Abstract
In Escherichia coli, RpoS, the general stress response sigma factor, regulates the activity of the specialized DNA polymerase DNA polymerase IV (Pol IV) both in stationary-phase and in exponential-phase cells. Because during exponential phase dinB, the gene encoding Pol IV, is transcribed independently of RpoS, RpoS must regulate Pol IV activity in growing cells indirectly via one or more intermediate factors. The results presented here show that one of these intermediate factors is SbcCD, an SMC-like protein and an ATP-dependent nuclease. By initiating or participating in double-strand break repair, SbcCD may provide DNA substrates for Pol IV polymerase activity.
In Escherichia coli and its relatives, DNA damage results in the induction of about 40 genes as part of the SOS response. Three of these genes encode specialized DNA polymerases: DNA polymerase II (Pol II; encoded by the polB gene), DNA Pol IV (encoded by the dinB gene), and DNA Pol V (encoded by the umuDC genes) (11, 35). Pol IV and Pol V are members of the Y family of DNA polymerases that are widely distributed in all domains of life. Y family polymerases can replicate damaged DNA, a process called translesion synthesis, but they replicate undamaged DNA with fidelities that are typically orders of magnitude lower than those achieved by replicative DNA polymerases (20, 48).
Pol V's cellular role is to aid in the bypass of DNA lesions that block replication, and it is highly proficient at replicating past a variety of such lesions (reviewed in reference 20). In contrast, the role of Pol IV is far less clear, since the number of DNA lesions that it can bypass is limited. Pol IV is particularly efficient at replicating past certain N2-deoxyguanosine adducts, including those produced by 4-nitroquiniline-1-oxide (4-NQO) and nitrofurazone (NFZ); consequently, resistance to these agents serves as an assay for Pol IV's lesion bypass activity (34). Pol IV is responsible for 50 to 80% of the Lac+ revertants, called adaptive mutations (4), that occur over several days when stationary-phase cells of the Lac− E. coli strain FC40 are incubated on lactose medium (17, 50). Because of this strong phenotype, adaptive mutation in FC40 is often used as an assay for Pol IV's mutagenic activity (e.g., see reference 23). Several factors existent in stationary-phase cells contribute to this high rate of Pol IV-dependent adaptive mutation: (i) transcription of the dinB gene is induced approximately 3-fold under the control of the stationary-phase sigma factor RpoS (42, 63), (ii) the Pol IV protein is stabilized by the chaperone GroEL (43), (iii) Pol IV activity is enhanced by cellular polyphosphate (65), and (iv) proposed inhibitors of Pol IV activity, such as UmuD, may be less active or abundant in stationary-phase cells (23). This growth phase regulation suggests that Pol IV's mutagenic activity may serve an important function during nutrient-limited conditions. In support of this hypothesis, after long-term culture, strains lacking Pol IV are poor competitors in mixed cultures with wild-type cells of E. coli (15, 77).
Pol IV and Pol V also differ in their degree of regulation in growing cells. As would be expected for an error-prone polymerase, the levels and activity of Pol V are tightly regulated to prevent unwanted mutagenic activity; indeed, in the absence of DNA damage, there is virtually no Pol V in the cell (53). In contrast, in normally growing cells there are about 250 molecules of Pol IV (36), a relatively high number compared to the 10 to 20 molecules of the replicase DNA Pol III (76). And yet, loss of Pol IV has little effect on mutation rates in growing cells, which means that Pol IV contributes little to growth-dependent spontaneous mutations that occur on the E. coli chromosome (40, 64, 75). However, overproduction of Pol IV increases the spontaneous mutation rates in a dose-dependent manner. For example, the presence of a copy of the dinB gene on the F′ episome in addition to the copy on the chromosome results in 4-fold more Pol IV and a 2- to 3-fold increase in mutation frequencies (22, 36). The presence of the dinB gene on a multicopy plasmid results in 10- to 20-fold more Pol IV (36, 73) and, depending on the mutational target, 5- to 200-fold increases in mutation frequencies (37, 39, 63, 65, 70, 73, 75). These observations strongly suggest that the mutagenic activity of Pol IV normally is tightly regulated in growing cells but that even a modest increase in abundance allows Pol IV to, at least partially, escape this regulation.
As mentioned above, independently of its regulation as part of the SOS response, Pol IV is regulated by RpoS (also referred to as σSand σ38), the stationary-phase and general stress response sigma factor (42). RpoS regulates over 100 genes during stationary phase and up to 500 genes in response to various other stresses (54, 68, 71). In addition, RpoS has been found by transcription microarray analysis to regulate directly or indirectly almost 300 genes in exponential-phase cells (14a). Recently, we found that RpoS drives the transcription of dinB in stationary-phase cells but not in exponential-phase cells; and yet, RpoS still affects Pol IV activity in exponential phase (63). In exponentially growing cells, overexpression of Pol IV from an RpoS-independent promoter increases the growth-dependent mutation rate 10-fold, but in cells lacking RpoS, this increase is only 4-fold, even though the amount of Pol IV is unchanged (63). Additionally, 4-NQO is more toxic to an rpoS mutant strain than to a wild-type strain even when Pol IV is overexpressed (63). These results indicate that during normal growth, RpoS affects both the mutagenic and the translesion synthesis activities of Pol IV via an indirect pathway.
The SbcCD complex is a nuclease with ATP-dependent double-stranded DNA exonuclease activity and ATP-independent single-stranded DNA endonuclease activity. SbcD is the nuclease subunit, and SbcC is the ATPase subunit (10). The genes encoding the two proteins are in an operon, sbcDC, and were originally identified as cosuppressors with sbcB of the recombination deficiency of recBC mutant strains (45). These results are commonly interpreted to mean that in the absence of RecBCD (exonuclease V), the elimination of both SbcB (exonuclease I) and SbcCD allows recombination substrates to be processed via the RecF recombination pathway (10).
SbcCD has a particular affinity for DNA hairpins and may be important in removing these replication-blocking structures to allow replication to be restarted by recombination (9, 25). SbcC is also a member of the structural maintenance of chromosomes (SMC) family of proteins, with strong structural homology to the Mre11/Rad50 complex involved in the repair of double-strand breaks (DSBs) in eukaryotes. The SbcCD protein complex may help to maintain chromosome structure during DNA repair, particularly during the recombination-dependent repair of DSBs that produces two double-strand ends (12, 25). Thus, SbcCD plays a positive role in both DNA repair and replication (9, 44).
We decided to investigate the role of SbcCD in Pol IV-dependent, growth-dependent spontaneous mutation for two reasons: (i) in nutrient-limited and stationary-phase cells, transcription of the sbcDC operon is under partial control of RpoS (13), and (ii) recombination plays an important role in the generation of Pol IV-dependent adaptive mutations in stationary-phase cells (4, 16, 30). However, we found, as did others, that SbcCD plays little, if any, role in Pol IV-dependent adaptive mutation (57; our unpublished results). In contrast, the results presented here show that SbcCD, RpoS, and RecB function in one genetic pathway that affects Pol IV-dependent spontaneous mutation in growing cells.
MATERIALS AND METHODS
Bacterial strains and media.
