Abstract
DifA is a methyl-accepting chemotaxis protein (MCP)-like sensory transducer that regulates exopolysaccharide (EPS) production in Myxococcus xanthus. Here mutational analysis and molecular biology were used to probe the signaling mechanisms of DifA in EPS regulation. We first identified the start codon of DifA experimentally; this identification extended the N terminus of DifA for 45 amino acids (aa) from the previous bioinformatics prediction. This extension helped to address the outstanding question of how DifA receives input signals from type 4 pili without a prominent periplasmic domain. The results suggest that DifA uses its N-terminus extension to sense an upstream signal in EPS regulation. We suggest that the perception of the input signal by DifA is mediated by protein-protein interactions with upstream components. Subsequent signal transmission likely involves transmembrane signaling instead of direct intramolecular interactions between the input and the output modules in the cytoplasm. The basic functional unit of DifA for signal transduction is likely dimeric as mutational alteration of the predicted dimeric interface of DifA significantly affected EPS production. Deletions of 14-aa segments in the C terminus suggest that the newly defined flexible bundle subdomain in MCPs is likely critical for DifA function because shortening of this bundle can lead to constitutively active mutations.
Myxococcus xanthus is a Gram-negative model bacterium for studies of multicellular development, gliding motility, and signal transduction (28, 42, 51). During vegetative growth, M. xanthus seeks nutrients and hunts other bacteria on solid surfaces by gliding motility. During development, hundreds of thousands of cells aggregate to form a multicellular fruiting body in response to starvation. Cells within fruiting bodies differentiate into dormant stress-resistant myxospores, which can germinate to reenter vegetative growth once conditions become favorable. Both vegetative and developmental cycles depend on gliding motility as well as a finely tuned signal transduction network. Without effective signal transduction, M. xanthus would be incapable of responding to environmental changes to undergo growth or development. Without motility, cells would not be able to actively seek nutrient sources for growth or to undergo developmental aggregation for fruiting body formation.
The Dif chemosensory pathway plays crucial roles in motility and its regulation in M. xanthus. This bacterium has two genetically distinct motility systems known as adventurous (A) and social (S) motility (21, 22). S motility is likely powered by the retraction of the type 4 pilus (TFP); exopolysaccharides (EPS) on the surface of a neighboring M. xanthus cell are proposed to trigger TFP retraction in S motility (32, 51). The Dif proteins, which are homologous to bacterial chemotaxis proteins, are crucial for S motility because they regulate EPS production (5, 8, 48, 50). Dif proteins are also involved in the chemotactic response to certain species of phosphatidylethanolamine (PE) (9, 26). It has been proposed that PE molecules may function as signals to distinguish between self and nonself during vegetative hunting and developmental aggregation (15, 26). The dual roles of Dif proteins in S motility and PE taxis may explain why EPS− dif mutants show no developmental aggregation (5, 8, 47, 48, 50); this also underscores the importance of the Dif pathway in the biology of M. xanthus.
DifA is the homolog of the methyl-accepting chemoreceptor protein (MCP) in the Dif pathway and is likely responsible for sensing input signals and transmitting them downstream (7, 48). However, the signaling mechanism utilized by DifA is not well understood, especially in regard to EPS regulation. Previous bioinformatics analysis predicted that DifA starts immediately with its first transmembrane helix (TM1), which is followed by a 10-residue periplasmic region, the second TM helix (TM2), and the cytoplasmic signaling domain (29, 48). The lack of an obvious signal sensory domain, whether at the cytoplasmic N terminus or in the periplasm, was perplexing. When it was later discovered that TFP likely acts upstream of Dif in EPS regulation (7), it was speculated that the TMs of DifA might interact with membrane proteins in the TFP assembly/disassembly complex to receive signals. However, all of our efforts to seek clues and evidence for such interactions have proved fruitless, and it remained a mystery how DifA senses and transmits signals in EPS regulation.
This report focuses on our study of the signaling mechanisms by DifA in EPS regulation. As an important first step, the DifA start codon was identified experimentally, which extended the DifA N terminus by an additional 48 amino acids (aa) from the previous prediction. Part of the picture emerging from this study is that DifA may use this newly discovered N terminus for signal reception. DifA also appears to require transmembrane signaling to relay its input signal to the output or signaling domain at the C terminus. Our results suggest that the basic functional unit of DifA may be a dimer. Its dimer interface and the flexible bundle subdomain (FBS) (1, 2) may both play critical roles in signaling in M. xanthus EPS regulation.
MATERIALS AND METHODS
Strains and growth conditions.
The M. xanthus strains used in this study are listed in Table 1. M. xanthus was grown at 32°C on either CTT (1% Casitone, 8 mM MgSO4, 10 mM Tris·HCl, 1 mM K2HPO4-KH2PO4, pH 7.6) or CYE (1% Casitone, 0.5% yeast extract, 0.1% MgSO4·7H2O, 10 mM MOPS [morpholinepropanesulfonic acid], pH 7.6) (13, 24). Escherichia coli strain XL1-Blue (Stratagene), used for routine cloning and plasmid construction, was grown at 37°C on Luria-Bertani (LB) medium (34). Liquid cultures were grown with a rotary shaker at 300 rotations per minute. Solid media for plates contained 1.5% agar. When necessary, kanamycin was added to media at 100 μg/ml.
TABLE 1.
