Abstract
Previous studies revealed that one species of methanogenic archaea, Methanocaldococcus jannaschii, is polyploid, while a second species, Methanothermobacter thermoautotrophicus, is diploid. To further investigate the distribution of ploidy in methanogenic archaea, species of two additional genera—Methanosarcina acetivorans and Methanococcus maripaludis—were investigated. M. acetivorans was found to be polyploid during fast growth (tD = 6 h; 17 genome copies) and oligoploid during slow growth (doubling time = 49 h; 3 genome copies). M. maripaludis has the highest ploidy level found for any archaeal species, with up to 55 genome copies in exponential phase and ca. 30 in stationary phase. A compilation of archaeal species with quantified ploidy levels reveals a clear dichotomy between Euryarchaeota and Crenarchaeota: none of seven euryarchaeal species of six genera is monoploid (haploid), while, in contrast, all six crenarchaeal species of four genera are monoploid, indicating significant genetic differences between these two kingdoms. Polyploidy in asexual species should lead to accumulation of inactivating mutations until the number of intact chromosomes per cell drops to zero (called “Muller's ratchet”). A mechanism to equalize the genome copies, such as gene conversion, would counteract this phenomenon. Making use of a previously constructed heterozygous mutant strain of the polyploid M. maripaludis we could show that in the absence of selection very fast equalization of genomes in M. maripaludis took place probably via a gene conversion mechanism. In addition, it was shown that the velocity of this phenomenon is inversely correlated to the strength of selection.
The existence of multiple copies of the genome in one cell is called polyploidy. If the genomes originate from several species, the resulting species is allopolyploid, while the multiplication of the chromosomes of one species leads to autopolyploidy. Many eukaryotes are polyploid, especially flowering plants, but also fish and amphibians. In evolution, the ploidy level can change in both directions, and it has been proposed that the diploid vertebrate genomes were derived by reduction from polyploid ancestors (49). The advantages and disadvantages of polyploidy have been discussed in several recent reviews (7, 16, 38). The advantages are more obvious for allopolyploids, in which alleles of two or more species are combined. They typically outperform their parent strains (heterosis effect). However, autoploidy also offers advantages, e.g., gene redundancy. Gene redundancy can be accompanied by higher resistance against DNA-damaging agents, and it offers the possibility of mutating one copy of a gene, while the wild-type information still remains available.
In contrast to eukaryotes, prokaryotes are usually thought to contain one copy of a circular chromosome. This is typically called “haploidy,” although the term “haploid” does not seem to make much sense in species that do not have a “diploid” stage. The term “monoploid” is probably more appropriate and will therefore be used here. It is also used for flowering plants with a C value of one (the C value expresses the haploid complement of the genome from parental contributions [see, for example, reference 31]). The best-studied bacterial species, Escherichia coli, is monoploid when it is grown under conditions where the doubling time is longer than the time to replicate the chromosome and segregate the products (4, 39). When E. coli is grown under optimal conditions in the laboratory, the generation time becomes shorter than the replication/segregation time, leading to reinitiation of replication before the previous replication round had been terminated. The number of replication origins per cell is then larger than the number of termini, and the cell becomes mero-oligoploid (4). However, it is not really clear whether these fast-growth conditions are relevant for E. coli growing in natural habitats. The best-studied Gram-positive bacterium, Bacillus subtilis, is also monoploid (47), as are several additional species. However, other bacterial species have been shown to be polyploid, e.g., Deinococcus radiodurans, Desulfovibrio gigas, and Borrelia hermsii (15, 19, 30). Since the number of “exceptions” has become greater than the number of species that adhere to the rule, it might be questioned whether monoploidy is really “typical” for bacteria. A review will summarize the current knowledge about ploidy levels and the possible evolutionary advantages of polyploidy in bacteria (J. Soppa, unpublished data).