All strains used in this study are E. coli K-12 derivatives and are listed in Table 1. Genetic manipulations were performed as previously described (51). Cultures were grown in Luria-Bertani (LB) liquid medium (51); agar medium contained 15 g/liter Bacto agar (LB); LB broth top agar contained 8 g/liter Bacto agar. When required, antibiotics were added at the following concentrations, in μg/ml: carbenicillin (Cb), 100; kanamycin (Kn), 30; chloramphenicol (Cm), 10; and tetracycline (Tc), 20.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotype | Reference |
|---|---|---|
| E. coli strains | ||
| FC36 | F−ara Δ(lac-proB)XIIIthi Rifr | 4 |
| FC40 | FC36/F′ Φ(lacI33-lacZ) Pro+ | 4 |
| FC722 | FC40 with a Tcs allele on the episome | 16 |
| FC1418 | FC40 lexA1 [LexA(Ind−)] | 42 |
| FC1419 | FC40 suA11 lexA71::Tn5 [LexA(Def)] | 42 |
| MC4100 | Δ(argF-lac)U169 rpsL150 relA1 araD139 flbB5301 deoC1 ptsF25 | 24 |
| GJ2770 | MC4100 [λkatE::lacZ (Kn)] | 56 |
| PFB236 | FC36 ΔdinB::Zeo | 73 |
| PFB665 | PFB236/F′ ΔdinB::Zeo with Tcs on the episome | 73 |
| PFG266 | FC722/pPFG96 | 65 |
| PFG419 | FC722 rpoS::Cm | This study |
| PFG420 | PFG419/pPFG96 | This study |
| PFG526 | GJ2770/pPFG96 | This study |
| PFG527 | GJ2770/pBAD24 | This study |
| PFG916 | FC722 ΔsbcD | This study |
| PFG947 | FC722 ΔsbcDC | This study |
| PFG948 | FC722 ΔsbcC | This study |
| PFG955 | PFG916/pPFG96 | This study |
| PFG956 | PFG948/pPFG96 | This study |
| PFG957 | PFG947/pPFG96 | This study |
| PFG988 | PFG419 ΔsbcDC | This study |
| PGF989 | PFG419 ΔsbcC/pPFG96 | This study |
| PFG990 | PFG419 ΔsbcD/pPFG96 | This study |
| PFG991 | PFG988/pPFG96 | This study |
| PFG1022 | FC722 ΔrecB::Kn | This study |
| PFG1024 | FC722 ΔrecB::Kn ΔsbcDC | This study |
| PFG1120 | PFG1022/pPFG96 | This study |
| PFG1121 | FC722 ΔrecB::Kn ΔsbcD/pPFG96 | This study |
| PFG1122 | PFG1024/pPFG96 | This study |
| PFG1123 | FC722 ΔrecB::Kn ΔsbcC/pPFG96 | This study |
| PFG1137 | FC722 ΔrecB::Kn rpoS::Cm | |
| PFG1138 | PFG1137/pPFG96 | This study |
| PFG1143 | FC722 ΔrecB::Kn rpoS::Cm ΔsbcDC/pPFG96 | This study |
| PFG1146 | PFB236 ΔrecB::Kn/F′ ΔdinB::Zeo with Tcs on the episome | This study |
| PFG1156 | FC722 ΔrecB::Kn rpoS::Cm ΔsbcDC | This study |
| Plasmids | ||
| pPFG96 | dinB cloned into pBAD24 | 65 |
| pBAD24 | Plasmid with pBR322 origin, araBAD promoter, and araC gene | 27 |
Plasmid pPFG96 (65) carries dinB cloned under the control of the araBAD promoter in pBAD24 (27); in strains carrying this plasmid, the levels of Pol IV are increased about 20-fold without arabinose induction (73). pPFG96 was transformed into the appropriate strains using the protocol previously described (6), selecting for Cbr. ΔsbcC::Kn, ΔsbcD::Kn, and ΔrecB::Kn mutant strains were constructed by P1vir transduction from the Keio collection of mutant E. coli strains (2), selecting for Knr. The ΔsbcDC::Kn double mutant strain was made using techniques described previously (14); the primer sequences were the same as the forward primer used to delete sbcD and the reverse primer used to delete sbcC in the Keio strains (2). The ΔsbcDC::Kn allele was then transduced into strains appropriate for this study. When required, the Kn resistance markers were removed from the genome as described previously (14). In all cases, ΔrecB::Kn was the last mutant allele added during strain construction.
Determination of growth rates.
Cultures of each strain were grown overnight to saturation in LB or LB plus Cb and then diluted 1:100 into LB or LB plus Cb and allowed to grow with shaking at 37°C. The optical density at 600 nm (OD600) of 100 μl of each culture was measured every 30 min until the cultures reached stationary phase. Growth rates were determined from the linear portion of the curve of ln(OD600) versus time in hours. Table 2 shows the growth rates calculated by combining these ln(OD600) values from two to four replicate cultures and determining the slope of the least-squares line and the standard error (SE) of the slope using the Excel function “Linest.”
TABLE 2.
Growth rates of strains with and without the Pol IV overexpression plasmid
| Strain | Relevant genotypea | Growth rate (h−1) | SEb |
|---|---|---|---|
| FC722 | Wild type | 1.61 | 0.04 |
| PFG266 | Wild type/pdinB++ | 1.64 | 0.11 |
| PFG419 | rpoS::Cm | 1.30 | 0.09 |
| PFG420 | rpoS::Cm/pdinB++ | 1.35 | 0.09 |
| PFG947 | Δ(sbcDC) | 1.65 | 0.04 |
| PFG957 | Δ(sbcDC)/pdinB++ | 1.66 | 0.05 |
| PFG1022 | Δ(recB::Kn) | 1.46 | 0.04 |
| PFG1120 | Δ(recB::Kn)/pdinB++ | 1.62 | 0.05 |
| PFG988 | rpoS::Cm Δ(sbcDC) | 1.57 | 0.06 |
| PFG991 | rpoS::Cm Δ(sbcDC)/pdinB++ | 1.68 | 0.07 |
| PFG1024 | Δ(recB::Kn) Δ(sbcDC) | 1.36 | 0.05 |
| PFG1122 | Δ(recB::Kn) Δ(sbcDC)/pdinB++ | 0.92 | 0.06 |
| PFG1137 | Δ(recB::Kn) rpoS::Cm | 1.18 | 0.07 |
| PFG1138 | Δ(recB::Kn) rpoS::Cm/pdinB++ | 1.03 | 0.06 |
| PFG1156 | Δ(recB::Kn) rpoS::Cm Δ(sbcDC) | 1.12 | 0.05 |
| PFG1143 | Δ(recB::Kn) rpoS::Cm Δ(sbcDC)/pdinB++ | 0.87 | 0.04 |
The designation “++” indicates that the gene is present in multiple copies.
SE, standard error of the slope of the least-squared line of ln(OD600) versus hours.
Determination of growth-dependent mutation rates.
Growth-dependent mutation rates were determined by fluctuation tests (18, 46). Cultures of each strain were grown overnight to saturation in LB plus appropriate antibiotics and then diluted 105-fold into the same medium. For the experiments whose results are shown in Fig. 2 and 3 below, 100-μl aliquots were distributed into 45 wells of a 96-well microtiter plate and incubated for 48 h at 37°C. Then, either 10 μl was diluted into 90 μl of saline (results are shown in Fig. 2) or the entire culture (results are shown in Fig. 3) was plated in LB top agar with Tc on LB agar-plus-Tc plates. For the remaining experiments, after the saturated cultures were diluted 10−5-fold, 1-ml aliquots were distributed into 30 to 40 culture tubes and incubated with shaking for 24 h at 37°C. Then, either 100 μl (results are shown in Fig. 6, 7A, 8A, and, for the rpoS sbcDC double mutant strain, Fig. 9) or 1 ml (results are shown in Fig. 7B, 8B, and for the rest of the strains, in Fig. 9) was plated onto LB agar plus Tc. Only colonies appearing on the LB-plus-Tc plates after 48 h were counted. The number of cells plated on the LB-plus-Tc plates was determined by plating appropriate dilutions from nine cultures of each strain on LB agar. The mean numbers of mutations per culture and their confidence limits were obtained with the Ma-Sandri-Sarkar (MSS) maximum-likelihood method (60) implemented with the FALCOR web tool found at http://www.mitochondria.org/protocols/FALCOR.html (28), corrected, when required, for plating only 1/10th of the culture. These values were divided by twice the total number of cells per culture to obtain the mutation rates per cell per generation and their confidence limits (18).
