621 |
Organism and system were selected according to BLAST scores, the presence of a putative coupling protein, genetic organization and function (see Materials and Methods). Systems shown to be conjugative are indicated by an asterisk (*).
The most similar homologs are indicated in boldface. -, No known ortholog.
Genes were carried on the chromosome (C) or plasmid (P).
Accession number of the genome or plasmid DNA sequence.
CLUSTAL W analysis of Campylobacter T4CPs were performed and revealed numerous stretches of amino acids with 100% identity within the protein sequences. High conservation extends into their shared C-terminal regions. The C. fetus protein carries additionally atypical insertions that are extremely rich in charged residues. Both the C. fetus subsp. venerealis and the C. hominis proteins carry long nonconserved C-terminal tails. In the case of F-like T4CPs, an atypical C-terminal extension is important for specificity of substrate recognition (55). A conserved domain database search facilitated assignment of the C. fetus subsp. venerealis VirD4 to the TraG, TraG/TraD family (pfam02534) of the P_loop_NTPase superfamily (cl09099). Presence of a virD4 homologue in C. fetus subsp. venerealis potentially links this T4SS to an ancestral or currently functional conjugative gene transfer system.
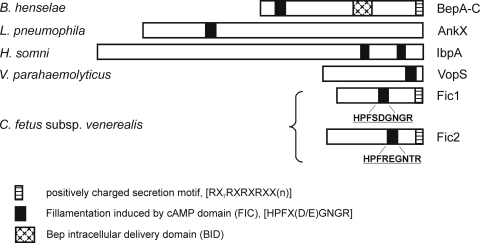
Understanding of bacterial T4SS has advanced dramatically in recent years. Accordingly, it becomes feasible to predict potential secretion substrates based on conserved protein domains, as well as gene organization (60). We identified two open reading frames (ORFs; orf38 and orf39; EU443150) downstream of the VirB/VirD4 locus that belong to the FIC (filamentation induced by cAMP) superfamily (cl00960). Related proteins are involved in cell division (36) and, importantly, are conserved among effector proteins secreted by type III (T3) and T4SS of Gram-negative bacteria (Fig. 1), including Bartonella species, Histophilus somni, Vibrio parahaemolyticus, and Legionella pneumophila (11, 53, 56, 60, 74, 76, 78). Fic1 of C. fetus subsp. venerealis was assigned to the FIC protein family COG3177 of unknown function, whereas Fic2 belongs to the FIC protein family COG2184, including proteins involved in cell division and chromosome partitioning. Fic1 (orf38) is 278 amino acids in length and the best BLASTP match (60% identity) was the Helicobacter cinaedi CCUC18818 hypothetical protein HcinC1-00045 (accession number ZP_03657318). Fic2 (orf39) is 306 amino acids in length and interestingly has no homology to proteins from H. cinaedi. The best BLASTP match (41% identity) is to the hypothetical protein CLOSS21_03156 (accession number ZP_02440650) from Clostridium sp. strain SS2/1. The presence of ORFs containing FIC domains 3′ to the VirB/VirD4 locus as part of the C. fetus subsp. venerealis chromosomal PAI supports the hypothesis that proteins Fic1 and Fic2 are translocated by the T4SS.
FIG. 1.
The FIC domain is conserved in T3 and T4SS substrates. Known effector proteins carrying the FIC domain and conserved motif [HPFx(D/E)GN(G/K)R] that contributes to AMPylation of host proteins. Where investigated, additional protein features required for T4CP recognition and translocation are illustrated as described previously (see below). Candidate secretion substrates of the C. fetus T4SS harboring the FIC signature are shown.
C. fetus subsp. venerealis mobilizes C. coli plasmid pIP1455 DNA to E. coli.
The T4SS of the C. fetus subsp. venerealis PAI is integrated in the genome. Since it is not carried by a self-transmissible conjugative plasmid, potential DNA transfer activity was tested via plasmid mobilization. Small cryptic plasmids often represent natural substrates for conjugative dissemination, since they encode a minimum of mobilization functions (a relaxase and nic site) and replication features supporting their spread and maintenance in bacterial populations. The plasmid profile of 60 C. fetus subsp. venerealis isolates carrying the T4 vir genes within our strain collection was analyzed, but small plasmids were generally absent (not shown). Given the strong conservation observed between components of the C. fetus subsp. venerealis PAI and those of the C. coli conjugative plasmid pCC31, we based the analysis on the cryptic C. coli plasmid pIP1455 (38). Vectors carrying the pIP1455 replicon can be propagated in C. fetus (33). Moreover, we noted that the pIP1455 backbone carries mobility features that might support its conjugative mobilization from a heterologous T4SS (Fig. 2). Test constructions for interspecies mobilization also carried selection markers for both species and the pBR322 origin for replication in E. coli (Fig. 2). The pIP1455-based shuttle vector pRYSS1 (33) was introduced to C. fetus subsp. venerealis strain 84-112 via conjugation. The capacity of this host to further transfer the test plasmid to the E. coli K-12 strain MT102 was investigated in a surface mating experiment. Suspensions of the potential donor and recipient bacteria were mixed and then collected on a filter, which was placed on blood agar for 24 h. C. fetus is not naturally competent. Nonetheless, filters were first soaked in buffer containing 1 mM MgCl2 without or with 300 μg of DNase I. At cell harvest, negative selection against C. fetus and positive selection for transconjugant E. coli was achieved by plating on LB agar. Presence of pRYSS1 in kanamycin- and chloramphenicol-resistant E. coli colonies was verified by plasmid isolation and restriction enzyme digestion (data not shown). Interspecies transfer occurred at frequencies of 3.16 × 10−6 ± 3.8 × 10−7 transconjugants per donor without DNase I and 4.03 × 10−6 ± 3.3 × 10−7 in the presence of DNase I; effectively ruling out transmission due to natural transformation. Parallel experiments using an alternative donor strain known to express the T4 vir genes, C. fetus subsp. venerealis ATCC 19438 (28), verified the transfer activity at equivalent frequencies (not shown). Conversely, substitution of the E. coli K-12 strain used as recipient in the experiment for E. coli MS411 or E. coli SAR18 gave rise to very low transfer frequencies (∼10−10 transconjugants per donor). All transconjugant colonies were routinely analyzed to verify that these indeed carried the mobilizable pRYSS1 plasmid. Uptake of plasmid DNA was confirmed in every case. The frequency of plasmid transfer was higher at 32°C than 37°C using E. coli recipient MT102 and the pIP1455-derived shuttle vector pRYGG1 (Fig. 3 A).