The situation is even less clear in archaea, because the number of species with an experimentally determined genome copy number is rather limited. A few crenarchaeal species from four different genera were all found to be monoploid [3, 24]). The number of euryachaeal species with an experimentally determined genome copy number is even smaller. A few species of haloarchaea from two genera have been shown to be polyploid under several different conditions (5, 6); therefore, it might be speculated that polyploidy is typical for and widespread in haloarchaea. Only two species of methanogenic archaea have been analyzed: Methanocaldococcus jannaschii was found to be polyploid (27), whereas the filamentous Methanothermobacter thermautotrophicus was found to be diploid (26). To better understand the situation in methanogenic archaea, one member of two additional genera from this group was analyzed. The ploidy levels were determined at different growth rates throughout culture growth. In addition, we made use of a heterozygous mutant strain that was recently constructed (44) to analyze the influence of different selection pressures on the velocity of gene conversion.
MATERIALS AND METHODS
Archaeal and bacterial strains and culture conditions.
Methanosarcina acetivorans C2A (DSM 2834 [43]) was cultivated as single cells in high-salt (HS) medium under strictly anaerobic conditions at 37°C as described previously (29). Either 125 mM methanol or 120 mM sodium acetate was used as the sole energy source. Methanococcus maripaludis S2 (DSM 14266 [48]) was grown anaerobically in McSe medium containing selenite (34), Casamino Acids, and acetate (48) at 37°C. The culture tubes were slightly agitated to prevent cell aggregation and to facilitate mass transfer at the gas-liquid interface. When grown autotrophically, M. maripaludis cultures were pressurized with 2 × 105 Pa of H2-CO2 (80:20); for growth on sodium formate (2% [wt/vol]), 0.5 × 105 Pa of N2-CO2 (80:20) was applied, and 80 mM morpholinepropanesulfonic acid (pH 6.8) was added to keep the pH constant. To exert various selective pressures for the presence of the codon-optimized pac (pacN, encoding the puromycin acetyltransferase [12] gene in M. maripaludis strain SkoD4 [44]), the medium was supplemented with different puromycin concentrations.
E. coli B was obtained from the German culture collection (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ], Braunschweig, Germany; DSMZ no. 2840) and grown either in complex medium (SOB+) or in M9 synthetic medium with 0.4% (wt/vol) succinate as a carbon and energy source (35).
Determination of cell densities and generation of lysates.
To derive growth curves, the optical density at 578 nm (OD578) of the cultures was determined by using a Spectronic 20 photometer (Thermo, Dreieich, Germany). Five independent cultures were grown, and the average OD578 values and their variances were calculated.
For every genome copy number determination, aliquots (0.5 or 1 ml, depending on the cell densities) were anaerobically withdrawn from each culture with sterile needles. The cell densities of the samples were determined by using a Neubauer counting chamber. After centrifugation (10 to 30 min, 16,000 × g, room temperature), the cell pellet was resuspended in distilled water (200 or 100 μl) and mixed thoroughly, resulting in cell lysis. Completeness of lysis was verified microscopically, and the integrity of genomic DNA was verified by agarose gel electrophoresis. Aliquots of the chromosomal DNA were dialyzed on membrane filters against distilled water. Serial dilutions were generated, and 5-μl aliquots were included as a template in real-time PCR analyses for quantification of genome copy numbers (see below).
Quantification of genome copy numbers using a real-time PCR method.
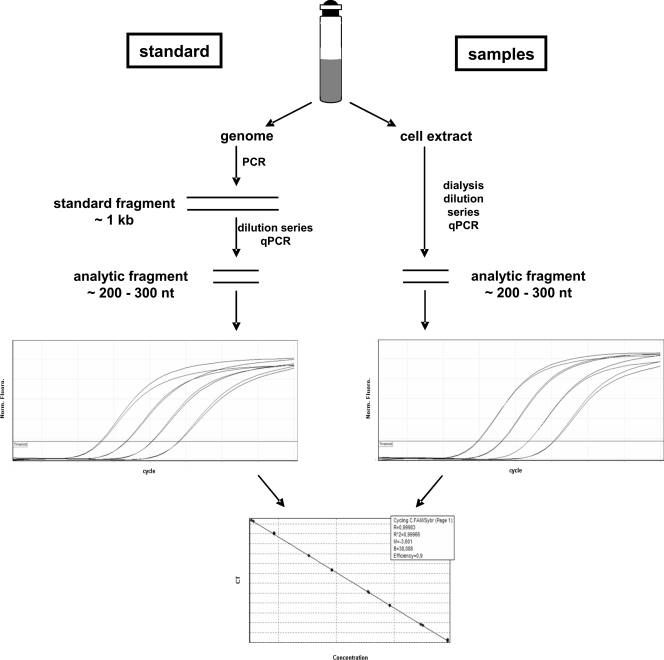