Western blots.
Western blotting was performed essentially as previously described (1). To measure the level of Pol IV protein in exponentially growing cells, strains were grown with shaking to an OD of ≈0.2 in LB plus appropriate antibiotics. One ml of each culture was centrifuged, and the pelleted cells washed in 1× M9 salts (51), resuspended in 1× SDS-PAGE sample loading buffer without bromophenol blue, and boiled for 15 min. The total amounts of protein in the samples were determined by Bradford assay (Bio-Rad Laboratories). An aliquot of each sample containing the desired amount of total cell protein (see the figure legends) was diluted into 1× SDS-PAGE sample loading buffer containing bromophenol blue and loaded onto a 12% SDS polyacrylamide gel. Proteins were separated by electrophoresis and then electrotransferred to an Immobilon-P membrane (pore size, 0.45 μm; Millipore Corp.). The membrane was probed with rabbit anti-Pol IV polyclonal antibody [obtained from H. Ohmori and clarified using acetone powder made from a ΔdinB strain as described in reference 29] and then with alkaline phosphatase-conjugated goat anti-rabbit antibody (Promega, Inc.) (results are shown in Fig. 6) or horseradish peroxidase-conjugated goat anti-rabbit antibody (Pierce-Thermo Scientific) (results are shown in Fig. 4 and 8). The bands were visualized using Western-Light chemiluminescent reagent (Applied Biosystems) (results are shown in Fig. 6) or Pierce ECL Western blotting substrate (Pierce-Thermo Scientific) (results are shown in Fig. 4 and 8). Band intensities were quantified using ImageJ software version 1.36b (58).
To detect the levels of RecA protein, cultures were grown with shaking to an OD600 of ≈0.3 in LB or in M9-glycerol minimal medium (51). Sample preparation, electrophoresis, and electrotransfer were as described above. RecA protein (obtained from New England Biolabs, Inc.) was included as a control. The membrane was probed with mouse anti-RecA monoclonal antibody (obtained from Stressgen Biotechnologies Corp.) and then with alkaline phosphate-conjugated rabbit anti-mouse antibody (Promega, Inc.) The bands were visualized using Western-Light chemiluminescent reagent (Applied Biosystems).
β-Galactosidase assays for katE expression.
Strain PFG526, which has a chromosomal katE::lacZ transcription fusion and carries plasmid pPFG96, and strain PFG527, the same strain but carrying the control plasmid pBAD24, were grown to saturation in LB plus antibiotics and then diluted 1:1,000 in the same medium and incubated with shaking at 37°C. Samples of each strain were taken at three points during growth, and the β-galactosidase activity was measured as previously described (51).
Assays for sensitivity to 4-NQO.
Cultures of each strain were grown overnight in LB plus appropriate drugs. Each culture was successively diluted 10-fold from 10−1 to 10−6, and 0.01 ml of each dilution was spotted on both an LB plate and an LB plate supplemented with 10 μM 4-nitroquiniline-1-oxide (4-NQO). Plates were incubated overnight at 37°C in the dark and then photographed. This assay is the same as previous published by us (63, 72) and similar to those reported by others for measuring sensitivity to 4-NQO and NFZ (3, 32, 52, 69). In all these assays, various dilutions of stationary-phase cells are plated on LB plates containing the agent and the numbers of CFU that subsequently appear are evaluated. In preliminary experiments, we found that cells are not killed by 4-NQO unless they are growing in its presence and that the plate-spotting procedure yields more reproducible results than can be obtained by growing the cells in liquid medium containing 4-NQO. Two 4-NQO concentrations were tested, and the one that gave the clearest results is presented in Fig. 5 below (although the results were not different). In addition, the experiment was repeated twice more to ensure that the results were reproducible.
RESULTS
Design of a strain with which to study Pol IV-dependent, growth-dependent mutation.
To study Pol IV's mutagenic activity in normally growing cells, we use an E. coli strain, FC722, that maximizes Pol IV's mutagenic phenotype (16). Pol IV preferentially makes frameshift mutations (38, 70); thus, the mutational target in FC722 is a tetA allele with a +1 frameshift mutation that renders the cell sensitive to tetracycline (Tcs). The reverting −1 frameshift mutation restores tetracycline resistance (Tcr). The frameshift is in a run of repeated G·C base pairs, a mutational target very similar to that in the lacI33-lacZ allele used for adaptive mutation (16). Pol IV also appears to preferentially make mutations on the F episome (40, 73), and so the mutant tetA allele in FC722 is carried on the F′ episome, where it reverts at a rate that is 5-fold higher than when it is carried on the chromosome (66, 73). We have hypothesized that this high level of mutation on the episome is due to a high frequency of DSBs, which allow Pol IV to gain access to DNA termini (73) (discussed further below).
During normal growth, strain FC722 has a mutation rate to Tcr of 1 to 5 mutations per 108 cells per generation, of which 50 to 70% are dependent on Pol IV (73) (see Fig. 7). This rate is increased about 10-fold when strains carry pPFG96 (63, 65, 73), a medium-copy-number plasmid with dinB under the control of the araBAD promoter on pBAD24 (27). Typically, we do not add arabinose to induce expression of the gene; under this condition, the levels of Pol IV are increased about 20-fold relative to the amount in FC722 without the plasmid (73), a level that is close to the estimated 10-fold increase that occurs after induction of the SOS response (36). This amount of Pol IV is only 1/5 of the level that has been reported to be lethal (67), but it is within the range that was reported to slow DNA replication (33). However, the presence of pPFG96 had no effect on the growth rate of most strains carrying it (Table 2). The exceptions were recB mutant strains carrying one or two additional mutant alleles; in these strains, the presence of pPFG96 reduced growth rates by 13 to 32% (Table 2).
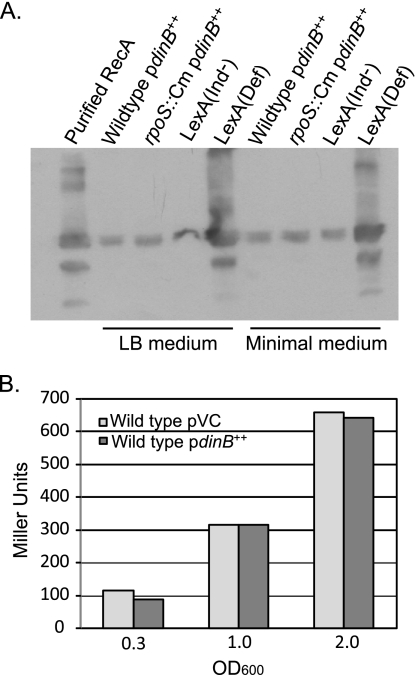
The presence of the dinB-carrying plasmid did not induce the RecA protein (Fig. 1 A), a sensitive assay for induction of the SOS response (26, 49), or the katE gene (Fig. 1B), an indicator of the level of RpoS activity (61, 62). In addition, exponential- and stationary-phase cells of all of the mutant strains listed in Table 2 were examined microscopically for filamentation, which also indicates induction of the SOS response (74). Almost all of the cultures, including those of the wild-type strain, contained some cells that were longer than two cell lengths, but in most cases, these did not exceed 1% of the total cell number. The exceptions were cultures of the recB rpoS, recB sbcDC, and recB rpoS sbcDC mutant strains, in which up to 5% of the cells were elongated. A low level of SOS induction is typical of cells defective in DNA repair functions (47) and has been previously reported for recB strains defective in additional recombination functions (5). However, in no case did the presence of the dinB-carrying plasmid significantly exacerbate the extent of filamentation.