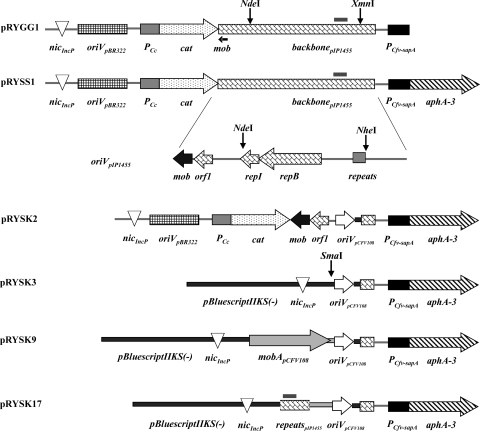
FIG. 2.
Constructions defining requirements for plasmid mobilization by the C. fetus subsp. venerealis T4SS. C. fetus cannot be readily transformed; thus, all vectors carry a P-type nic site to introduce the plasmid to donor cells. Selection in Campylobacter relies on a kanamycin resistance cassette (aphA-3) expressed from the C. fetus sapA promoter. Regions of the C. coli plasmid pIP1455 (backbone) are shown with functional modules identified in the expanded view below, including a putative mob gene (mob) and a potential P-type nic site (repeats). The C. fetus replicon (oriVpCFV108) and putative relaxase gene mobA from pCFV108 (gray arrow) are shown.
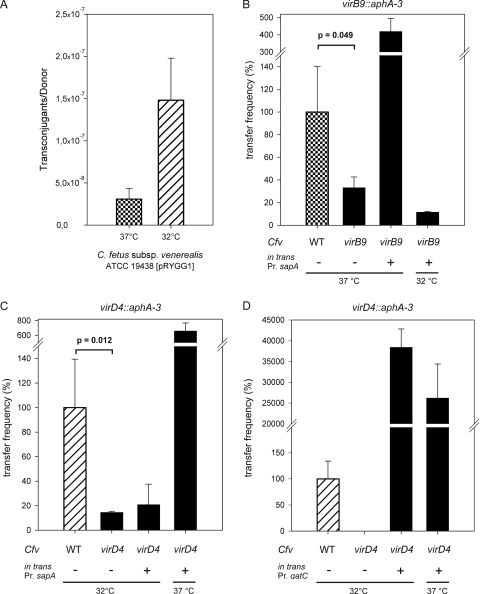
FIG. 3.
Efficient plasmid DNA translocation from C. fetus subsp. venerealis to E. coli requires the T4SS components VirB9 and VirD4 and moderate temperature. (A) Transfer of pRYGG1 DNA from C. fetus subsp. venerealis ATCC 19438 to E. coli MT102 was measured at 37 and 32°C (expressed as transconjugants relative to donor cells). (B to D) Comparison of the transfer frequencies of pRYGG1 from wild-type C. fetus subsp. venerealis ATCC 19438 (expressed as 100%) and either the isogenic virB9 mutant 3-18 (B) or that of pRYSS1 from the virD4 mutant SK1 (C and D). For complementation wild-type virB9 was expressed in 3-18 (B) and virD4 in mutant SK1 (C and D) from the promoter indicated (below) and at the experimental temperatures shown. The frequencies shown are expressed relative to the wild-type strain at the temperature indicated. The data are from at least three experiments. Statistically significant differences (unpaired Student t test) between wild-type and mutant conjugation frequencies are indicated.
We next confirmed that mobilization of the plasmid was due to features carried by the pIP1455 backbone by measuring transfer of a different plasmid with replication functions of the C. fetus plasmid pCFV108 (33). No transfer from C. fetus subsp. venerealis ATCC 19438 or 84-112 to E. coli MT102 was observed for this replicon (pRYSK3, Fig. 2), even with the additional presence of the putative relaxase gene mobA from pCFV108 on the construction (pRYSK9) (not shown). In an effort to identify the genetic elements present in pIP1455 that support mobilization from C. fetus subsp. venerealis donors, we eliminated all pIP1455 DNA in the test plasmid except the putative mob gene and the adjacent orf1 of unknown function resulting in pRYSK2 (Fig. 2). A 427-bp fragment of pIP1455 carrying a putative oriT was excised to create pRYSK17 (Fig. 2). We found that division of the pIP1455 DNA into distinct putative mobilization modules failed to support detectable mobilization of pRYSK2 and pRYSK17 under our conditions (not shown). In summary, interspecies transfer from C. fetus subsp. venerealis to E. coli MT102 occurred at detectable levels only when the mobilization substrate carried all of the C. coli plasmid pIP1455 sequences provided in pRYSS1. The mechanism of plasmid DNA transmission was unaffected by externally applied nuclease, supporting a conclusion of direct cell transfer.
Interspecies plasmid DNA transfer is attenuated in vir-deficient C. fetus subsp. venerealis mutants.