To determine genome copy numbers, a recently developed real-time PCR approach was applied (5). A schematic overview of the method is given in Fig. 1. At first, fragments of ∼1 kbp were amplified using standard PCRs with isolated genomic DNA of M. acetivorans or M. maripaludis as templates. A list of primers used is given in Table 1 . The fragments were purified by using preparative agarose gel electrophoresis and an AxyPrepDNA gel extraction kit (Axygen Biosciences). The DNA mass concentrations were determined photometrically, and the concentrations of DNA molecules were calculated using the molecular weights computed with “oligo calc” (www.basic.northwestern.edu/biotools). For each standard fragment, a dilution series was generated and used for real-time PCR analysis in parallel with the dilution series of the respective cell extract. The “analysis fragments” were ∼300 bp and were internal to the standard fragments. Real-time PCR analyses were performed as previously described (5). By comparison of the threshold cycle (CT) differences of the different dilutions it was verified that the PCR was exponential at least up to the threshold DNA concentration used for the analysis (i.e., a 10-fold dilution corresponds to a CT difference of ∼3.32). A standard curve was generated and used to calculate the genome copy numbers present in the dilutions of the cell extract. In combination with the cell density, the number of genome copies per cell can be calculated.
FIG. 1.
Overview of the method for the quantification of genome copy numbers. Details are explained in the text.
TABLE 1.
Sequences and applications of oligonucleotides
| Oligonucleotide | Sequence (5′-3′) | Application |
|---|---|---|
| S_5′-msaceti_DM | GTGCTGGAAGAACATCGGGAACAGTTAAGG | Synthesis of real-time PCR standard of M. acetivorans |
| S_3′-msaceti_DM | GGTCACCCAGGAAATCAGCGACAATAAACC | Synthesis of real-time PCR standard of M. acetivorans |
| A_5′-msaceti_DM | GCCGGTCTTGCCGGAATAATCTGCAATAGC | Detection of genome copies in M. acetivorans |
| A_3′-msaceti_DM | ACCAGACATTGCCGGTACATCGTCTCAAGC | Detection of genome copies in M. acetivorans |
| S_5′-mcmari_DM | GCAGGTTCGATTCTTTCGTGGCCATAAGGG | Synthesis of real-time PCR standard of M. maripaludis |
| S_3′-mcmari_DM | GCAGGTCAGGATACCGACATTGACATATGC | Synthesis of real-time PCR standard of M. maripaludis |
| A_5′-mcmari_DM | CAGCAACCTCCTTCAATACCCTCGATTTCG | Detection of genome copies in M. maripaludis |
| A_3′-mcmari_DM | ACAAGGATTGTTGGAGAACATGGCGACAGC | Detection of genome copies in M. maripaludis |
| S_5′-mcmari_WT | GTACCCTGCAAATACTGCAACCATAGCTACAGCCAG | Synthesis of real-time PCR standard of M. maripaludis wt gene conversion |
| S_3′-mcmari_WT | CCACGATTATTGGCGGGCATACCATTTTAAATC | Synthesis of real-time PCR standard of M. maripaludis wt gene conversion |
| A_5′-mcmari_WT1 | CCCAAAATACCAAATCCTGTGACATCAGTCATTGC | Detection of genome copies in M. maripaludis wt gene conversion |
| A_3′-mcmari_WT1 | GCTATCAAGGGTTACCGAGGAATTTGAAGATTTAATCG | Detection of genome copies in M. maripaludis wt gene conversion |
| S_5′-mcmari_Mut | GCAGGAGGTGATCATATGACAGAATACAAACCAACAG | Synthesis of real-time PCR standard of M. maripaludis mutant gene conversion |
| S_3′-mcmari_Mut | CCTTTTCCACTGCAAAGAGCATGTCCAAGCTAG | Synthesis of real-time PCR standard of M. maripaludis mutant gene conversion |
| A_5′-mcmari_Mut | GTAAAGTTTGGGTAGCTGATGATGGTGCTGCTG | Detection of genome copies in M. maripaludis mutant gene conversion |
| A_3′-mcmari_Mut | CAGCTGCTTCTACACCAGGTAATACAACTGCTGAACC | Detection of genome copies in M. maripaludis mutant gene conversion |
The following points have to be optimized for every new species under investigation and have been optimized for the two species of methanogenic archaea used here (M. acetivorans and M. maripaludis): (i) the cell density has to be quantified with a very low variance; (ii) the method of cell disruption has to be ca. 100% effective, yet leaving the genomic DNA intact; and (iii) the real-time PCR has to be truly exponential, i.e., the differences in CT values of 10-fold dilutions of the templates (standard fragment and cell extract) have to be ∼3.32.