FIG. 1.
Overexpression of Pol IV under the control of the araBAD promoter does not induce the SOS regulon or the RpoS regulon. (A) Western blot probed with anti-RecA antibody. The lanes on the left side show cultures grown to mid-exponential phase in LB, and the lanes on the right side show cultures grown to mid-exponential phase in minimal medium. In the LexA(Def) strain, all SOS genes are constitutively expressed, while in the LexA(Ind−) strain, all SOS genes are repressed. Forty micrograms of total cell protein was loaded in each lane of the polyacrylamide gel, with the exception of the first lane, in which purified RecA protein was loaded. Wild type/pdinB++, strain PFG266; rpoS::Cm/pdinB++, strain PFG420; LexA(Ind−), strain FC1418; LexA(Def), strain FC1419. (B) Results of β-galactosidase assays showing expression from the RpoS-regulated katE promoter in wild-type strains carrying the vector control pBAD24 (VC; strain PFG527, light bars) and pPFG96, with Pol IV overexpressed under the control of the araBAD promoter (strain PFG526, dark bars). Different points during growth of the cultures are shown as follows: mid-exponential phase, OD600 = 0.3; early stationary phase, OD600 = 1.0; stationary phase, OD600 = 2.0.
Taken together, our results indicate that neither wild-type nor mutant cells are unduly stressed by the levels of Pol IV produced by plasmid pPFG96. Thus, strain FC722 carrying this plasmid allows us to investigate the mechanisms by which Pol IV produces mutations in growing cells when it is at its fully induced state without the complications that would result from simultaneous induction of the entire suite of SOS gene products. In addition, cellular events involving Pol IV that normally occur at undetectable frequencies may be revealed when Pol IV is at its maximum levels.
Pol IV-dependent, growth-dependent mutation is decreased in strains lacking SbcC, SbcD, or SbcCD.
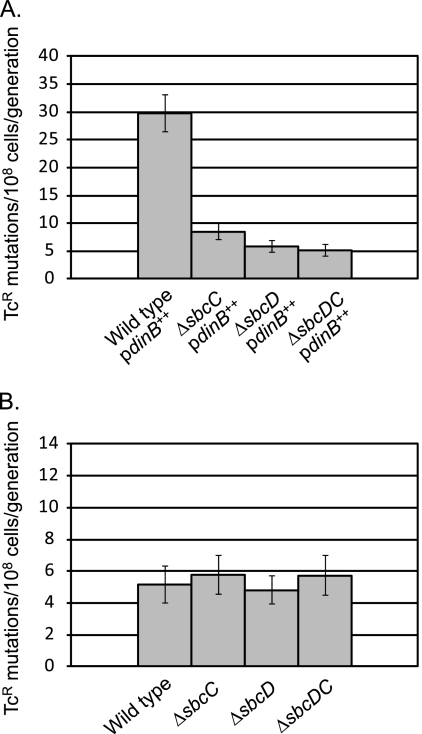
SbcCD is encoded by the sbcDC operon; to analyze the phenotypes of the complex and each subunit separately, we constructed nonpolar deletions of each gene and a deletion that removed both genes. As shown in Fig. 2 A, deletion of sbcC, sbcD, or both genes together resulted in a 3- to 4-fold decrease in the growth-dependent mutation rate when Pol IV was overexpressed from the araBAD promoter. In strains with wild-type (not overexpressed) levels of Pol IV, deletion of sbcC, sbcD, or both genes had no effect (Fig. 2B). These results indicate that SbcCD specifically enhances Pol IV-dependent, growth-dependent spontaneous mutation and that strains lacking SbcC, SbcD, or SbcCD have the same phenotype.
FIG. 2.
Pol IV-dependent, growth-dependent mutation is decreased in strains lacking SbcC, SbcD, or SbcCD. Mutation rates were determined from 36-culture fluctuation tests and were calculated by the MSS maximum-likelihood method; bars show 95% confidence levels (28, 60). (A) Results for strains carrying pPFG96 with Pol IV overexpressed under the control of the araBAD promoter. Wild type, PFG266; ΔsbcC, PFG956; ΔsbcD, PFG955; ΔsbcDC, PFG957. (B) Results for strains without excess Pol IV. Wild type, FC722; ΔsbcC, PFG948; ΔsbcD, PFG916; ΔsbcDC, PFG947.
RpoS and SbcCD are in the same pathway affecting Pol IV-dependent, growth-dependent mutation.
In order to determine if SbcCD and RpoS are involved in the same pathway that regulates Pol IV-dependent, growth-dependent mutation, we tested strains lacking both RpoS and SbcCD. With Pol IV overexpressed from the araBAD promoter, the mutation rate to Tcr of the strain lacking both RpoS and SbcCD was the same as that of the strain lacking only RpoS (Fig. 3) and that of the strain lacking only SbcCD (compare the rate shown in Fig. 2). That the phenotype of the rpoS sbcDC double mutant strain was not more severe than that of either single mutant strain (i.e., loss of RpoS and loss of SbcCD are mutually epistatic) means that RpoS and SbcCD act in the same pathway affecting Pol IV mutagenic activity.
FIG. 3.
Loss of SbcCD and loss of RpoS are mutually epistatic for Pol IV-dependent, growth-dependent mutation. Mutation rates were determined from 36-culture fluctuation tests and were calculated by the MSS maximum-likelihood method; bars show 95% confidence levels (28, 60). All strains carried pPFG96 with Pol IV overexpressed under the control of the araBAD promoter. rpoS::Cm/pdinB++, PFG420; rpoS::Cm ΔsbcC/pdinB++, PFG989; rpoS::Cm ΔsbcD/pdinB++, PFG990; rpoS::Cm ΔsbcDC/pdinB++, PFG991.
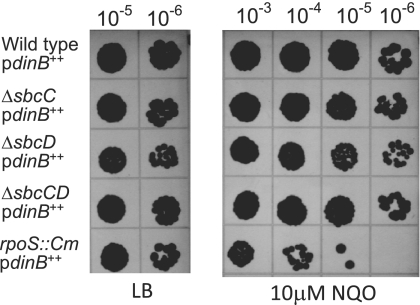
Loss of SbcCD does not significantly affect the levels of Pol IV or resistance to 4-NQO.
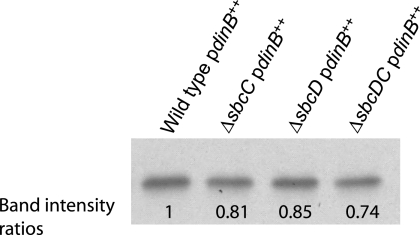
Western blot analysis revealed only a slight reduction in Pol IV levels in strains overexpressing Pol IV but lacking SbcC, SbcD, or SbcCD (Fig. 4). It is unlikely that this decrease, which is less than 30%, is responsible for the 4-fold reduction in the growth-dependent spontaneous mutation rate seen in these strains (Fig. 2A). Additionally, mutations in sbcDC did not affect survival during 4-NQO exposure when Pol IV was overexpressed (Fig. 5) or when it was not (data not shown). In contrast, mutations in RpoS cause a decrease in survival during 4-NQO exposure whether or not Pol IV is overexpressed (Fig. 5) (63). This difference indicates that the role that RpoS plays in Pol IV-dependent translesion synthesis is independent of SbcCD and that the regulation of Pol IV-dependent translesion synthesis and spontaneous mutagenesis are not the same.
FIG. 4.