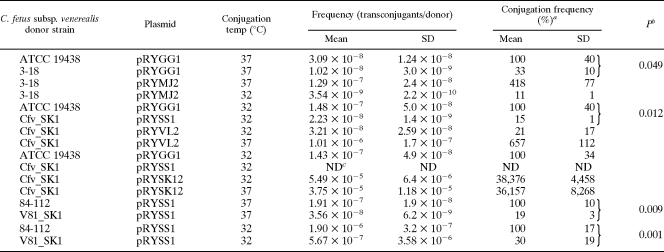
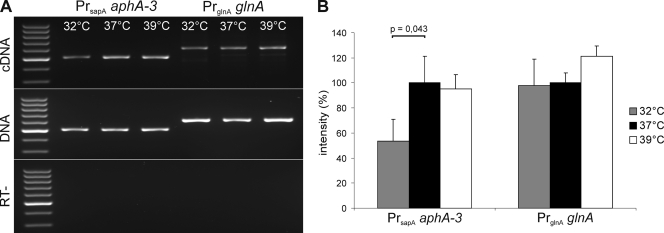
To establish a functional link between the T4SS and DNA transfer, mutants carrying gene disruptions in virB9 or virD4 were investigated. The mutant vir derivatives of C. fetus subsp. venerealis ATCC 19438 strains were shown in our earlier work to attenuate virulence during infection of cultured human cells (28). Plasmid transfer efficiencies were compared when the wild-type parental strains and mutants C. fetus subsp. venerealis 3-18 (ΔvirB9) or C. fetus subsp. venerealis SK1 (ΔvirD4) served as plasmid donors. Test plasmid pRYGG1 contained the same pIP1455 backbone elements as pRYSS1 but lacked the kanamycin expression cassette also used for insertion disruption of the vir genes in the mutant donors. To complement the mutant phenotypes in trans, the mobilizable (pRYGG1-based) vector carried also a wild-type copy of the affected vir gene(s). Each experiment was performed in triplicate with recipient E. coli MT102 at both 32 and 37°C. For technical reasons, each experiment compared a maximum of four donor strains. The results are summarized in Fig. 3 and Table 5. The plasmid transfer frequencies were expressed relative to the parental strain at one temperature (100%), as indicated. For the wild-type strains, the frequencies were higher at 32°C than at 37°C (as illustrated generally in Fig. 3A). In the absence of virB9, transfer of the plasmid from C. fetus subsp. venerealis was reduced significantly to one third of the wild-type activity (Fig. 3B and Table 5). The mobilizable complementation vector pRYMJ2 (33) provided in trans virB9 plus all downstream genes from the operon: virB10, virB11, and virD4, under the control of the sapA promoter. Interestingly, although a higher transfer efficiency was expected at 32°C, maximal complementation could only be achieved at 37°C. Similar results were obtained when the C. fetus subsp. venerealis host deficient in virD4 was used as a donor. Inactivation of virD4 reduced plasmid transfer significantly to 15% of the wild-type level. Again, complementation using the sapA promoter-dependent vector pRYVL2 was only successful at 37°C (Fig. 3C and Table 5). Since conjugation frequencies were generally higher at the lower temperature (Fig. 3A), we hypothesized that the failure of complementation at 32°C was most likely due to low expression levels from the complementation constructions. In the C. fetus chromosome, the sapA promoter controls synthesis of the surface layer proteins, which mediate serum resistance and antigenic variation in the mammalian host (64). It is conceivable that expression of this locus is temperature dependent. To confirm this, we isolated total RNA from C. fetus subsp. venerealis ATCC 19438 carrying plasmid pRYSS1 after cultivation at 32, 37, or 39°C. pRYSS1 carries aphA-3 under the control of the sapA promoter. RT-PCR was used to convert aphA-3 transcripts to cDNA for a quantitative comparison of sapA promoter activity as a function of temperature. For each preparation of RNA, cDNA was also generated from transcripts of the housekeeping gene glnA. Amplification of the initial yield of glnA cDNA templates resulted in equivalent amounts of product regardless of the cultivation temperature. In contrast the yield of aphA-3 product was consistently weaker from RNA obtained after 32°C growth than at the higher temperatures (Fig. 4). This significant difference implies that the sapA promoter is less active at 32°C.
TABLE 5.
virB9 and virD4 are required for efficient DNA mobilization by C. fetus subsp. venerealis
a Conjugation frequency is expressed as the mean percent relative to the corresponding wild-type strain at the indicated temperature.
b The statistical significance (P) between wild-type and vir mutant strains (unpaired t test) is given.
c ND, not detected.
FIG. 4.
Expression of aphA-3 controlled by PrsapA is significantly reduced at 32°C. (A) RNA of Campylobacter fetus subsp. venerealis ATCC 19438(pRYSS1) was isolated after cultivation at the temperatures shown. cDNA of aphA-3 and glnA was produced with reverse transcriptase (RT) from an equivalent portion of total RNA and amplified, and the products were analyzed on a 2% agarose TAE gel (upper panel). Plasmid DNA (aphA-3) and genomic DNA (glnA) templates served as positive controls for the amplification conditions (middle panel). Negative controls for DNA contamination (RT−) lacked reverse transcriptase (lower panel). (B) The intensities of products of amplified cDNAs were measured with ImageJ 1.43 (http://rsbweb.nih.gov/ij/), and the yield for each gene is expressed relative to the value obtained after cultivation at 37°C. Significantly lower expression of the aphA-3 marker controlled by PrsapA was measured at 32°C compared to 37°C growth, as indicated (unpaired t test). Standard deviations from three independent experiments at each growth temperature are shown.
To eliminate the observed PrsapA temperature dependence in complementation experiments, housekeeping genes were identified in the C. fetus subsp. fetus 82-40 genome sequence (GenBank accession number NC_008599) and used to amplify putative constitutively expressed promoter elements from the chromosome. The C. fetus gene gatC is predicted to encode an aspartyl/glutamyl-tRNA amidotransferase. We replaced the sapA promoter on pRYVL2 with the gatC promoter to generate the complementation vector pRYSK12. Gene transfer experiments using the virD4 mutant were then repeated. In this set of experiments (Fig. 3D), the mutant's ability to mobilize plasmid DNA was reduced below the detection limit at 32°C. The expression of wild-type virD4 under the control of the gatC promoter not only restored plasmid mobilization from this host at 37 and 32°C but actually led to >100-fold enhancement of transfer compared to the wild type. For our analysis of the virulence attributes of C. fetus subsp. venerealis, we additionally created a gene disruption in virD4 of a particularly virulent field isolate C. fetus subsp. venerealis 84-112 (28). Comparison of the DNA mobilization capacity of this mutant and the parent strain verified the importance of virD4 in gene transfer (Table 5). In summary, we conclude that transmission of plasmid DNA from C. fetus subsp. venerealis to E. coli involves at least two functional components of the T4SS: VirB9 and VirD4. The data also show that the sapA promoter was more active at 37°C than at 32°C. Replacement of this regulatory element with a more constitutively expressed promoter led to dramatically enhanced DNA translocation frequencies. This finding implies that the wild-type level of expression of the chromosomally encoded T4SS under these laboratory conditions is low. As a result, the level of secretion activity observed thus far is apparently well below the capacity of the system.
IncPα plasmid-mediated surface exclusion disrupts DNA delivery by the C. fetus subsp. venerealis T4SS.