Quantification of genome copy numbers using a spectroscopic approach.
This approach was used for M. maripaludis, as well as for E. coli B. For E. coli, cells from 2-ml portions of an exponentially growing culture were harvested by centrifugation and resuspended in 0.2 ml of distilled water. The cells were lysed by adding lysozyme to a final concentration of 0.2 mg/ml, followed by incubation for 2 h at room temperature. By counting the cell density before lysozyme addition and after incubation, it was verified that more than 99% of the cells had been lysed. Cell debris was removed by centrifugation. DNase-free RNase (Applichem, Darmstadt, Germany) was added to a final concentration of 10 μg/ml to the supernatant. It was incubated for 1 h at room temperature to ensure RNA digestion. DNA was precipitated by adding a 1/10 volume of potassium acetate (3 M, pH 5.5) and 2.5 volumes of ethanol, followed by a 20-min incubation at −25°C. The precipitated DNA was pelleted by centrifugation (20 min, 4°C, 13,000 × g) and washed with 70% ethanol. The DNA was dissolved in 0.1 ml of Tris-EDTA by incubation at 65°C for several hours.
Genomic DNA of M. maripaludis was isolated by adopting a method published by Eikmanns et al. (10), which includes a treatment with protease and RNase, protein removal, and subsequent precipitation of genomic DNA. Cells from 1.5 ml of an exponential M. maripaludis S2 culture (cell density of 3.5 × 108 to 3.9 × 108 cells/ml) from formate medium were harvested by centrifugation and suspended in 566 μl of 850 mM sucrose with 80 mM NaHCO3. Then, 1 μl of 1 mg of RNase A/ml, 30 μl of 10% (wt/vol) sodium dodecyl sulfate, and 3 μl of 20 mg of proteinase K/ml were added, and the sample was incubated for 2 h at 60°C. After this, 150 μl of saturated NaCl was added, the proteins were mixed thoroughly and pelleted by a 30-min centrifugation at 2,000 × g, and the clear supernatant was transferred into a new reaction cup. The genomic DNA was precipitated by adding a 0.6 volume of isopropanol, followed by 1 h of incubation at −20°C and 30 min of centrifugation at 14,000 × g and 4°C. The DNA pellet was washed with 250 μl of 70% (vol/vol) ethanol, dried, and dissolved in 100 μl of bidistilled water.
The DNA samples of both species were used to record spectra from 220 to 340 nm. The spectra had the typical shapes of nucleic acids spectra and E260/E280 quotients typical for pure nucleic acids.
Determination of cell sizes.
To analyze the cell size of M. acetivorans, cells were grown to exponential phase on methanol or sodium acetate, respectively. Cell sizes were determined with a microscope (Axioskop 40; Zeiss, Oberkochen, Germany) using an ocular micrometer, and average sizes were calculated. In addition, the average cell size of M. maripaludis was determined using an exponential culture grown on formate. Average cell sizes were used to calculate approximate cell volumes under the assumption that the cells were (nearly) spherical.
Analysis of gene conversion.