M. xanthus strains in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| DK1622 | Wild type | 24 |
| YZ601 | ΔdifA | 47 |
| YZ606 | ΔdifA/difA(M49A) | This study |
| YZ619 | ΔdifA/difA+ | 10 |
| YZ707 | ΔdifA/difA(E183D) | 10 |
| YZ708 | ΔdifA/difA(E428D) | 10 |
| YZ709 | ΔdifA/difA (−138GTC) | This study |
| YZ710 | ΔdifA/difA (+1GTC) | This study |
| YZ711 | ΔdifA/difA (+1ATG) | This study |
| YZ712 | ΔdifA/difA(E158D) | 10 |
| YZ713 | ΔdifA/difA(Q394D) | 10 |
| YZ714 | ΔdifA/difA(E400D) | 10 |
| YZ715 | ΔdifA/difA(E407D) | 10 |
| YZ745 | ΔdifA/difA(E183A) | This study |
| YZ746 | ΔdifA/difA(Q394A) | This study |
| YZ747 | ΔdifA/difA(E400A) | This study |
| YZ748 | ΔdifA/difA(E407A) | This study |
| YZ749 | ΔdifA/difA(E183Q) | This study |
| YZ750 | ΔdifA/difA(Q394E) | This study |
| YZ751 | ΔdifA/difA(E400Q) | This study |
| YZ752 | ΔdifA/difA(E407Q) | This study |
| YZ753 | ΔdifA/difA(Q394N) | This study |
| YZ756 | ΔdifA/difA(Q429D) | This study |
| YZ758 | ΔdifA/difA(E158A) | This study |
| YZ759 | ΔdifA/difA(E158Q) | This study |
| YZ760 | ΔdifA/difA(E428A) | This study |
| YZ761 | ΔdifA/difA(E428Q) | This study |
| YZ762 | ΔdifA/difA(Q429A) | This study |
| YZ763 | ΔdifA/difA(Q429E) | This study |
| YZ764 | ΔdifA/difA(Q429N) | This study |
| YZ776 | ΔdifA/difA (Δ2-40) | This study |
| YZ777 | ΔdifA/difA (ΔI) | This study |
| YZ778 | ΔdifA/difA (ΔIV) | This study |
| YZ779 | ΔdifA/difA [Δ(I + IV)] | This study |
| YZ780 | ΔdifA/difA (ΔII) | This study |
| YZ781 | ΔdifA/difA (ΔIII) | This study |
| YZ782 | ΔdifA/difA [Δ(II + III)] | This study |
| YZ1710 | ΔdifA/difA (Δ10-35) | This study |
| YZ1711 | ΔdifA/difA (Δ2-8) | This study |
| YZ1712 | ΔdifA/difA (Δ9-18) | This study |
| YZ1713 | ΔdifA/difA (Δ19-28) | This study |
| YZ1714 | ΔdifA/difA (Δ29-38) | This study |
| YZ1820 | ΔdifA/difA(G45A) | This study |
| YZ1821 | ΔdifA/difA(Y46C) | This study |
| YZ1822 | ΔdifA/difA(L38P) | This study |
| YZ1823 | ΔdifA/difA(P96L) | This study |
Plasmid construction.
The plasmids used in this study, which all contain the Mx8 phage attachment site (attP), are listed in Table 2. All plasmids with mutations in difA are derivatives of pWB210 or pWB230; both contain full-length difA and its upstream promoter. They differ in that pWB230 harbors two silent mutations to engineer two restriction sites for the convenience of introducing targeted mutations (Table 2, Fig. 1) (10). pHN001 through pHN004, as well as pXQ707 and pXQ708, all contain aspartate (D) substitutions of E158, E183, Q394, E400, E407, and E428 (10). The plasmids constructed in this study were similarly generated, as previously described (10). pXQ731 through pXQ749 contain amino acid substitutions of E158, E183, Q394, E400, E407, E428, and Q429 in DifA, as specified in Table 2. pXQ752, pXQ754 through pXQ759, and pXQ769 through pXQ773 all contain various in-frame deletions either at the N-terminal cytoplasmic region or of C-terminal (insertion/deletion) indel segments as indicated (Table 2). pXQ709 contains a GTG-to-GTC mutation at −138 upstream of the difA coding region (Fig. 1). The GTG start codon of difA was mutated to GTC and ATG in pXQ710 and pXQ711, respectively. pWB126 contains the M49A mutation (Table 2). All plasmids constructed in this study were confirmed by restriction digestion and DNA sequencing.
TABLE 2.
Plasmids used in this study
| Designation | Description | Source or reference |
|---|---|---|
| pWB126 | difA(M49A) in pWB210; Kanr | This study |
| pWB200 | Mx8 phage ATT site; ccdB for lethal selection; Kanr | 47 |
| pWB210 | Full-length difA with promoter in pWB200; Kanr | 10 |
| pWB230 | pWB210 with 2 silent point mutations in difA (CGC to CGG at codon 34 to introduce an SmaI site and CTG to CTC at codon 151 to introduce an SacI site) | 10 |
| pHN001 | difA(E158D) in pWB230; Kanr | 10 |
| pHN002 | difA(Q394D) in pWB230; Kanr | 10 |
| pHN003 | difA(E400D) in pWB230; Kanr | 10 |
| pHN004 | difA(E407D) in pWB230; Kanr | 10 |
| pXQ707 | difA(E183D) in pWB230; Kanr | 10 |
| pXQ708 | difA(E428D) in pWB230; Kanr | 10 |
| pXQ709 | pWB210 with the −138GTC mutation; Kanr | This study |
| pXQ710 | pWB210 with the difA start codon GTG mutated to GTC; Kanr | This study |
| pXQ711 | pWB210 with the difA start codon GTG mutated to ATG; Kanr | This study |
| pXQ731 | difA(E183A) in pWB230; Kanr | This study |
| pXQ732 | difA(Q394A) in pWB230; Kanr | This study |
| pXQ733 | difA(E400A) in pWB230; Kanr | This study |
| pXQ734 | difA(E407A) in pWB230; Kanr | This study |
| pXQ735 | difA(E183Q) in pWB230; Kanr | This study |
| pXQ736 | difA(Q346E) in pWB230; Kanr | This study |