Conjugation systems typically express one or more entry or surface exclusion functions, which minimize the likelihood that the host cell participates in mating with a second bacterium harboring the same or a closely related plasmid (25, 77). Restriction of lateral DNA transmission to a plasmid-carrying host due to surface exclusion is indicative, therefore, of functional relatedness between the competing transfer machineries. To functionally classify the C. fetus subsp. venerealis T4SS components, we tested whether the efficiency of plasmid mobility from a wild-type strain was measurably reduced by the established presence of alternative T4SS in the recipient population. Paradigm conjugation systems drawn from plasmid incompatibility groups Inc-P, -W, -I, and -F were introduced to the E. coli recipient strain. No variation in the frequency of plasmid transmission to E. coli MT102 was observed when the strain additionally harbored Inc-W, -I, or -F conjugative prototypes (Table 6). In contrast, transfer from C. fetus subsp. venerealis was substantially diminished when the recipient population carried the IncPα plasmid RP4. The observed exclusion index (EI) (∼32-fold) indicates that the C. fetus subsp. venerealis T4 pathway is functionally related to P-like conjugation proteins.
TABLE 6.
Exclusion index values for prototypic incompatibility groups in C. fetus subsp. venerealis conjugative mobilization
| E. coli recipient strain | Inc group | Frequency (transconjugants/donor) |
Conjugation frequency (%) |
Pa | EIb | ||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||||
| MT102 | None | 1.26 × 10−5 | 1.6 × 10−6 | 100 | 13 | ||
| MT102(RP4) | IncP | 3.96 × 10−7 | 4.2 × 10−8 | 3 | 0 | 0.006 | 31.7 |
| MT102(pSU2007) | IncW | 2.36 × 10−5 | 8.9 × 10−6 | 188 | 70 | 0.158 | 0.5 |
| MT102(pAR119) | IncF | 7.41 × 10−6 | 3.4 × 10−7 | 59 | 3 | 0.028 | 1.7 |
| MT102(pLG211) | IncI | 1.11 × 10−5 | 1.0 × 10−6 | 89 | 8 | 0.278 | 1.1 |
P values refer to the statistical significance distinguishing transfer frequencies to plasmid-free recipients and plasmid-carrying E. coli (unpaired t test).
The exclusion index (EI) expresses the fold difference in transfer frequency to plasmid-free recipients versus plasmid-carrying E. coli.
Intraspecies mobilization of plasmid DNA in C. fetus.
Determination of the C. fetus subsp. venerealis T4SS host range is cumbersome due to the lack of suitable shuttle vectors that can be maintained in both C. fetus donors and a diversity of potential hosts. At present, vectors adapted for E. coli-C. fetus applications have proven to be largely species specific among Campylobacters (33). Moreover, C. jejuni and C. coli are poor recipients due to the presence of closely related conjugative plasmids pTet and pCC31, respectively, and due to natural competence. Alternatively, the C. fetus subsp. venerealis cryptic plasmid pCFV108 that we have used for vector development carried a repE homologue most closely related to replication initiation proteins of enterococcal plasmids pS68, pEF47, and pAMα1 (33). Attempts to stably transform Enterococcus faecalis with shuttle vectors based on the pCFV108 repE gene with or without the replicative iterons in cis proved negative, however (K. Weaver, unpublished data). In the absence of more universally applicable plasmid substrates, the testable range of mobilization by the C. fetus subsp. venerealis T4SS was limited to intraspecies transfer. Selection for C. fetus subsp. fetus transconjugants was facilitated by the ciprofloxacin-resistant phenotype of the human blood isolate F12 (37). Triplicate experiments revealed that pRYSS1 was mobilized from C. fetus subsp. venerealis 84-112 to C. fetus subsp. fetus F12 with a frequency of 2.84 × 10−6 ± 5.02 × 10−7 in the absence of DNase I and 2.89 × 10−6 ± 4.02 × 10−7 with DNase I. C. fetus subsp. fetus strains in our collection (n = 43) lack detectable homologues to the C. fetus subsp. venerealis T4SS (28). Accordingly, a subsequent transmission of the plasmid from C. fetus subsp. fetus F12 transconjugants was not observed. Further experiments combining C. fetus subsp. fetus ATCC 27374(pRYGG1) or C. fetus subsp. fetus 82-40(pRYEL1) as potential plasmid donors with E. coli MT102 did not yield transconjugants. Finally, to assess whether DNA can be exchanged within the subspecies we inserted the resistance marker aphA-3 into the C. fetus subsp. venerealis cdtB gene (outside of the PAI) to obtain the selectable recipient strain JL1. In the subsequent mating experiments, either C. fetus subsp. venerealis ATCC 19438 harboring pRYGG1 or strain 84-112 harboring pRYEL1 served as donors. A strong exclusion phenotype was expected and, indeed, the obtained frequencies were lower than 3 × 10−9 transconjugants per donor. Nonetheless, plasmid uptake was confirmed in all colonies we tested. Taken together, these results demonstrate that C. fetus subsp. venerealis supports intra- and interspecies gene transfer via a mechanism that is independent of transformation, requires the T4SS, and is sensitive to the surface or entry exclusion functions of RP4, as well as C. fetus subsp. venerealis strains expressing an identical system.
Transcriptional organization of C. fetus subsp. venerealis PAI region harboring the fic genes.
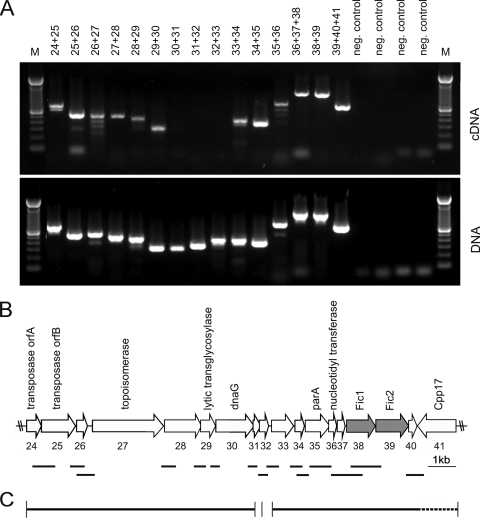
The genome region distal to the virB/virD4 module contains two ORFs carrying the FIC signature motif common to secreted effector proteins of several bacterial pathogens. We investigated the transcriptional organization of this genome region using RT-PCR. Primers were placed to detect readthrough of transcripts into the adjacent or further downstream genes. One large transcriptional unit spanning 7.5 kb was mapped from the transposase orfA, or gene 24 of the PAI sequence (28), and the putative primase gene dnaG (Fig. 5). No transcripts were detected for the small ORFs 31 and 32 depicted in Fig. 5. A second large transcript spanning 6 kb, including the fic1 and fic2 genes, was delineated. Transcription originating in this region is not effectively terminated, since readthrough into the downstream, oppositely oriented gene cpp17 was observed.