Loss of SbcCD does not significantly affect the amount of Pol IV in strains overexpressing Pol IV from an exogenous promoter. A Western blot probed with anti-Pol IV antibody is shown. Twenty micrograms of total cell protein was loaded in each lane of the polyacrylamide gel. Bands were quantified using ImageJ software (58), and the ratios of the intensities are presented. All strains carried pPFG96 with Pol IV overexpressed under the control of the araBAD promoter. Wild-type/pdinB++, PFG266; ΔsbcC/pdinB++, PFG956; ΔsbcD/pdinB++, PFG955; ΔsbcDC/pdinB++, PFG957.
FIG. 5.
Loss of SbcCD does not affect sensitivity to 4-NQO. Spot tests were performed as described in Materials and Methods; uninformative lower dilutions are not shown. All strains carried pPFG96 with Pol IV overexpressed under the control of the araBAD promoter. Wild-type/pdinB++, PFG266; ΔsbcC/pdinB++, PFG956; ΔsbcD/pdinB++, PFG955; ΔsbcDC/pdinB++, PFG957; rpoS::Cm/pdinB++, PFG420.
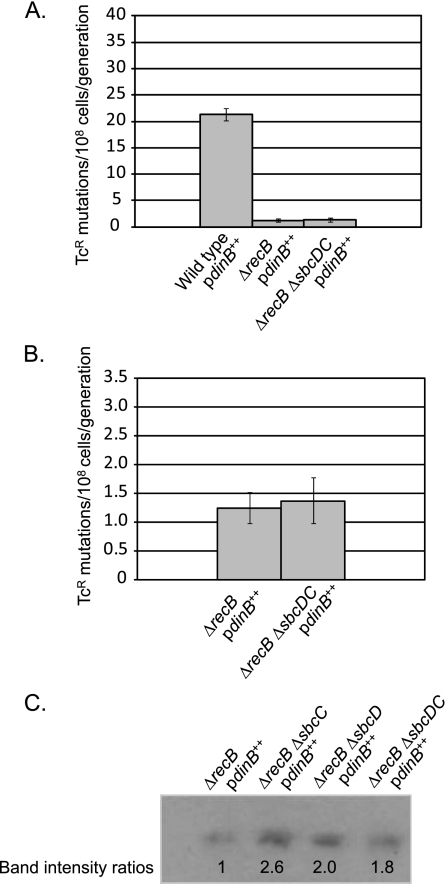
RecB and SbcCD are in the same pathway affecting Pol IV-dependent, growth-dependent mutation.
The Western blot and 4-NQO assay results led us to conclude that SbcCD specifically affects the ability of Pol IV to produce mutations. DSBs are an established substrate for Pol IV, and increasing the frequency of DSBs results in an increase in Pol IV-dependent mutagenesis (31, 55, 59). Because many SMC-like proteins, including SbcCD, have been shown to participate in DSB repair (12, 25), we hypothesized that SbcCD might affect Pol IV mutagenic activity via this pathway. We tested this hypothesis by measuring the spontaneous mutation rate to Tcr of ΔrecB mutant strains, which lack the RecBCD exonuclease (exonuclease V) that initiates the major DSB repair pathway in E. coli. As shown in Fig. 6, the ΔsbcDC ΔrecB mutant strain had the same low mutation rate as the ΔrecB mutant strain (i.e., loss of RecB is epistatic to loss of SbcCD), indicating that SbcCD and RecB function in the same pathway leading to Pol IV-dependent, growth-dependent mutation. Interestingly, there was more Pol IV in strains lacking both SbcCD and RecB than in the strain lacking only RecB (Fig. 6C), but this extra Pol IV clearly did not result in a higher rate of growth-dependent mutation (Fig. 6A).
FIG. 6.
Loss of SbcCD has no effect on Pol IV-dependent, growth-dependent mutation in a strain lacking RecB. (A) Mutation rates were determined by 36-culture fluctuation tests and were calculated by the MSS maximum-likelihood method; bars show 95% confidence levels (28, 60). (B) The same data as in panel A are shown but plotted on a different scale. (C) Western blot probed with anti-Pol IV antibody; 10 μg of total cell protein was loaded in each lane of the polyacrylamide gel. All strains carried pPFG96 with Pol IV overexpressed under the control of the araBAD promoter. Wild-type/pdinB++, PFG266; ΔrecB/pdinB++, PFG1120; ΔrecB ΔsbcC/pdinB++, PFG1123; ΔrecB ΔsbcD/pdinB++, PFG1121; ΔrecB ΔsbcDC/pdinB++, PFG1122.
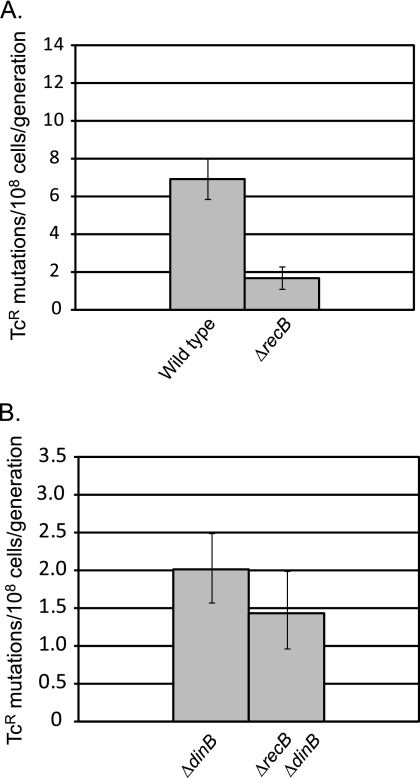
Since the mutation rates of the strains missing RecB, whether or not they were also missing SbcCD, were significantly lower than those of sbcDC mutant strains (compare Fig. 2A and 6A), it would appear that RecB is involved in both an SbcCD-dependent and an SbcCD-independent pathway for Pol IV mutagenesis. The effect of RecB is dependent on Pol IV but not on overexpression of Pol IV; the results in Fig. 7 show that loss of RecB reduced growth-dependent mutations in a strain with wild-type levels of Pol IV but had no effect in a ΔdinB mutant strain lacking Pol IV entirely.
FIG. 7.
The effect of RecB on growth-dependent mutation is dependent upon Pol IV. (A) Mutation rates were determined by 30-culture fluctuation tests and were calculated by the MSS maximum-likelihood method; bars show 95% confidence levels (28, 60). (B) Mutation rates were determined as described for panel A but in a separate experiment with 10 times more cells plated. Pol IV was not overexpressed in these strains. Wild type, FC722; ΔrecB, PFG1022; ΔdinB, PFB665; ΔrecB ΔdinB, PFG1146.
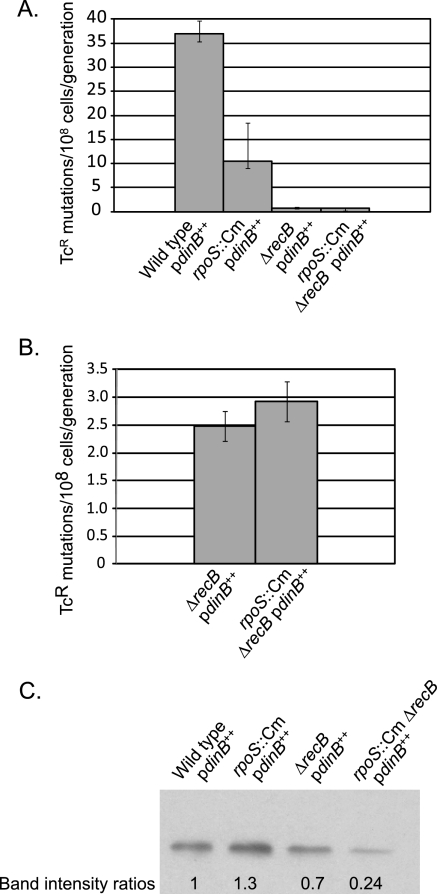
RecB and RpoS are in the same pathway affecting Pol IV-dependent, growth-dependent mutation.