| pXQ737 | difA(E400Q) in pWB230; Kanr | This study |
| pXQ738 | difA(E407Q) in pWB230; Kanr | This study |
| pXQ739 | difA(Q346N) in pWB230; Kanr | This study |
| pXQ740 | difA(Q429D) in pWB230; Kanr | This study |
| pXQ742 | difA(E158A) in pWB230; Kanr | This study |
| pXQ743 | difA(E158Q) in pWB230; Kanr | This study |
| pXQ744 | difA(E428A) in pWB230; Kanr | This study |
| pXQ746 | difA(E428Q) in pWB230; Kanr | This study |
| pXQ747 | difA(Q429A) in pWB230; Kanr | This study |
| pXQ748 | difA(Q381E) in pWB230; Kanr | This study |
| pXQ749 | difA(Q381N) in pWB230; Kanr | This study |
| pXQ752 | difA (Δ2-40) in pWB230; Kanr | This study |
| pXQ754 | difA (ΔI) in pWB230; Kanr | This study |
| pXQ755 | difA (ΔIV) in pWB230; Kanr | This study |
| pXQ756 | difA [Δ(I+IV)] in pWB230; Kanr | This study |
| pXQ757 | difA (ΔII) in pWB230; Kanr | This study |
| pXQ758 | difA (ΔIII) in pWB230; Kanr | This study |
| pXQ759 | difA [Δ(II + III)] in pWB230; Kanr | This study |
| pXQ769 | difA (Δ10-35) in pWB230; Kanr | This study |
| pXQ770 | difA (Δ2-8) in pWB230; Kanr | This study |
| pXQ771 | difA (Δ9-18) in pWB230; Kanr | This study |
| pXQ772 | difA (Δ19-28) in pWB230; Kanr | This study |
| pXQ773 | difA (Δ29-38) in pWB230; Kanr | This study |
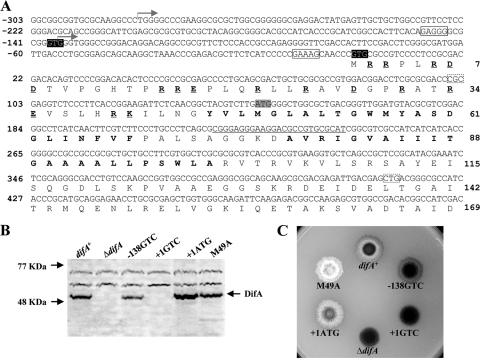
FIG. 1.
Identification of DifA start codon. (A) The upstream and 5′ coding sequence of difA with deduced amino acids. The oligonucleotide used for primer extension (see Fig. S1 in the supplemental material) is complementary to the underlined DNA sequence centered around the codon for D77. The two transcription initiation sites identified by primer extension are marked by two forward arrows, respectively. The two potential GTG start codons for DifA at −138 and +1 are shaded in black, with their consensus ribosomal binding sites boxed with solid lines. The ATG proposed by bioinformatics as the start codon for DifA is shaded in gray. The amino acids in bold are the two predicted transmembrane helices TM1 and TM2. The charged residues preceding TM1 are in bold and underlined. Codons 34 (CGC) and 138 (CTG), boxed by dashed lines, were changed to CGG and CTC as silent mutations to introduce two restriction sites in pWB230 (Table 2). (B and C) Immunoblot analysis (B) and EPS production (C) of strains with mutations of the potential start codons. For immunoblotting, whole-cell lysates from 5 × 108 cells for each strain were separated by SDS-PAGE and probed with anti-DifA antibodies. For EPS analysis, 5 μl of cells at approximately 5 × 109 cells/ml was spotted onto CTT plates containing calcofluor white at 50 μg per ml; the plates were photographed under UV illumination after 5 days of incubation at 32°C. The diameter of the plates shown is ∼9 cm. Strains: difA+, YZ619; ΔdifA, YZ601; −138GTC, YZ709; +1GTC, YZ710; +1ATG, YZ711; and M49A, YZ606.
Construction of M. xanthus mutants.
YZ601, a difA deletion mutant (8), was used as the parental strain for all mutants constructed in this study. The plasmids with difA mutations (Table 2) were electroporated (25) into YZ601 (ΔdifA) to construct the various strains with specified difA mutations (Table 1); these include all strains listed in Table 1 from YZ606 to YZ1714.
Strains YZ1820 through YZ1823 (Table 1) were generated from random mutagenesis of DifA. Error-prone PCR (31) was performed targeting the codons 35 to 136 of DifA by using the attP integrative plasmid pWB230 (Table 2) as the template and the oligonucleotides dAEP5SmaIF (5′-GACCTCGCGCGACCCGGGAGG-3′) and dAEP3SacIR (5′-GCCCACGAGCTCGCGCAG-3′) as primers. The PCR products were cloned into the SmaI and SacI sites of pWB230 after digestion with the same enzymes to generate a mutant library which was electroporated (25) into YZ601 (ΔdifA). The resulting transformants were plated onto CTT agar containing 60 μg/ml Congo red to identify EPS− colonies, as indicated by the lack of Congo red binding (8). The EPS− transformants that produced full-length DifA protein were identified by immunoblotting using anti-DifA antibodies (47). The region targeted for mutagenesis from these transformants was PCR amplified and sequenced to identify mutations. The PCR fragments with single point mutations were recloned into the SmaI and SacI sites of pWB230; the resulting plasmids were electroporated (25) into YZ601 to generate a second round of transformants for the confirmation of the association of individual mutations with the EPS− phenotype. The second generation of transformants gave rise to YZ1820 through YZ1823, which harbor G45A, Y46C, L38P, and P96L substitution mutations, respectively (Table 1).
Cellular fractionation and immunoblotting.