FIG. 5.
Transcriptional organization of C. fetus subsp. venerealis PAI region harboring the fic genes. (A) Gene organization of the region extending from transposase orfA to the putative nickase gene downstream of the fic genes was analyzed in strain ATCC 19438 by ORF-spanning amplification of cDNA produced with reverse transcriptase PCR (upper panel). Genomic DNA served as positive controls for each primer combination (lower panel). Numbers above the lanes indicate the ORFs common to the amplified cDNA according to the gene designations shown in panel B. Negative controls lacked reverse transcriptase (upper panel) or template (lower panel). (B) The 3′ region of the genomic island of C. fetus subsp. venerealis ATCC 19438 is illustrated. Gene numbers assigned previously (28) (below) and putative functional assignments are shown (above). fic genes are shaded gray. (C) Transcription map depicts two distinct mRNA fragments encoding ORFs 24 to 30 and ORFs 33 to 40, respectively (solid lines). ORFs 31 and 32 of unknown function are not transcribed with the other genes nor each other. Transcripts initiated within units 33 to 40 extend into the putative relaxase gene cpp17 in reverse orientation (dotted line). The separately amplified cDNA regions shown in panel A are indicated schematically (above).
Test for interbacterial protein translocation.
Currently, the most promising approach available to identify potential effector proteins recognized and secreted by T4SS is the Cre recombinase assay for translocation (CRAfT) developed by Vergunst et al. (67, 68). The assay enables translocation events to be detected via fusion of the candidate protein to the Cre recombinase. Cre alone is not transferable, but when fused to a true T4 substrate, it gains access to the T4 pathway via the T4CP. Target cells are engineered to indicator strains with a reporter gene flanked by loxP recombination sites. Uptake of the T4 substrate-Cre fusion protein by recipients is detected via recombination of the reporter cassette and the emergence of a heritable phenotypic difference.
As described above, the Fic1 and Fic2 proteins encoded by the C. fetus subsp. venerealis PAI are possible secretion substrates. We have shown that E. coli is a suitable recipient for C. fetus subsp. venerealis T4-mediated secretion of nucleoprotein substrates. It is conceivable, therefore, that if the function of the Campylobacter Fic proteins normally requires recognition by the T4CP, then those interactions should also enable the putative effectors to be transferred via this pathway to E. coli. To test this hypothesis, fusions of Fic1 and Fic2 to Cre recombinase were cloned in the C. fetus expression vector, pRYSK3, which cannot be mobilized by the T4SS. C. fetus subsp. venerealis strains ATCC 19438 and 84-112, each expressing Cre alone (pRYSK15), or the fusions Cre-Fic1 (pRYSK13), or Cre-Fic2 (pRYSK14) were combined with E. coli indicator strains under conditions that maximally supported DNA mobilization via the C. fetus T4SS. The E. coli recipient cells were plated onto chloramphenicol containing LB agar to detect recombinant progeny. The frequency of recombination was measurably higher in every experiment involving the Cre-Fic2 fusion protein compared to Cre alone (n = 4), but the difference was not statistically significant (data not shown). Drawing on our results with the T4-mediated plasmid transmission from C. fetus to E. coli, we varied the temperature, the species ratio, and overall cell density. Again, Cre-Fic2 expressing cells led to 2-fold-higher recombination frequencies in E. coli than in cultures expressing Cre alone. Despite these steps toward optimization, the low frequencies we observed were not sufficiently distinct from background levels to be statistically significant. We have used CRAfT to map the protein translocation signals present on the TraI protein recognized by the F-like T4SS in E. coli (39). In our experience with F-like systems, even under conditions that support very efficient nucleoprotein transfer (>1 transfer events per donor cell), the frequency of concomitant protein translocation leading to productive recombination is typically limited to 3 to 0.3% of the transconjugant cells. It follows that the frequency of DNA transfer we are currently able to obtain in C. fetus-E. coli mating (10−6 to 10−7 transconjugants/donor) is too low for the subsequent Cre-based recombination events to occur at readily detectable levels.
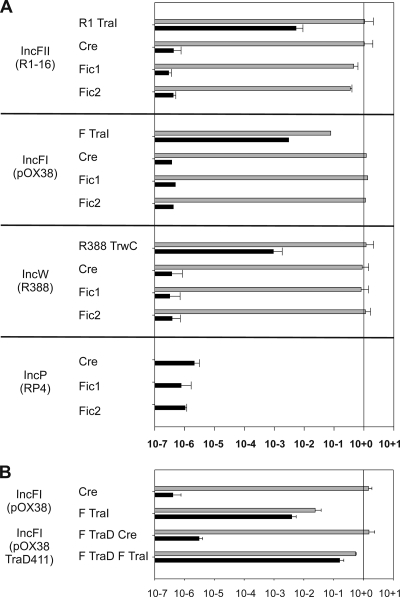
The C. fetus subsp. venerealis Fic proteins display conserved motifs shared by diverse T4S substrates, suggesting that heterologous T4SS might support productive interactions with the Fic proteins. To test this possibility, we chose the F and R1 conjugation systems since we have investigated protein translocation by these paradigms in detail (39). The T4CPs of the broad-host-range plasmids R388 and RP4 are more permissive for interactions with heterologous plasmids, and, importantly, RP4 is apparently most functionally related to the C. fetus components (Table 6). To provide positive controls for translocation of the Cre-protein fusions, relaxase genes were fused 3′ to cre in the CRAfT reporter vector CFP B (49). Protein translocation was measured indirectly by the frequency of Cre-catalyzed recombination in the recipient cell population. Simultaneous monitoring of plasmid transfer provided a measure for the secretion activity of the system in every experiment. A summary of the translocation data is shown in Fig. 6. High frequencies of DNA (self) transfer were detected for the F-like systems R1-16 (1/donor) and pOX38 (10−1/donor), as well as R388 (1/donor). By contrast, interbacterial transfer of the C. fetus Fic proteins was not detectable with any conjugation system. The high exclusion index mediated by IncP plasmid RP4 (Table 6) against the C. fetus transfer machinery implies functional relatedness for these systems. Nevertheless, the RP4 transfer proteins expressed by the S17 λ pir donor did not support detectable Fic protein transfer. The RP4 conjugation genes are integrated in the S17 λ pir chromosome, and thus an internal standard for conjugation proficiency is lacking. Given that we used this strain routinely for delivery of all shuttle vectors into C. fetus strains throughout the study, we propose that the negative results for Fic1 and Fic2 translocation indicate a true lack of productive interactions with the RP4 substrate receptor, TraG.