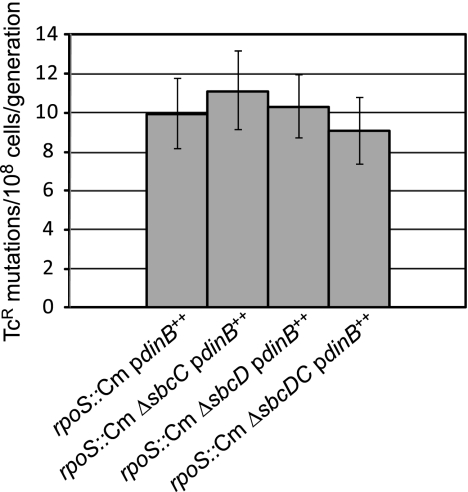
The epistatic relationships between RpoS and SbcCD and between RecB and SbcCD have one of two explanations: (i) all three of these proteins function in the same pathway affecting Pol IV-dependent, growth-dependent mutation, or (ii) SbcCD is involved in two distinct pathways affecting Pol IV-dependent, growth-dependent mutation, one that is RpoS dependent and another that is RecB dependent. To determine which of these alternatives is true, we tested the relationship between RpoS and RecB. As shown in Fig. 8, loss of RpoS did not further reduce the low mutation rate of a recB mutant strain (i.e., loss of RecB is epistatic to loss of RpoS), indicating that RecB and RpoS act in the same pathway affecting Pol IV mutagenic activity. Although the strain lacking both RpoS and RecB had less than half the amount of Pol IV as a strain lacking only RecB (Fig. 8C), this reduction did not result in a lower rate of Pol IV-dependent, growth-dependent spontaneous mutation (Fig. 8B). As shown in Fig. 9, the triple mutant strain lacking RecB, RpoS, and SbcCD did not have an additional reduction in mutation rate compared to any other strain lacking RecB. This result provides further confirmation that RecB, RpoS, and SbcCD act in one pathway leading to Pol IV-dependent, growth-dependent mutation.
FIG. 8.
Loss of RpoS has no effect on the Pol IV-dependent, growth-dependent mutation rate in a strain lacking RecB. (A) Mutation rates were determined by 36-culture fluctuation tests and were calculated by the MSS maximum-likelihood method; bars show 95% confidence levels (28, 60). (B) Data from a repeat experiment in which only the ΔrecB strain and the rpoS::Cm ΔrecB strain were tested. (C) Western blot probed with anti-Pol IV antibody; 20 μg of total cell protein was loaded in each lane of the polyacrylamide gel. All strains carried pPFG96 with Pol IV overexpressed under the control of the araBAD promoter. Wild type/pdinB++, PFG266; rpoS::Cm/pdinB++, PFG420; ΔrecB/pdinB++, PFG1120; ΔrecB rpoS::Cm/pdinB++, PFG1138.
FIG. 9.
Loss of SbcCD, RpoS, or both does not further reduce the growth-dependent mutation rate of strains lacking RecB. Mutation rates were determined by 36-culture fluctuation tests and were calculated by the MSS maximum-likelihood method; bars show 95% confidence levels (28, 60). All strains carried pPFG96 with Pol IV overexpressed under the control of the araBAD promoter. rpoS::Cm ΔsbcDC/pdinB++, PFG991; ΔrecB ΔsbcDC/pdinB++, PFG1122; ΔrecB rpoS::Cm/pdinB++, PFG1138; ΔrecB rpoS::Cm ΔsbcDC/pdinB++, PFG1143.
That the mutation rate of any strain missing RecB is significantly lower than those of strains missing only RpoS, only SbcCD, or both RpoS and SbcCD (compare the rates in Fig. 2A, 8A, and 9) suggests that RecB has an additional, RpoS-SbcCD-independent role in Pol IV-dependent, growth-dependent mutation. Taken together, our results suggest that RecBCD processing of DSBs is required for at least two pathways, one that is RpoS-SbcCD dependent and one that is not, both of which lead to Pol IV-dependent mutations.
DISCUSSION
RpoS is required for the maximum levels of two Pol IV-dependent mutational processes: (i) adaptive mutation in stationary-phase cells (42) and (ii) spontaneous mutation in growing cells when Pol IV is overexpressed (63). Because RpoS is a transcription factor, an obvious way that it could regulate Pol IV is by regulating the transcription of the dinB gene encoding Pol IV. RpoS does indeed drive the transcription of dinB, but only in stationary-phase cells (63). Thus, RpoS must affect Pol IV in growing cells indirectly by regulating another gene (or genes) whose product in turn affects Pol IV activity. We have determined that SbcCD, which is regulated by RpoS (13), is likely to be this intermediate factor (Fig. 2A and 3). We and others have found that SbcCD has little role in adaptive mutation (57; our unpublished results), and so, the RpoS-dependent regulation of Pol IV-dependent mutation in stationary-phase cells and in growing cells is fundamentally different.
It has recently been suggested that our assay for growth-dependent spontaneous mutation, reversion of the mutant tetA allele in strain FC722, also detects some adaptive mutation events (8). Indeed, we previously found that reversion to Tcr in FC722 during lactose selection occurred at the same rate and had the same genetic requirements as adaptive reversion to Lac+ (16). However, for the experiments presented here, we counted only the Tcr colonies present 2 days after plating, when the great majority of the colonies are the result of mutations that occur during growth of the cultures prior to plating (7, 16). In addition, as mentioned above, adaptive mutation is not significantly affected by loss of SbcCD (57; our unpublished results).
The results presented here were obtained in cells that were overproducing Pol IV at levels comparable to the levels present in cells induced for the SOS response (36, 73). These levels are well below those that have been reported to be lethal (67). In our experiments, Pol IV overexpression had no or only modest effects on growth rates and did not result in the induction of major stress responses (Table 2, Fig. 1). In addition, transcriptional microarray analyses of these strains revealed no pattern of induction of other stress responses (our unpublished results). Our results are relevant to the mechanisms by which Pol IV produces mutations in growing cells when it is at its fully induced state. In addition, when Pol IV is at its maximum levels, cellular events involving Pol IV that normally occur at undetectable frequencies may be revealed.
Pol IV translesion synthesis, as measured by resistance to 4-NQO, is enhanced by RpoS but independent of SbcCD (Fig. 5) (63). This result leads to two conclusions: (i) there is at least one other factor regulated by RpoS that plays a role in Pol IV-dependent translesion synthesis, and (ii) although RpoS plays a role in each, Pol IV-dependent translesion synthesis and mutagenesis in growing cells are regulated by different mechanisms. Our results thus add to previously published results (23, 69) supporting the hypothesis that Pol IV translesion synthesis and mutagenesis are mechanistically and genetically distinct. Indeed, of three phenotypes for Pol IV in vivo, resistance to 4-NQO, stationary-phase or adaptive mutation, and growth-dependent spontaneous mutation when Pol IV is overexpressed, all involve RpoS but SbcCD is important only for the last. Thus, the mechanisms of these three Pol IV-dependent phenomena are, at least in some ways, different.