DifA membrane localization was examined by a fractionation procedure similar to that described previously (33, 46). Cultures were grown in CYE to approximately 4 × 108 cells/ml. Cells were harvested by centrifugation at 10,000 × g, washed, and resuspended to 5 × 109 cells/ml in 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer (pH 7.2). Cells were disrupted by sonication, and whole-cell lysates were obtained by centrifugation at 15,000 × g for 15 min. The lysates were quantified by protein content determined by the Bradford assay (11). Whole-cell lysates were centrifuged at 100,000 × g for 1 h, the resulting supernatant was recovered as the soluble fraction, and the pellet was resuspended in an equivalent volume of HEPES buffer as the membrane fraction. Whole-cell lysates and different fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblotting with anti-DifA polyclonal antibodies (45) by standard protocols (37).
Modeling of DifA structure.
The cytoplasmic domain of DifA (DifAC) shares 29% identity and 49% similarity with Thermotoga maritima MCP1143 C terminus (MCP1143C) (35) For modeling, DifAC was analyzed against the T. maritima MCP1143C dimer (Protein Data Bank [PDB] code 2CH7) with the Oligomer Modeling program of SWISS-MODEL (3, 18, 38). The resulting DifAC dimer model was visualized and analyzed with VMD software (23).
Miscellaneous techniques.
To examine EPS production, M. xanthus strains were first grown in CYE liquid medium to ∼1.0 × 108 to ∼2.0 × 108 cells/ml. Cells were washed and resuspended in MOPS (morpholinepropanesulfonic acid) buffer (10 mM MOPS, 2 mM MgSO4, pH 7.6) at approximately 5.0 × 109 cells per ml. Five microliters of these cell suspensions was spotted onto the surface of CTT plates with the fluorescent dye calcofluor white at 50 μg/ml. These plates were incubated at 32°C for 5 days before documentation under the UV illumination at ∼365 nm.
RESULTS
Identification of the start codon for DifA.
DifA was previously predicted to be a 43.5-kDa protein with an ATG start codon (Fig. 1A) (10, 17, 29, 48). However, its apparent molecular mass is about 50 kDa, as estimated by SDS-PAGE and immunoblotting (Fig. 1B). This indicated that the actual start for DifA could be about 50 codons upstream. Indeed, the complementation of a difA mutation required more DNA at the 5′ end than expected (10). We mutated this suspected ATG start codon to the alanine codon GCA. This mutation, designated M49A (also see later sections), showed no obvious effect on the level of DifA protein (Fig. 1B) but resulted in substantial increases in EPS level over the wild type (WT) (Fig. 1C). These results indicated that this ATG is a methionine codon within the difA coding region instead of the start codon.
To identify the actual start codon for DifA, primer extension was used to first find the transcription initiation site upstream of difA. This analysis consistently revealed two distinct transcription start sites (see Fig. S1 in the supplemental material), one at a thymidine and another at a guanine 427 and 280 bases upstream of the ATG codon (Fig. 1A), respectively. The start codon for DifA must reside in the region between this thymidine and the ATG codon for methionine. Although no in-frame ATGs exist in this region, there are two in-frame GTGs (Fig. 1A) that could be used as start codons in M. xanthus (43, 44). When the upstream GTG was mutated to GTC, the resulting mutation (−138GTC) did not eliminate or alter the size of DifA protein (Fig. 1B) and EPS production was still detected (Fig. 1C). In contrast, when the second or downstream GTG was mutated to a GTC, the resulting mutation (+1GTC) led to the elimination of DifA protein (Fig. 1B), EPS production (Fig. 1C), development, and S motility (data not shown). When this GTG was mutated to an ATG, this mutation (+1ATG) resulted in increases in DifA protein as well as EPS levels (Fig. 1B and C). The calculated molecular mass of the polypeptide initiating from this GTG is 49.1 kDa, consistent with that of DifA estimated from SDS-PAGE (Fig. 1B). We concluded that the start codon for DifA is the second GTG, which extends the DifA N terminus an additional 48 amino acids from the previous prediction (Fig. 1A).
The cytoplasmic N terminus of DifA is required for EPS production.
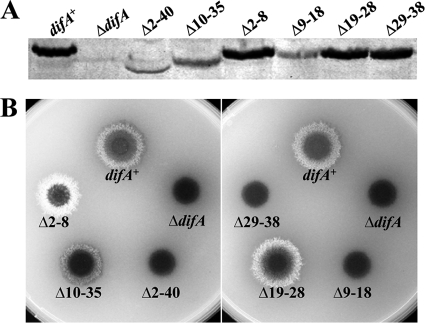
A reanalysis using TMHMM (27) predicted DifA to have two transmembrane (TM) helices near the N terminus (Fig. 1A), with both its N and C termini in the cytoplasm. The predicted topology of DifA is reminiscent of the E. coli aerotaxis receptor Aer (6, 36, 41) in two ways. On one hand, it has a longer cytoplasmic N-terminus extension (NTE; amino acids 1 to 45) than the more canonical chemoreceptors. On the other, its predicted periplasmic region of nine residues is much shorter (amino acids 69 to 77) than those in other receptors. As such, its cytoplasmic NTE could be critical in sensing and transducing signals (12). To examine this possibility, amino acids 2 to 40 and 10 to 35 were deleted from DifA, respectively (Fig. 2A). The first deletion (Δ2-40) resulted in undetectable levels of EPS (Fig. 2B) but it also severely reduced the level of DifA, as analyzed by immunoblotting (Fig. 2A). The second deletion (Δ10-35) diminished DifA levels to a lesser extent (Fig. 2A) and resulted in an intermediate EPS phenotype (Fig. 2B). Since these results were inconclusive, smaller deletion mutants encompassing amino acids 2 to 8, 9 to 18, 19 to 28, and 29 to 38 were constructed. The Δ9-18 mutation resembled the first large deletion (Δ2-40) and led to significantly reduced levels of DifA and undetectable EPS (Fig. 2). However, the other three mutations (Δ2-8, Δ19-28, and Δ29-38) had no obvious effect on DifA levels (Fig. 2A) while resulting in a spectrum of EPS levels (Fig. 2B). The Δ2-8 mutant clearly overproduced EPS, the Δ19-28 mutant had EPS levels similar to or slightly above that of the difA+ control, and the Δ29-38 mutant produced no detectable EPS (Fig. 2B). These results (Fig. 2) indicate that NTE is critical for DifA function in EPS regulation, likely due to its ability to influence the signaling properties of this M. xanthus sensory transducer.