For the analysis of gene conversion the recently constructed M. maripaludis strain SkoD4 was used, which harbors different genomes that contain either the selD gene or the pacN gene at the selD locus (44). In the presence of selection using puromycin, the copy number of genomes with the puromycin resistance gene pacN is very high, whereas only very few copies of the selD-containing genomes remain in the cell. A single colony of the strain SkoD4 was used to inoculate a preculture, which was grown in 5 ml of liquid medium on H2+CO2 to stationary phase. The preculture was used to inoculate a first experimental culture with a starting OD578 of 0.05. The culture was incubated without mixing at room temperature overnight, which led to the adaptation of the cells to the fresh medium but negligible growth, and transferred to 37°C in the morning. The test tubes were incubated with slight agitation to inhibit formation of cell aggregates and to enhance mass transfer of the gaseous substrates into the liquid medium. The generation time of all cultures was ca. 4 h. After two generations, the culture was used to inoculate a new culture, which was treated as described above and had a starting OD578 of ∼0.05. By serial dilutions M. maripaludis was grown for 20 generations, in each case starting the culture with an OD578 of 0.05 and growing it for two generations. Aliquots were removed regularly, so that samples were present for every few generations. At the time of aliquot removal the cell density was determined microscopically using a counting chamber. Selected aliquots were used to quantify the copy numbers of the wild-type and the pacN-containing genomes using the real-time PCR approach described above (for the primers, see Table 1).
RESULTS
Growth-phase-dependent ploidy of M. acetivorans grown on two different energy substrates.
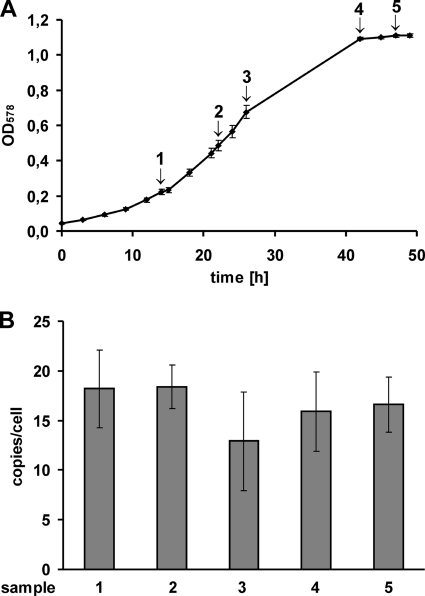
Five independent cultures of M. acetivorans were grown on methanol as the sole energy source with a doubling time of ∼6 h in the exponential phase. An average growth curve and the variance between the cultures are shown in Fig. 2 A. At the five indicated time points, aliquots were removed and used for the quantification of the ploidy levels. The results are shown in Fig. 2B. During fast growth on methanol M. acetivorans was found to be highly polyploid with an average genome copy number of about 17 per cell, independent of the growth phase.
FIG. 2.
Growth-phase-dependent ploidy of fast-growing M. acetivorans. (A) An average growth curve of M. acetivorans grown on methanol is shown that was derived from five independent cultures. Standard deviations are included. Numbers and arrows indicate the times at which aliquots were removed for quantification of the genome copy numbers. The arithmetic instead of the semilogarithmic representation (which is standard for physiological experiments concentrating on exponential phase) was chosen to better visualize the transition between exponential and stationary phase. (B) Genome copies per cell of M. acetivorans grown on methanol. Average values and SDs from five independent experiments for the following growth phases are shown: 1, early exponential phase; 2, mid-exponential phase; 3, late exponential phase; 4, early stationary phase; and 5, late stationary phase.
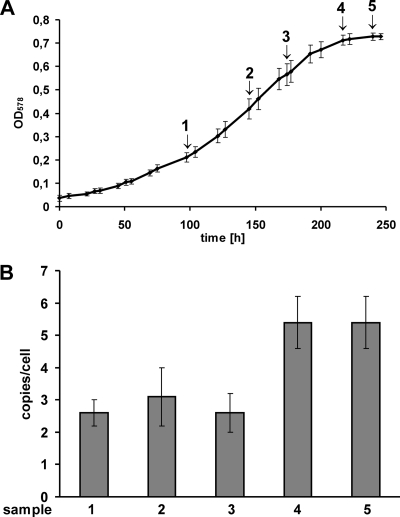
To determine whether the ploidy level of M. acetivorans is influenced by the substrate and thus the growth rate (directly or indirectly), cells growing slowly on acetate were also analyzed. To this end, five independent cultures were grown on acetate as the sole energy source, with a doubling time of 49 h in the exponential phase. The average growth curve and variance between the cultures are shown in Fig. 3 A. At the five indicated time points, aliquots were removed, and the genome copy numbers were quantified. The results are shown in Fig. 3B. It turned out that during very slow growth M. acetivorans is oligoploid, with about three genome copies per cell during the exponential phase and about five genome copies per cell during the stationary phase.