RpoS, SbcCD, and RecB act in a common pathway that stimulates Pol IV-dependent, growth-dependent mutation (Fig. 2, 3, and 6 to 9) (63). Because the phenotype of loss of RecB is much more severe than the phenotypes of loss of RpoS, SbcCD, or both, RecB must have an additional RpoS- and SbcCD-independent role in growth-dependent mutation. In addition, the large effect of RecB is manifest even in cells that are not overexpressing Pol IV (Fig. 7), which is not the case for RpoS (63) or SbcCD (Fig. 2B); the weaker phenotypes of deleting rpoS or sbcDC are difficult to demonstrate in cells expressing Pol IV at wild-type levels. Nevertheless, the role of RecB in growth-dependent mutation appears to be entirely dependent on Pol IV, and vice versa, as the mutation rate of a ΔrecB ΔdinB double mutant strain was not significantly different from that of a ΔdinB mutant strain or that of a ΔrecB mutant strain (Fig. 7).
It is unlikely that SbcCD is involved in the actual regulation of Pol IV; rather, it probably affects Pol IV's ability to access DNA. Because of SbcCD's known role in DSB repair (12, 25) and because SbcCD is in a RecB-dependent pathway that promotes Pol IV-dependent mutations, we hypothesize that SbcCD participates in the generation or repair of DSBs via a particular pathway that maximizes Pol IV's mutagenic activity.
Adaptive reversion of the episomal lacI33-lacZ allele requires both recombination and conjugal functions but not actual conjugation (4, 19, 21, 30). It has been proposed that a DSB is created when a replication fork collapses upon reaching the persistent nick produced by TraI activity at the episome's conjugal origin, oriT (41). We have further proposed that when the replication machinery is reestablished, it initially contains either DNA polymerase II or DNA polymerase IV instead of the normal replicative polymerase, DNA polymerase III. When Pol II is utilized, the synthesis is mostly error free, but when Pol IV is utilized, it produces the errors that lead to adaptive mutations (17, 59). We hypothesize that the same mechanism generates the Pol IV-dependent, growth-dependent mutations observed here, but in contrast to the conditions that pertain in nongrowing cells, in normally growing cells, other sources of DSBs can result in Pol IV gaining access to the replication fork. In support of this hypothesis, a DSB created near the tetA mutational target enhanced reversion of the mutant tetA allele in exponentially growing cells only if the cells were expressing RpoS (55). We suggest that RpoS was required in these experiments, at least in part, to upregulate SbcCD.
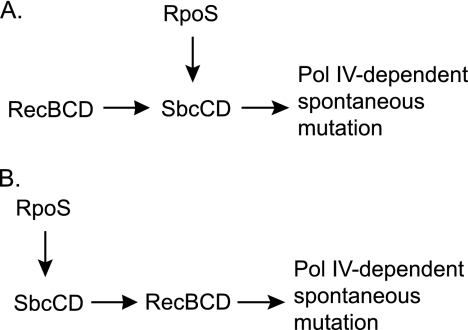
Two models can explain how RpoS, RecB, and SbcCD act in the same pathway to stimulate Pol IV-dependent spontaneous mutation. In both models, RpoS regulates the expression of SbcCD, but the roles of SbcCD and RecB are different. In the first model, a spontaneous DNA DSB, RecBCD resects the double-strand break, initiating homologous recombination. SbcCD acts as an SMC complex in a fashion similar to the eukaryotic Mre11/Rad50 complex, maintaining proper chromosome arrangement for processing and repair (Fig. 10 A). In the second model, SbcCD acts first in the pathway, making a DSB by cleaving a hairpin in the DNA; RecBCD then resects the DSB to initiate homologous recombination (Fig. 10B). In both models, Pol IV initiates DNA synthesis from recombination intermediates as part of the repair process, generating mutations. Neither model is excluded by our data, although the first, in which SbcCD plays a supporting but not an essential role, appears to better fit the results.
FIG. 10.
Two models for the roles of SbcCD, RecB, and RpoS in Pol IV-dependent spontaneous mutation. In both models, RpoS upregulates SbcCD and RecBCD initiates DNA DSB repair via homologous recombination. (A) A DNA DSB occurs, repair is initiated, and SbcCD acts as an SMC complex, maintaining the proper orientation of the intermediates for repair. (B) SbcCD acts as a nuclease, producing a DSB by cleaving a DNA hairpin. In both models, Pol IV initiates DNA synthesis from recombination intermediates, generating mutations. The arrows indicate a positive interaction either on a physical or on a genetic level (see Discussion).
Acknowledgments
We thank D. Boyd, M. J. Casadaban (deceased), J. H. Miller, R. W. Simons, G. C. Walker, and the National BioResource Project (Japan) for bacterial strains, bacteriophage, and plasmids. We are grateful to members of our laboratory for advice and patience.
This work was supported by USPHS NIH grant GM065175 to P.L.F. and U.S. NSF IGERT training grant 0504627/206251A to K.A.M.S.
Footnotes
Published ahead of print on 3 December 2010.
REFERENCES
- 1.Ausubel, F. M., et al. 1988. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Baba, T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beuning, P. J., S. M. Simon, V. G. Godoy, D. F. Jarosz, and G. C. Walker. 2006. Characterization of Escherichia coli translesion synthesis polymerases and their accessory factors. Methods Enzymol. 408:318-340. [DOI] [PubMed] [Google Scholar]
- 4.Cairns, J., and P. L. Foster. 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, K. L., Y. K. Mak, and P. Oliver. 1993. Expression of the recA gene in recombination-deficient (rec−) strains of Escherichia coli. Biochimie 75:775-783. [DOI] [PubMed] [Google Scholar]
- 6.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U. S. A. 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, S. E., V. G. Godoy, and G. C. Walker. 2009. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J. Bacteriol. 191:665-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, S. E., and G. C. Walker. 2010. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr. Biol. 20:80-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connelly, J. C., L. A. Kirkham, and D. R. F. Leach. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. U. S. A. 95:7969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connelly, J. C., and D. R. F. Leach. 1996. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells 1:285-291. [DOI] [PubMed] [Google Scholar]
- 11.Courcelle, J., A. Khodursky, B. Peter, P. O. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cromie, G. A., and D. R. Leach. 2001. Recombinational repair of chromosomal DNA double-strand breaks generated by a restriction endonuclease. Mol. Microbiol. 41:873-883. [DOI] [PubMed] [Google Scholar]
- 13.Darmon, E., et al. 2007. SbcCD regulation and localization in Escherichia coli. J. Bacteriol. 189:6686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Dong, T., M. G. Kirchhof, and H. E. Schellborn. 2008. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol. Genet. Genomics 279:267-277. [DOI] [PubMed] [Google Scholar]
- 15.Finkel, S. E. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4:113-120. [DOI] [PubMed] [Google Scholar]
- 16.Foster, P. L. 1997. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J. Bacteriol. 179:1550-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster, P. L. 2000. Adaptive mutation in Escherichia coli. Cold Spring Harb. Symp. Quant. Biol. 65:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, P. L. 2006. Methods for determining spontaneous mutation rates. Methods Enzymol. 409:195-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, P. L., and J. M. Trimarchi. 1995. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc. Natl. Acad. Sci. U. S. A. 92:5487-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs, R. P., S. Fujii, and J. Wagner. 2004. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv. Protein Chem. 69:229-264. [DOI] [PubMed] [Google Scholar]
- 21.Galitski, T., and J. R. Roth. 1995. Evidence that F plasmid transfer replication underlies apparent adaptive mutation. Science 268:421-423. [DOI] [PubMed] [Google Scholar]
- 22.Gawel, D., P. T. Pham, I. J. Fijalkowska, P. Jonczyk, and R. M. Schaaper. 2008. Role of accessory DNA polymerases in DNA replication in Escherichia coli: analysis of the dnaX36 mutator mutant. J. Bacteriol. 190:1730-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godoy, V. G., et al. 2007. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28:1058-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gowrishankar, J. 1985. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J. Bacteriol. 164:434-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graumann, P. L., and T. Knust. 2009. Dynamics of the bacterial SMC complex and SMC-like proteins involved in DNA repair. Chromosome Res. 17:265-275. [DOI] [PubMed] [Google Scholar]