FIG. 2.
Deletions in the N-terminus extension of DifA affect EPS production. Shown are immunoblot analysis (A) and EPS production (B) of mutants with deletions in the predicted cytoplasmic N terminus of DifA. Experiments were performed as described for Fig. 1B and C. Strains: difA+, YZ619; ΔdifA, YZ601; Δ2-40, YZ776; Δ10-35, YZ1710; Δ2-8, YZ1711; Δ9-18, YZ1712; Δ19-28, YZ1713; and Δ29-38, YZ1714.
DifA transmembrane helices participate in signal transduction.
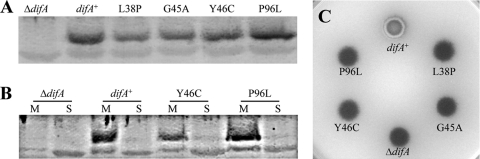
Random mutagenesis was used to probe whether the predicted TM1 and TM2 (residues 46 to 68 and 78 to 100) (Fig. 1) are important for signaling by DifA. The codons for amino acids 35 to 136 were subjected to error-prone PCR mutagenesis (Fig. 1) (Materials and Methods) using a plasmid that is able to complement difA deletions (10). Mutagenized plasmids were transformed into a difA deletion mutant (YZ601), and the transformants were screened on plates with the dye Congo red to detect abnormalities in EPS production (7). Two mutations preceding the first TM helix, L38P and G45A, were found to eliminate EPS production (Fig. 3C), with minor effects on the level of DifA (Fig. 3A); this further validated the above conclusion that the NTE is crucial for DifA function in EPS regulation. More importantly, the Y46C and P96L mutations in TM1 and TM2, respectively (Fig. 1A), were found to eliminate EPS production, without obvious reduction in DifA (Fig. 3A) or alteration in its membrane association (Fig. 3B). Since the M49A mutation, which led to a drastic increase in EPS production (Fig. 1C), is predicted to be in TM1 (Fig. 1A), these results indicate that the DifA TM helices are likely active participants in signal transduction rather than merely passive membrane anchors for DifA.
FIG. 3.
Point mutations in the N terminus of DifA influence EPS production. (A) DifA protein levels by immunoblot analysis. Four micrograms of whole-cell lysates from each strain was analyzed by SDS-PAGE and probed with polyclonal anti-DifA antibodies. (B) DifA membrane localization. The membrane (M) and the soluble (S) fractions normalized to whole-cell lysates were analyzed by immunoblotting as in panel A. (C) EPS production of the point mutants was examined as described for Fig. 1C. Strains: difA+, YZ619; ΔdifA, YZ601; L38P, YZ1822; G45A, YZ1820; Y46C, YZ1821; and P96L, YZ1823.
Indels are critical for DifA function.
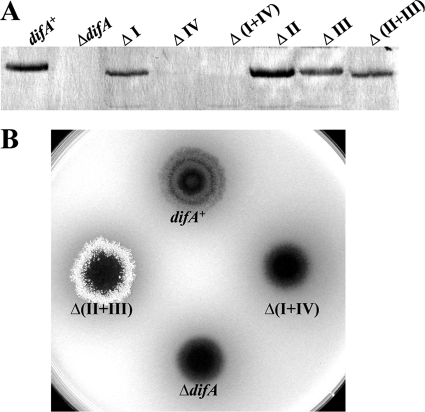
MCPs are categorized into seven major classes based on the multiples of seven amino acid (7-aa) heptads in their cytoplasmic domains (2). Classes 44H, 40H, and 36H among them were recognized earlier based on the presence of two pairs of 14-aa insertion/deletion (indel) segments (30). DifA belongs to 44H with four 14-aa indels (see Fig. S2 in the supplemental material). These indels in DifA can be designated I, II, III, and IV from the N to the C termini (30, 40) (Fig. S2). The pair I and IV are absent from both 36H and 40H while the pair II and III are absent in 36H but present in 40H (2, 40). Each pair of these indels (I plus IV and II plus III) was deleted from DifA to examine their effects on EPS regulation by DifA. As shown in Fig. 4B, the Δ(I+IV) mutant had no detectable EPS or DifA (Fig. 4). In contrast, the Δ(II+III) mutant had drastically elevated levels of EPS, with slightly reduced amounts of DifA protein (Fig. 4). The deletions of single indels, regardless of their effect on DifA protein level (Fig. 4A), all led to an EPS− phenotype (data not shown). The results here indicated that the simultaneous deletion of indels II and III converts DifA into an actively signaling conformation.
FIG. 4.
Deletions of DifA indels impact EPS production. Shown are immunoblot analysis (A) and EPS production (B) of indicated deletions of indel segments. See Fig. 1 for experimental details. Strains: difA+, YZ619; ΔdifA, YZ601; ΔI, YZ777; ΔIV, YZ778; Δ(I+IV), YZ779; ΔII, YZ780; ΔIII, YZ781; and Δ(II+III), YZ782.
Five glutamate (E) and glutamine (Q) residues are critical for DifA function in EPS regulation.