FIG. 6.
Efficient T4 secretion is sensitive to the relative concentration of T4CP and substrate protein. (A) Heterologous conjugation systems were tested for recognition and transfer of the C. fetus Fic proteins. The protein translocation frequencies (recombination events per donor) supported by R1-16, pOX38, R388, and RP4 conjugation systems are shown (black bars, right) for Cre alone or Cre fused to the full-length proteins indicated (left). DNA self-transfer frequencies for each plasmid system are indicated with gray bars. Standard deviations are shown. (B) DNA self-transfer of pOX38 is inhibited by cre-traI overexpression in trans (gray bars). By comparison, frequencies of gene and protein transfer are markedly increased when F traD and the cre-traI are both overexpressed in trans to traD-null derivative pOX38traD411.
Relative concentrations of a substrate protein and the T4CP alters secretion efficiency.
The C. fetus subsp. venerealis virB/virD4 genes are organized as an operon, and in an earlier study we demonstrated that the operon is expressed under laboratory conditions (28). The dramatic stimulation in gene transfer obtained when virB9 and the downstream genes virB10, virB11, and virD4—or virD4 alone—were expressed in trans from plasmid vectors on the virB9 and virD4 mutant backgrounds (Fig. 3 and Table 5) suggests that controlled gene regulation is limiting the translocation activity observed. Nonetheless, virD4 is cotranscribed with upstream virB genes (data not shown); thus, the intracellular concentration of this protein would be expected to be similar to the other coregulated T4 components. Regulation of the virB/virD4 operon promoter has not been studied. We sought to determine whether in trans expression of virD4 might be part of a positive-feedback loop for operon upregulation. Expression of virB genes upstream of virD4 and downstream PAI genes in C. fetus subsp. venerealis V81_SK1 was compared with or without the virD4 expression vector pRYSK12 in trans. No significant change in mRNA levels was apparent based on the yield of amplified cDNA (data not shown). A VirD4-dependent positive regulation of transporter components was therefore ruled out under these conditions.
Balanced intracellular concentrations of the T4CP VirD4 and secretion substrates may be important to transporter activity. The identity of the conjugative relaxase involved in DNA mobilization by C. fetus subsp. venerealis is not yet known. To evaluate the principle of how imbalanced expression of substrate and receptor genes might affect T4 secretion, we instead returned to the well-characterized F system. Overexpression of the F traI gene in trans to the wild-type conjugation genes causes a negative dominant phenotype for conjugation (30). That negative effect is also visible in our experiments where pOX38 transfer was reduced by 66-fold in the presence of a plasmid expressing Cre-TraIF compared to Cre alone (Fig. 6A). We tested the effect of simultaneous overexpression of both F traD and traI from plasmids in trans (Fig. 6B). Under these conditions, the efficiencies of conjugative DNA transfer increased by 22-fold, and protein translocation was raised 40-fold in comparison to traI expression alone. We conclude that in the F system the relative concentrations of the T4CP and substrate protein are indeed decisive for efficient transfer. C. fetus gene transfer was markedly increased when virD4 was expressed in trans from plasmid vectors (Fig. 3 and Table 5). Based on these observations, we propose that a lack of coordinate regulation of the conjugative relaxase with the virB/virD4 components under our laboratory conditions may lead to the low levels of conjugative transfer observed with this organism thus far.
DISCUSSION
This study demonstrates that the C. fetus subsp. venerealis VirB/VirD4 T4SS supports inter- and intraspecies mobilization of DNA. The frequencies of plasmid transfer observed under these conditions were low. The VirB/VirD4 T4SS is also necessary for C. fetus subsp. venerealis to efficiently invade and induce cytolethal effects in cultured human epithelial cells in vitro (28). The question then arises whether the capacity to translocate DNA represents an important contribution to the fitness and pathogenicity of C. fetus or whether the (remaining) capacity for gene transfer observed here represents a vestige of the system's evolutionary origin as a conjugation system. In any case, this report is the first description of conjugative plasmid transfer in C. fetus.
We currently know little about regulation of the vir genes. Conjugation was measured over the temperature range from 25 to 39°C with a modest (5-fold) gain in the activity maximum observed at 32°C. Whether this reflects a temperature dependence in vir gene regulation or the activity optima of the proteins involved is unknown. Controlled synthesis of conjugative pili on the bacterial cell surface probably alters motility and would be expected to initiate formation of stabilizing multicellular structures such as microcolonies and biofilm similarly to many Gram-negative bacteria (6, 26, 46, 51, 71). Thus, the elaboration of pili may provide an advantage in the successful colonization of the bovine genital tract also in the absence of macromolecular transport. Given this diversity of potential functions, the virB/virD4 gene region is probably subject to complex patterns of gene regulation that respond to both environmental and host-dependent cues.
Current knowledge of genome composition and dynamics for this pathogen is rudimentary, but laterally acquired genes are predicted to contribute to the distinct niche preferences exhibited by this taxon (2, 34). Experimental evidence for natural transformation is still lacking (8); thus, knowledge of gene mobilization via conjugative mechanisms is important in analyzing the emerging genome sequences of the C. fetus subspecies. Moreover, knowledge that lateral gene transfer occurs between the C. fetus subspecies has significant implications for the design of genotyping schemes that reliably differentiate the subspecies (1). C. fetus subsp. venerealis is the causative agent of a contagious venereal disease causing severe reproductive problems in cattle. Bovine genital campylobacteriosis is a notifiable disease of the World Organization for Animal Health (OIE) and is a substantial burden for the international trade of animals and animal products. The microbiological and molecular differentiation of C. fetus subsp. venerealis from C. fetus subsp. fetus is extremely difficult (72). A single phenotypic test is recommended by the OIE for typing the subspecies (66); thus, international efforts currently focus on optimizing genotypic tests. The present study demonstrated that genes are exchanged between the C. fetus subspecies via conjugation. Clearly, knowledge of lateral DNA transfer should factor into the selection of diagnostic targets.