- 26.Gudas, L. J., and D. W. Mount. 1977. Identification of the recA (tif) gene product of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 74:5280-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall, B. M., C. X. Ma, P. Liang, and K. K. Singh. 2009. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25:1564-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Harris, R. S., S. Longerich, and S. M. Rosenberg. 1994. Recombination in adaptive mutation. Science 264:258-260. [DOI] [PubMed] [Google Scholar]
- 31.He, A. S., P. R. Rohatgi, M. N. Hersh, and S. M. Rosenberg. 2006. Roles of E. coli double-strand-break-repair proteins in stress-induced mutation. DNA Repair (Amst.) 5:258-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heltzel, J. M., R. W. Maul, S. K. Scouten Ponticelli, and M. D. Sutton. 2009. A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc. Natl. Acad. Sci. U. S. A. 106:12664-12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Indiani, C., L. D. Langston, O. Yurieva, M. F. Goodman, and M. O'Donnell. 2009. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc. Natl. Acad. Sci. U. S. A. 106:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarosz, D. F., V. G. Godoy, J. C. Delaney, J. M. Essigmann, and G. C. Walker. 2006. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439:225-228. [DOI] [PubMed] [Google Scholar]
- 35.Kenyon, C. J., and G. C. Walker. 1980. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 77:2819-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, S. R., K. Matsui, M. Yamada, P. Gruz, and T. Nohmi. 2001. Roles of chromosomal and episomal dinB genes encoding DNA Pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266:207-215. [DOI] [PubMed] [Google Scholar]
- 37.Kim, S. R., et al. 1997. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. U. S. A. 94:13792-13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi, S., M. R. Valentine, P. Pham, M. O'Donnell, and M. F. Goodman. 2002. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 277:34198-34207. [DOI] [PubMed] [Google Scholar]
- 39.Kuban, W., et al. 2005. Mutator phenotype resulting from DNA polymerase IV overproduction in Escherichia coli: preferential mutagenesis on the lagging strand. J. Bacteriol. 187:6862-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuban, W., et al. 2004. Role of Escherichia coli DNA polymerase IV in in vivo replication fidelity. J. Bacteriol. 186:4802-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16:373-384. [DOI] [PubMed] [Google Scholar]
- 42.Layton, J. C., and P. L. Foster. 2003. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol. Microbiol. 50:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Layton, J. C., and P. L. Foster. 2005. Error-prone DNA polymerase IV is regulated by the heat shock chaperone GroE in Escherichia coli. J. Bacteriol. 187:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leach, D. R. 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16:893-900. [DOI] [PubMed] [Google Scholar]
- 45.Lloyd, R. G., and C. Buckman. 1985. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164:836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCool, J. D., et al. 2004. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol. Microbiol. 53:1343-1357. [DOI] [PubMed] [Google Scholar]
- 48.McCulloch, S. D., and T. A. Kunkel. 2008. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18:148-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McEntee, K. 1977. Protein X is the product of the recA gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 74:5275-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenzie, G. J., P. L. Lee, M. J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579. [DOI] [PubMed] [Google Scholar]
- 51.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Ona, K. R., C. T. Courcelle, and J. Courcelle. 2009. Nucleotide excision repair is a predominant mechanism for processing nitrofurazone-induced DNA damage in Escherichia coli. J. Bacteriol. 191:4959-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel, M., Q. Jiang, R. Woodgate, M. M. Cox, and M. F. Goodman. 2010. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit. Rev. Biochem. Mol. Biol. 45:171-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580-591. [DOI] [PubMed] [Google Scholar]
- 55.Ponder, R. G., N. C. Fonville, and S. M. Rosenberg. 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19:791-804. [DOI] [PubMed] [Google Scholar]
- 56.Rajkumari, K., and J. Gowrishankar. 2002. An N-terminally truncated RpoS (σS) protein in Escherichia coli is active in vivo and exhibits normal environmental regulation even in the absence of rpoS transcriptional and translational control signals. J. Bacteriol. 184:3167-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramirez-Santos, J., V. Garcia-Mata, S. Poggio, L. Camarena, and M. C. Gomez-Eichelmann. 2009. Role of single-strand DNA 3′-5′ exonuclease ExoI and nuclease SbcCD in stationary-phase mutation in Escherichia coli K-12. Arch. Microbiol. 191:185-190. [DOI] [PubMed] [Google Scholar]
- 58.Rasband, W. S. 1997. ImageJ. U.S. National Institutes of Health, Bethesda, MD.
- 59.Rodriguez, C., J. Tompkin, J. Hazel, and P. L. Foster. 2002. Induction of a DNA nickase in the presence of its target site stimulates adaptive mutation in Escherichia coli. J. Bacteriol. 184:5599-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarkar, S., W. T. Ma, and G. H. Sandri. 1992. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica 85:173-179. [DOI] [PubMed] [Google Scholar]
- 61.Schellhorn, H. E. 1995. Regulation of hydroperoxidase (catalase) expression in Escherichia coli. FEMS Microbiol. Lett. 131:113-119. [DOI] [PubMed] [Google Scholar]
- 62.Schellhorn, H. E., and H. M. Hassan. 1988. Transcriptional regulation of katE in Escherichia coli K-12. J. Bacteriol. 170:4286-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Storvik, K. A., and P. L. Foster. 2010. RpoS, the stress-response sigma factor, plays a dual role in the regulation of Escherichia coli's error-prone DNA polymerase IV. J. Bacteriol. 192:3639-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strauss, B. S., R. Roberts, L. Francis, and P. Pouryazdanparast. 2000. Role of the dinB gene product in spontaneous mutation in Escherichia coli with an impaired replicative polymerase. J. Bacteriol. 182:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stumpf, J. D., and P. L. Foster. 2005. Polyphosphate kinase regulates error-prone replication by DNA polymerase IV in Escherichia coli. Mol. Microbiol. 57:751-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stumpf, J. D., A. R. Poteete, and P. L. Foster. 2007. Amplification of lac cannot account for adaptive mutation to Lac+ in Escherichia coli. J. Bacteriol. 189:2291-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uchida, K., et al. 2008. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol. Microbiol. 70:608-622. [DOI] [PubMed] [Google Scholar]
- 68.Vijayakumar, S. R., M. G. Kirchhof, C. L. Patten, and H. E. Schellhorn. 2004. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J. Bacteriol. 186:8499-8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wagner, J., H. Etienne, R. P. Fuchs, A. Cordonnier, and D. Burnouf. 2009. Distinct beta-clamp interactions govern the activities of the Y family Pol IV DNA polymerase. Mol. Microbiol. 74:1143-1151. [DOI] [PubMed] [Google Scholar]
- 70.Wagner, J., and T. Nohmi. 2000. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J. Bacteriol. 182:4587-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams, A. B., K. M. Hetrick, and P. L. Foster. 2010. Interplay of DNA repair, homologous recombination, and DNA polymerases in resistance to the DNA damaging agent 4-nitroquinoline-1-oxide in Escherichia coli. DNA Repair 9:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Williams, A. B., and P. L. Foster. 2007. The Escherichia coli histone-like protein HU has a role in stationary phase adaptive mutation. Genetics 177:723-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Witkin, E. M. 1976. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol. Rev. 40:869-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolff, E., M. Kim, K. Hu, H. Yang, and J. H. Miller. 2004. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J. Bacteriol. 186:2900-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu, Y. H., M. A. Franden, J. R. Hawker, Jr., and C. S. McHenry. 1984. Monoclonal antibodies specific for the alpha subunit of the Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 259:12117-12122. [PubMed] [Google Scholar]
- 77.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. U. S. A. 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]