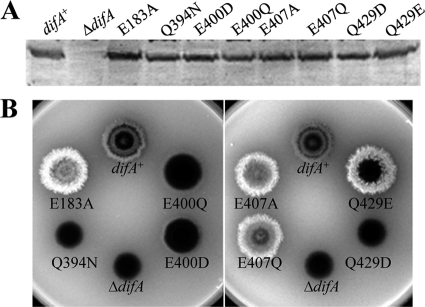
DifA is likely unmethylated despite its homology to MCP transducers (10, 45). Yet, DifA still contains five doublets and one triplet of glutamate (E) and/or glutamine (Q) in its predicted methylation subdomains (10) (see Fig. S2 in the supplemental material). This is surprising because the second residue in such an E/Q pair can be methylated in many known MCPs (40). The presence of these pairs in DifA suggests that selective pressure other than methylation may be at work for DifA. In this study, mutations were constructed at E158, E183, Q394, E400, and E407, the second residue of the five E/Q pairs (Fig. S2). In addition, E428 and Q429, the second and the third residues in a QEQ triplet (Fig. S2), were also targeted for mutagenesis. These seven residues were all mutated to aspartate (D) and alanine (A). Reciprocal changes were also made between E and Q (Table 3). The two glutamines (Q346 and Q381) were additionally mutated to asparagine (N). None of the mutations was found to impact the protein level or the apparent electrophoretic mobility of DifA (selected data shown in Fig. 5A), confirming the lack of methylation from previous studies (10, 45). Mutations of five of the seven residues (E183, Q394, E400, E407, and Q429) were found to alter EPS production, as summarized in Table 3 and Fig. 5. When replaced by an alanine (A), three of the five residues (E183, E407 and Q429) resulted in increased EPS production. When replaced by others, their mutations resulted in various phenotypes from no EPS production to EPS overproduction (Table 3 and Fig. 5B). The replacement of the other two residues (Q394 and E400) only led to decreases in EPS production. These results indicate that these five residues (E183, Q394, E400, E407, and Q429) likely play critical roles in the signaling function of DifA in EPS regulation.
TABLE 3.
EPS production of the mutants with single substitutions
| Residue | Result for substitutiona: |
||||
|---|---|---|---|---|---|
| D | Q | E | A | N | |
| E158 | + | + | NA | + | NA |
| E183 | +/− | + | NA | ++ | NA |
| Q394 | − | NA | − | + | − |
| E400 | +/− | +/− | NA | − | NA |
| E407 | + | ++ | NA | ++ | NA |
| E428 | + | + | NA | + | NA |
| Q429 | − | NA | ++ | ++ | + |
+, the strain produces a similar level of EPS to that of YZ619 (ΔdifA/difA+); −, no detectable EPS production; ++, EPS production is significantly increased over the wild type; +/−, EPS production is significantly reduced. NA, not applicable.
FIG. 5.
Mutations of certain E/Q residues impact EPS production. Immunoblot analysis (A) and EPS production (B) of indicated glutamate (E) or glutamine (Q) substitution mutants. See the legend to Fig. 1 for experimental details. Strains: difA+, YZ619; ΔdifA, YZ601; E183A, YZ745; Q394N, YZ745; E400D, YZ714; E400Q, YZ751; E407A, YZ748; E407Q, YZ738; and Q429E, YZ763.
DISCUSSION
Here we focused on dissecting the signaling mechanisms of DifA, an MCP-like signal transducer essential for S motility and development of M. xanthus. The experimental identification of the start codon extends the N terminus of DifA for an additional 48 amino acids from previous computational predictions (Fig. 1). Bioinformatics predicts DifA to have two TM segments and a short periplasmic region with both its N and C termini in the cytoplasm. Figure 6A depicts the predicted membrane topology of DifA and provides a summary of the mutational analysis in this study. Our results suggest that DifA may use its cytoplasmic N-terminal extension (NTE) to sense signals in the cell (Fig. 2), which sheds light on how DifA could sense input signals for EPS regulation without a prominent periplasmic domain. In addition, transmembrane signaling appears critical for the signal transmission by DifA from its N-terminal sensory domain to the C-terminal output or signaling domain (Fig. 1 and 2).
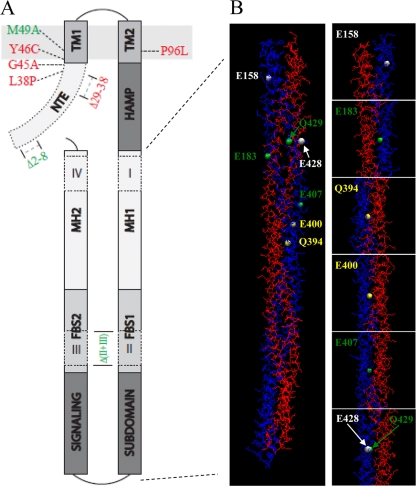
FIG. 6.
(A) Predicted topology of DifA and the effect of selected mutations on EPS production. The N-terminal extension (NTE) is cytoplasmic. It is followed by two transmembrane helices, TM1 and TM2, which are separated by nine residues in the periplasm. The HAMP domain connects TM2 to the C-terminal signaling domain, which forms a coiled-coil hairpin with three subdomains: the signaling subdomain, flexible bundle subdomain (FBS1 and FBS2), and the methylation helices (MH1 and MH2). Indels I and IV are in the MHs, whereas II and III are in the FBSs. Also indicated in the drawing are the positions of selected mutations and their effects on EPS production. Only mutations with minimum impact on DifA protein level are shown, and those in green led to increased EPS production, while those in red resulted in reduced or undetectable EPS levels. (B) The modeled structure of a DifAC dimer and the position of seven mutated E/Q residues in Fig. 5. The DifAC dimer is shown on the left. The two chains of the dimer are labeled blue (chain A) and red (chain B), respectively. The positions of the mutated E/Q residues in chain A are shown as white, green, or yellow spheres, corresponding to their mutations with either no phenotype, reduced EPS level only, or both increased and decreased EPS levels, respectively (see text). For a clearer illustration of the dimer interface, the panels on the right show the zoomed-in view of residues that were mutated. The same E/Q residues in chain B of the dimer are located in similar positions to those in chain A, although they are not in perfect symmetry.