FIG. 3.
Growth-phase-dependent ploidy of slow-growing M. acetivorans. (A) An average growth curve of M. acetivorans grown on acetate is shown that was derived from five independent cultures. For further explanation, see the legend to Fig. 2. (B) Genome copies per cell of M. acetivorans grown on sodium acetate. For further explanation, see the legend to Fig. 2.
When the cell densities were determined under the microscope using a counting chamber, it became apparent that the sizes of fast-growing and slow-growing M. acetivorans cells differ from each other. Therefore, the ploidy levels should be normalized to the cell volume for a better comparison of intracellular genome concentrations. Microscopic determination of the average cell diameters revealed 2.3 ± 0.2 μm for cells grown on methanol and 1.8 ± 0.2 μm for cells grown on acetate, corresponding to average cell volumes of 6.2 ± 1.8 fl and 3.3 ± 0.9 fl, respectively, during exponential growth. Therefore, fast-growing M. acetivorans cells contain about 2.7 genomes per fl, while slow-growing cells contain ∼0.9 genomes per fl.
Growth-phase-dependent ploidy of M. maripaludis.
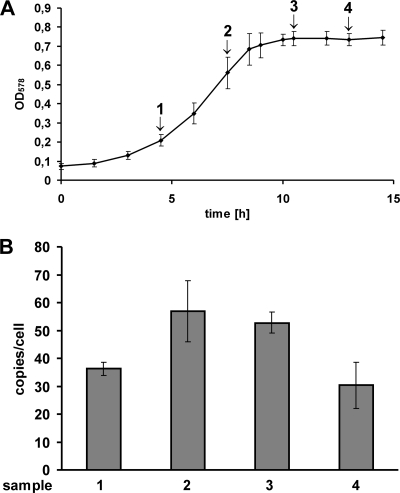
M. maripaludis was chosen as a representative of another genus of methanogenic archaea. Five independent cultures were grown on formate as sole energy source, resulting in a doubling time of 2 h in the exponential phase. The average growth curve is shown in Fig. 4 A. At the four indicated time points, aliquots were removed and used to quantify the genome copy number. M. maripaludis turned out to be highly polyploid, with up to more than 50 genomes per cell (Fig. 4B). The ploidy level is somewhat higher during late exponential phase and transition phase than during early exponential and stationary phase, but the highest ploidy level of all methanogenic archaea analyzed until now persists throughout all phases. The average cell size of M. maripaludis growing exponentially on formate was determined to be 1.5 ± 0.2 μm, corresponding to a cell volume of ca. 1.5 ± 0.2 fl. Therefore, exponentially growing M. maripaludis cells contain ∼33 genomes per fl, a >10-fold-higher value than for M. acetivorans.
FIG. 4.
Growth-phase-dependent ploidy of M. maripaludis. (A) An average growth curve of M. maripaludis grown on formate is shown that was derived from five independent cultures. For further explanation, see the legend to Fig. 2. (B) Genome copies per cell of M. maripaludis grown on sodium formate. Average values and SDs from five independent experiments for the following growth phases are shown: 1, early exponential phase; 2, late exponential phase; 3, early stationary phase; and 4, late stationary phase.
Validation of the real-time PCR approach.
The real-time PCR method had already been validated against several other methods (see the Discussion). Still, the ploidy level of M. maripaludis is the highest determined with this method thus far, and we wanted to verify the results and validate the method against an additional independent method, i.e., quantification of genomic DNA making use of its absorbance at 260 nm. When it is taken into account that the E. coli genome is nearly 3-fold larger than the M. maripaludis genome, the real-time PCR results imply that M. maripaludis contains nearly 10-fold more DNA nucleotides per cell compared to slowly growing E. coli cells. E. coli B was grown in synthetic medium with succinate as the sole carbon source. Under these conditions E. coli has a doubling time of ∼100 min during exponential growth. For cultures with this growth rate, average genome copy numbers of 1.6 (4) and 2.5 (29a) have been reported. At a cell density of 2.8 × 108 cells/ml, an aliquot was removed and used for DNA isolation (see Materials and Methods). Three independent M. maripaludis cultures were grown, and at cell densities of about 3.7 × 108 cells/ml, aliquots were removed and used for DNA isolation. The DNA samples were applied to record spectra from 220 nm to 340 nm, yielding typical spectra and E260/E280 quotients of pure DNA. The following parameters were used to calculate the average genome copy number of the two species from the E260 values and the cell densities: genome sizes of 1.66 Mbp (M. maripaludis S2) and 4.6 Mbp (E. coli B), an average molecular mass of 660 g/mol for one base pair, an E260 of 1 for a DNA concentration of 50 ng/μl, and the Avogadro number. For E. coli, the calculation led to the value of two genomes per cell, and for the M. maripaludis cultures the average value was 48 genomes per cell (standard deviation [SD] = 0.8). Therefore, these results are in excellent agreement with the results of the real-time PCR approach and underscore that M. maripaludis is highly polyploidy.