The FIC domain-containing proteins of C. fetus are potential effector proteins translocated by the T4 machinery to mammalian cells. Transmission of bacterial Fic proteins to the eukaryotic cytosol regulates host processes important to pathogen survival and replication. AnkX protein of Legionella pneumophila alters the microtubule-dependent transport of vesicles (47). The FIC domain of VopS covalently modifies Rho GTPase threonine with AMP disrupting downstream signal transduction in the host cell (76). The AMPylation domain is shared by doc toxins, FIC, and the type III effector AvrB (35). IbpA protein of Histophilus somni adenylylates and inactivates Rho GTPases of epithelial target cells ultimately leading to collapse of the cellular cytoskeleton (74, 78). The VirB/VirD4-translocated substrates of Bartonella henselae, BepA-BepG, are required for invasion, proinflammatory activation and antiapoptotic protection of vascular endothelial cells (56, 60). BepA, BepB, and BepC share an FIC domain. In light of the genetic organization of the virB/virD4/bep PAI of Bartonella (57), the distal proximity of the fic1 and fic2 genes to the C. fetus vir operon supports the hypothesis that these proteins are delivered as effector molecules to target cells. Experimental approaches to demonstrate translocation of a putative bacterial secretion substrate to a recipient cell have been vastly improved by molecular reporter systems based on Cre recombinase or the calmodulin-dependent adenylate cyclase (Cya) (45, 56, 62). Application of the Cre recombinase reporter assay to the Fic proteins of C. fetus was chosen to evaluate whether the cognate or heterologous systems would support detectable levels of secretion to bacterial target cells. The current results neither support nor rule out the potential for Fic proteins to perform relevant pathogenic functions in animal cell infection. Our efforts now focus on applications of the Cya reporter in cultured human cells invaded by C. fetus.
Acknowledgments
This study was financed by FWF grants P20479-B05 and P18607-B12 and a Hygiene Fund Young Scientist grant of the Medical University of Graz (to S.K.).
We thank K. Weaver for testing plasmid replication in E. faecalis, M. Llosa and R. J. Meyer for providing plasmids, and B. Munk for his contribution to this study.
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1.Abril, C., I. Brodard, and V. Perreten. 2010. Two novel antibiotic resistance genes, tet(44) and ant(6)-Ib, are located within a transferable pathogenicity island in Campylobacter fetus subsp. fetus. Antimicrob. Agents Chemother. 54:3052-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abril, C., et al. 2007. Discovery of insertion element ISCfe1: a new tool for Campylobacter fetus subspecies differentiation. Clin. Microbiol. Infect. 13:993-1000. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Martinez, C. E., and P. J. Christie. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen, J. B., et al. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9:207-217. [DOI] [PubMed] [Google Scholar]
- 6.Barrios, A. F., R. Zuo, D. Ren, and T. K. Wood. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188-200. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, M. J. 1998. Campylobacter fetus-emerging infection and model system for bacterial pathogenesis at mucosal surfaces. Clin. Infect. Dis. 27:256-258. [DOI] [PubMed] [Google Scholar]
- 8.Blaser, M. J., D. G. Newell, S. A. Thompson, and E. L. Zechner. 2008. Pathogenesis of Campylobacter fetus infections, p. 401-428. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, D.C.
- 9.Boschiroli, M. L., et al. 2002. Type IV secretion and Brucella virulence. Vet. Microbiol. 90:341-348. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan-Wollaston, V., J. E. Passiatore, and F. Cannon. 1987. The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature 328:172-175. [Google Scholar]
- 11.Cambronne, E. D., and C. R. Roy. 2006. Recognition and delivery of effector proteins into eukaryotic cells by bacterial secretion systems. Traffic 7:929-939. [DOI] [PubMed] [Google Scholar]
- 12.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandler, M., and D. J. Galas. 1983. Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J. Mol. Biol. 170:61-91. [DOI] [PubMed] [Google Scholar]
- 14.Chatfield, L. K., E. Orr, G. J. Boulnois, and B. M. Wilkins. 1982. DNA primase of plasmid ColIb is involved in conjugal DNA synthesis in donor and recipient bacteria. J. Bacteriol. 152:1188-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, I., P. J. Christie, and D. Dubnau. 2005. The ins and outs of DNA transfer in bacteria. Science 310:1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 19.Ding, Z., K. Atmakuri, and P. J. Christie. 2003. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 11:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Lopez, R., et al. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30:942-966. [DOI] [PubMed] [Google Scholar]
- 21.Fischer, W., R. Haas, and S. Odenbreit. 2002. Type IV secretion systems in pathogenic bacteria. Int. J. Med. Microbiol. 292:159-168. [DOI] [PubMed] [Google Scholar]
- 22.Fischer, W., et al. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 23.Franco, I. S., H. A. Shuman, and X. Charpentier. 2009. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 11:1435-1443. [DOI] [PubMed] [Google Scholar]
- 24.Frank, A. C., C. M. Alsmark, M. Thollesson, and S. G. Andersson. 2005. Functional divergence and horizontal transfer of type IV secretion systems. Mol. Biol. Evol. 22:1325-1336. [DOI] [PubMed] [Google Scholar]
- 25.Garcillan-Barcia, M. P., and F. de la Cruz. 2008. Why is entry exclusion an essential feature of conjugative plasmids? Plasmid 60:1-18. [DOI] [PubMed] [Google Scholar]
- 26.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 27.Goebel, W., W. Lindenmaier, H. Schrempf, R. Kollek, and D. Blohm. 1977. Dissociation and recombination of fragments with defined functions of the antibiotic resistance factor R1, p. 261-275. In J. Drews and G. Högenauer (ed.), Topics in infectious diseases, vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 28.Gorkiewicz, G., et al. 2010. A genomic island defines subspecies-specific virulence features of the host-adapted pathogen Campylobacter fetus subsp. venerealis. J. Bacteriol. 192:502-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandoso, G., M. Llosa, J. C. Zabala, and F. de la Cruz. 1994. Purification and biochemical characterization of TrwC, the helicase involved in plasmid R388 conjugal DNA transfer. Eur. J. Biochem. 226:403-412. [DOI] [PubMed] [Google Scholar]