We propose the following working model for DifA signaling. First, the N terminus of DifA receives input signals through protein-protein interactions with TFP components or other unknown partners upstream. The sensory information received by DifA is then relayed to the C-terminal signaling domain. We suggest that the intramolecular signal transmission by DifA from its N to C termini passes through the two TM helices to cause a piston-like movement of TM2, as proposed for classical MCPs (19, 20). We speculate that the activity of DifA to stimulate DifE kinase downstream is modulated by the conformation of its flexible bundle subdomain (FBS) (2) and by interactions at the dimer interface of DifA. The rest of the Discussion section focuses on experimental results here and elsewhere that are supportive of or consistent with this working model.
The results of computational and mutational analyses of the N-terminus extension of DifA suggest that this region may detect upstream signals through protein-protein interactions. Experimentally, the N-terminal lesions of Δ2-8, Δ29-38, L38P, and G45A in DifA either abolished or increased EPS production without major effects on the level of DifA protein (Fig. 3 and 6A). In other words, the cytoplasmic N terminus of DifA can produce both signal-on and signal-off mutations similarly to the PAS domain or the signal detection domain of Aer (12). This and the overall structural similarity between Aer and DifA, together with the understanding of the functional features of MCP, argue that DifA likely uses its N terminus, instead of any other regions, to receive input signals. Previous studies indicated that TFP likely mediates signal input into the Dif pathway either directly or indirectly (7). It is known that Pil proteins form a TFP complex that transverses the cell envelope (4). Some Pil proteins (such as PilC) are transmembrane with extensive cytoplasmic domains, while others (such as PilT) are entirely cytoplasmic (4). Since computational analysis of the 45-aa-long DifA N terminus identified no homology to other proteins and predicted no prominent secondary structure (not shown), and since its 16 residues are charged amino acids (Fig. 1), this short region is unlikely structured as or folds into an actual domain on its own. We speculate, without experimental proof, that the DifA N terminus interacts with Pil proteins either directly or indirectly for signal perception in EPS regulation.
We favor a transmembrane signaling mechanism for the communication between the NTE and the output or signaling domain of DifA. Two mutations, Y46C and P96L, in the predicted TM1 and TM2, respectively, were found to abolish EPS production without drastic effects on DifA level or its membrane association (Fig. 3 and 6A). More importantly, located in TM1 is the M49A mutation that greatly increased EPS production (Fig. 1C and 6A). It has been demonstrated that signal transmission from the input to the output domain in E. coli chemoreceptors involves a piston-like movement of TM2 or the signaling helix (16, 19, 20). In our opinion, if the TMs were bypassed by DifA for signal transmission, the presence of a signal-on or constitutively active mutation in TM1 as well as the signal-off mutations in both TM1 and TM2 (Fig. 6A) would be more difficult to explain. Since these EPS− mutations did not alter DifA membrane localization, it is likely that the TMs are not merely membrane anchors but rather active participants in signal transmission. We speculate that a conformational change in the N terminus of DifA is transmitted through TM1, resulting in movement of TM2 similar to that of the signaling helix in E. coli MCPs. This speculative TM1-to-TM2 signal transmission could involve the short periplasmic region or be mediated by direct interactions between the TMs. The Dif pathway was previously engineered to respond to nitrate by use of a NarX-DifA chimera (47). Recent structural studies of the NarX sensory module clearly demonstrated a piston-like displacement in its transmembrane signaling helix or TM2 with and without nitrate (14). Since the NarX-DifA chimera included the entire NarX sensory module, including TM1 and TM2 (47), our previous observation is in agreement with a transmembrane-dependent signaling mechanism by DifA (14, 39, 47). More definitive and prudent conclusions regarding the various mechanisms of intramolecular signal transmission by DifA require additional experimentation.
DifA and other 44H class MCP transducers (2) have the longest C-terminal signaling domain (10, 47, 48). It is known that this domain forms a coiled-coil hairpin that folds into a four-helix bundle in a receptor dimer (Fig. 6). The most striking phenotype from our indel deletions was the increased EPS production by the Δ(II+III) mutant (Fig. 6A and 4B). Since indels II and III are in the flexible bundle subdomain (FBS) (2), the observation here is supportive of FBS playing critical roles in MCP signal transmission, as put forth by Alexander and Zhulin (2). It has been proposed that reduced flexibility in the MCP signaling domain corresponds with the kinase-on state (16, 19, 20). We suggest that the shortening of FBS reduces the flexibility of DifA, which in turn leads to activation of the downstream kinase and stimulation of EPS production.
For transmembrane signaling by MCPs, a receptor dimer is the basic functional unit because it is sufficient to transmit extracellular signals to conformational changes in the signaling domain (19). As previous studies provided evidence of DifA dimer formation (49), a dimer of the C terminus of DifA (DifAC) was generated by molecular modeling (see Materials and Methods and see Fig. S2 in the supplemental material,) to examine the positions of the mutated glutamates (E) and glutamines (Q) in the C terminus (Fig. 5 and Table 3). For this purpose, the dimer of T. maritima MCP1143C (35), a 44H MCP (Fig. S2) (2, 40), was used as the template for modeling with SWISS-MODEL (3, 18, 38). Figure 6 is a representation of the modeled DifAC dimer. E183, Q394, E400, E407, and Q429, whose mutations affected EPS production (Fig. 4 and Table 3), are all located at the dimer interface, and there are no other residues hindering their interactions with the other monomer (Fig. 6). In contrast, E158 is quite distant from the dimer interface (Fig. 6; see Fig. S3 in the supplemental material). Similarly, the side chain of E428 is not as close to the other monomer as the neighboring amino acids (Fig. S3). While the modeled structure of DifA should be viewed with caution, it suggested that the important E/Q residues may correlate with their proximity to the dimer interface. Since the stabilization/strengthening and destabilization/weakening of the dimer interface of chemoreceptors likely regulate the activity of the downstream kinases (19), our observations are consistent with the dimeric interface of DifA affecting the activity of downstream targets in a similar fashion.
Supplementary Material
Acknowledgments
This work was supported by grant GM071601 and ARRA supplements from the National Institutes of Health to Z.Y. H.M.N. was a MEAMP fellow at Virginia Tech.
We thank Florian Schubot for helpful discussions.
Footnotes
Published ahead of print on 3 December 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexander, R. 2007. Evolutionary genomics of methyl-accepting chemotaxis proteins. Georgia Institute of Technology, Atlanta, GA.
- 2.Alexander, R. P., and I. B. Zhulin. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Nat. Acad. Sci. U. S. A. 104:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 4.Ayers, M., P. L. Howell, and L. L. Burrows. 2010. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5:1203-1218. [DOI] [PubMed] [Google Scholar]
- 5.Bellenger, K., X. Ma, W. Shi, and Z. Yang. 2002. A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis. J. Bacteriol. 184:5654-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, W. P., Q. Xu, and Z. Yang. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61:447-456. [DOI] [PubMed] [Google Scholar]
- 8.Black, W. P., and Z. Yang. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J. Bacteriol. 186:1001-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonner, P. J., and L. J. Shimkets. 2006. Phospholipid directed motility of surface-motile bacteria. Mol. Microbiol. 61:1101-1109. [DOI] [PubMed] [Google Scholar]
- 10.Bonner, P. J., et al. 2005. The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol. Microbiol. 57:1499-1508. [DOI] [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Campbell, A. J., K. J. Watts, M. S. Johnson, and B. L. Taylor. 2010. Gain-of-function mutations cluster in distinct regions associated with the signalling pathway in the PAS domain of the aerotaxis receptor, Aer. Mol. Microbiol. 77:575-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campos, J. M., and D. R. Zusman. 1975. Regulation of development in Myxococcus xanthus: effect of 3′:5′-cyclic AMP, ADP, and nutrition. Proc. Natl. Acad. Sci. U. S. A. 72:518-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, J., and W. A. Hendrickson. 2009. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure 17:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis, P. D., R. Geyer, D. C. White, and L. J. Shimkets. 2006. Novel lipids in Myxococcus xanthus and their role in chemotaxis. Environ. Microbiol. 8:1935-1949. [DOI] [PubMed] [Google Scholar]
- 16.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldman, B. S., et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 19.Hazelbauer, G. L., J. J. Falke, and J. S. Parkinson. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazelbauer, G. L., and W. C. Lai. 2010. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr. Opin. Microbiol. 13:124-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171:167-176. [Google Scholar]
- 22.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus: two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 23.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:27-28, 33-38. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 26.Kearns, D. B., and L. J. Shimkets. 2001. Lipid chemotaxis and signal transduction in Myxococcus xanthus. Trends Microbiol. 9:126-129. [DOI] [PubMed] [Google Scholar]
- 27.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 28.Kroos, L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13-39. [DOI] [PubMed] [Google Scholar]
- 29.Lancero, H., et al. 2002. Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J. Bacteriol. 184:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568-585. [DOI] [PubMed] [Google Scholar]
- 31.Leung, D. W., E. Chen, and D. V. Goeddel. 1989. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11-15. [Google Scholar]
- 32.Mauriello, E. M., T. Mignot, Z. Yang, and D. R. Zusman. 2010. Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol. Mol. Biol. Rev. 74:229-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride, M. J., T. Kohler, and D. R. Zusman. 1992. Methylation of FrzCD, a methyl-accepting taxis protein of Myxococcus xanthus, is correlated with factors affecting cell behavior. J. Bacteriol. 174:4246-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Park, S.-Y., et al. 2006. Reconstruction of the chemotaxis receptor-kinase assembly. Nat. Struct. Mol. Biol. 13:400-407. [DOI] [PubMed] [Google Scholar]
- 36.Rebbapragada, A., et al. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. U. S. A. 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart, V., and L.-L. Chen. 2010. The S helix mediates signal transmission as a HAMP domain coiled-coil extension in the NarX nitrate sensor from Escherichia coli K-12. J. Bacteriol. 192:734-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szurmant, H., and G. W. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor, B. L. 2007. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 65:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Velicer, G. J., and M. Vos. 2009. Sociobiology of the myxobacteria. Annu. Rev. Microbiol. 63:599-623. [DOI] [PubMed] [Google Scholar]
- 43.Ward, M. J., H. Lew, and D. R. Zusman. 2000. Disruption of aldA influences the developmental process in Myxococcus xanthus. J. Bacteriol. 182:546-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 45.Xu, Q., W. P. Black, C. L. Cadieux, and Z. Yang. 2008. Independence and interdependence of Dif and Frz chemosensory pathways in Myxococcus xanthus chemotaxis. Mol. Microbiol. 69:714-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, Q., W. P. Black, E. M. Mauriello, D. R. Zusman, and Z. Yang. 2007. Chemotaxis mediated by NarX-FrzCD chimeras and nonadapting repellent responses in Myxococcus xanthus. Mol. Microbiol. 66:1370-1381. [DOI] [PubMed] [Google Scholar]
- 47.Xu, Q., W. P. Black, S. M. Ward, and Z. Yang. 2005. Nitrate-dependent activation of the Dif signaling pathway of Myxococcus xanthus mediated by a NarX-DifA interspecies chimera. J. Bacteriol. 187:6410-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, Z., Y. Geng, D. Xu, H. B. Kaplan, and W. Shi. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 30:1123-1130. [DOI] [PubMed] [Google Scholar]
- 49.Yang, Z., and Z. Li. 2005. Demonstration of interactions among Myxococcus xanthus Dif chemotaxis-like proteins by the yeast two-hybrid system. Arch. Microbiol. 183:243-252. [DOI] [PubMed] [Google Scholar]
- 50.Yang, Z., et al. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zusman, D. R., A. E. Scott, Z. Yang, and J. R. Kirby. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat. Rev. Microbiol. 5:862-872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.