- 30.Haft, R. J., et al. 2006. General mutagenesis of F plasmid TraI reveals its role in conjugative regulation. J. Bacteriol. 188:6346-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu, L., and D. J. Kopecko. 2000. Interactions of Campylobacter with eukaryotic cells: gut luminal colonization and mucosal invasion mechanisms, p. 191-215. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 32.Juhas, M., D. W. Crook, and D. W. Hood. 2008. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 10:2377-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kienesberger, S., et al. 2007. Development of experimental genetic tools for Campylobacter fetus. Appl. Environ. Microbiol. 73:4619-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kienesberger, S., G. Gorkiewicz, H. Wolinski, and E. L. Zechner. 2010. New molecular microbiology approaches in the study of Campylobacter fetus. Microb. Biotechnol., in press. doi: 10.1111/j.1751-7915.2010.00173.x. [DOI] [PMC free article] [PubMed]
- 35.Kinch, L. N., M. L. Yarbrough, K. Orth, and N. V. Grishin. 2009. Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One 4:e5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komano, T., R. Utsumi, and M. Kawamukai. 1991. Functional analysis of the fic gene involved in regulation of cell division. Res. Microbiol. 142:269-277. [DOI] [PubMed] [Google Scholar]
- 37.Krause, R., et al. 2002. Recurrent septicemia due to Campylobacter fetus and Campylobacter lari in an immunocompetent patient. Infection 30:171-174. [DOI] [PubMed] [Google Scholar]
- 38.Lambert, T., G. Gerbaud, P. Trieu-Cuot, and P. Courvalin. 1985. Structural relationship between the genes encoding 3′-aminoglycoside phosphotransferases in Campylobacter and in gram-positive cocci. Ann. Inst. Pasteur Microbiol. 136B:135-150. [DOI] [PubMed] [Google Scholar]
- 39.Lang, S., et al. 2010. Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol. Microbiol., in press. [DOI] [PubMed]
- 40.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 41.Lin, T. S., and C. I. Kado. 1993. The virD4 gene is required for virulence while virD3 and orf5 are not required for virulence of Agrobacterium tumefaciens. Mol. Microbiol. 9:803-812. [DOI] [PubMed] [Google Scholar]
- 42.Lu, J., et al. 2008. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol. 70:89-99. [DOI] [PubMed] [Google Scholar]
- 43.Maneewannakul, K., et al. 1996. Construction of derivatives of the F plasmid pOX-tra715: characterization of traY and traD mutants that can be complemented in trans. Mol. Microbiol. 22:197-205. [DOI] [PubMed] [Google Scholar]
- 44.Martinez, E., and F. de la Cruz. 1988. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol. Gen. Genet. 211:320-325. [DOI] [PubMed] [Google Scholar]
- 45.Nagai, H., et al. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. U. S. A. 102:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 47.Pan, X., A. Luhrmann, A. Satoh, M. A. Laskowski-Arce, and C. R. Roy. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pansegrau, W., et al. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids: compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 49.Parker, C., and R. J. Meyer. 2007. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. Mol. Microbiol. 66:252-261. [DOI] [PubMed] [Google Scholar]
- 50.Perez-Perez, G. I., M. J. Blaser, and J. H. Bryner. 1986. Lipopolysaccharide structures of Campylobacter fetus are related to heat-stable serogroups. Infect. Immun. 51:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 52.Reisner, A., S. Molin, and E. L. Zechner. 2002. Recombinogenic engineering of conjugative plasmids with fluorescent marker cassettes. FEMS Microbiol. Ecol. 42:251-259. [DOI] [PubMed] [Google Scholar]
- 53.Roy, C. R., and S. Mukherjee. 2009. Bacterial FIC proteins AMP up infection. Sci. Signal 2:pe14. [DOI] [PubMed] [Google Scholar]
- 54.Salgado Pabon, W., et al. 2010. Increased expression of the type IV secretion system in piliated Neisseria gonorrhoeae variants. J. Bacteriol. 192:1912-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sastre, J. I., E. Cabezon, and F. de la Cruz. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 180:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmid, M. C., et al. 2006. A translocated bacterial protein protects vascular endothelial cells from apoptosis. PLoS Pathog. 2:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schroder, G., and C. Dehio. 2005. Virulence-associated type IV secretion systems of Bartonella. Trends Microbiol. 13:336-342. [DOI] [PubMed] [Google Scholar]
- 58.Schroder, G., and E. Lanka. 2005. The mating pair formation system of conjugative plasmids: a versatile secretion machinery for transfer of proteins and DNA. Plasmid 54:1-25. [DOI] [PubMed] [Google Scholar]
- 59.Schulein, R., and C. Dehio. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46:1053-1067. [DOI] [PubMed] [Google Scholar]
- 60.Schulein, R., et al. 2005. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. U. S. A. 102:856-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infections, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 62.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 63.Thompson, S. A., and M. J. Blaser. 2000. Pathogenesis of Campylobacter fetus infections, p. 321-347. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 64.Tummuru, M. K., and M. J. Blaser. 1993. Rearrangement of sapA homologs with conserved and variable regions in Campylobacter fetus. Proc. Natl. Acad. Sci. U. S. A. 90:7265-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Bergen, M. A., et al. 2005. Clonal nature of Campylobacter fetus as defined by multilocus sequence typing. J. Clin. Microbiol. 43:5888-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Bergen, M. A., et al. 2005. Amplified fragment length polymorphism based identification of genetic markers and novel PCR assay for differentiation of Campylobacter fetus subspecies. J. Med. Microbiol. 54:1217-1224. [DOI] [PubMed] [Google Scholar]
- 67.Vergunst, A. C., et al. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 68.Vergunst, A. C., et al. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. U. S. A. 102:832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 70.Voth, D. E., and R. A. Heinzen. 2009. Coxiella type IV secretion and cellular microbiology. Curr. Opin. Microbiol. 12:74-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willoughby, K., et al. 2005. A multiplex polymerase chain reaction to detect and differentiate Campylobacter fetus subspecies fetus and Campylobacter fetus subspecies venerealis: use on UK isolates of C. fetus and other Campylobacter spp. J. Appl. Microbiol. 99:758-766. [DOI] [PubMed] [Google Scholar]
- 73.Woodcock, D. M., et al. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Worby, C. A., et al. 2009. The fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34:93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 76.Yarbrough, M. L., et al. 2009. AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323:269-272. [DOI] [PubMed] [Google Scholar]
- 77.Zechner, E. L., et al. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, Netherlands.
- 78.Zekarias, B., et al. Histophilus somni IbpA DR2/Fic in virulence and immunoprotection at the natural host alveolar epithelial barrier. Infect. Immun. 78:1850-1858. [DOI] [PMC free article] [PubMed]