Experimental heterozygosity of M. maripaludis and the velocity of gene conversion under different selection pressures.
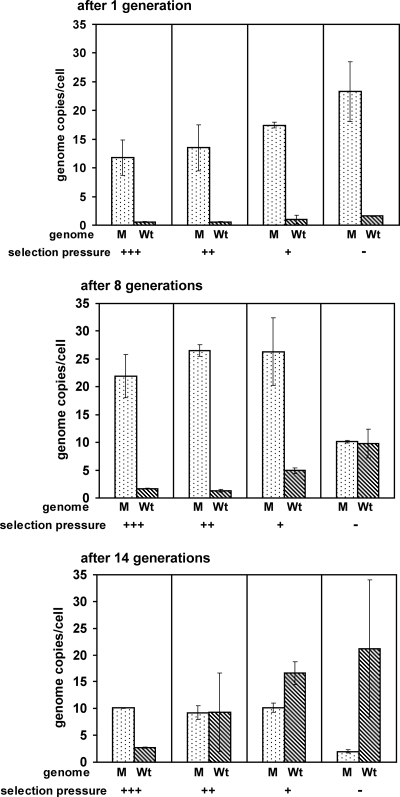
At first glance, the ease in which nonessential genes can be deleted from the chromosomes of several species of methanogenic archaea (33) seems to contradict the finding that they are polyploid. However, polyploidy and the ease of mutant construction would be congruent if methanogenic archaea would employ a mechanism, such as gene conversion, to equalize the many genome copies. Therefore, an experiment was designed to clarify whether gene conversion exists in methanogenic archaea and how efficient it is in genome equalization. We made use of a heterozygous mutant that was recently constructed (44). The attempt to replace the selenophosphate synthetase-encoding gene (selD) of M. maripaludis S2 with a puromycin resistance cassette (pacN) revealed that the selD gene is essential and cannot be completely removed from the cell. Therefore, selection in the presence of puromycin yielded a strain (designated SkoD4) that contained two different genomes; the majority of genome copies carried the pacN gene at the selD locus, while a few genome copies retained the native selD gene at this site (44). The strain was used to inoculate four different cultures, which were incubated under four different conditions: (i) in the presence of the “normal” puromycin concentration (2.5 μg ml−1), which is totally inhibitory for the wild-type (45); (ii and iii) in the presence of two lower puromycin concentrations (0.6 and 0.15 μg ml−1, respectively); and (iv) in the absence of puromycin. These conditions represent three different selection pressures, as well as the total lack of selection pressure to retain the pacN-containing genome. All four cultures were grown for about 20 generations by serial dilutions into fresh medium, and aliquots were removed every few generations (see Materials and Methods). Using the real-time PCR approach described above, the average copy numbers of pacN-containing genomes, as well as of wild-type genomes, were quantified. After the first generation, all cells contained approximately two wild-type genomes and a much higher number of pacN-containing genomes (Fig. 5, upper panel). After eight generations, the situation remained unchanged in cells that were experiencing the full selection pressure and the second-highest puromycin concentration (Fig. 5, middle panel). In stark contrast, the cells cultivated in the absence of selection now contained an equal number of genomes of both types, whereas the number of wild-type genome copies had increased to five in cells incubated with the lowest puromycin concentration. The quantification of the genomes of both types after 14 generations is shown in the lower panel of Fig. 5. In the absence of selection, the pacN-containing genomes have been nearly completely lost, and the cells contain almost exclusively wild-type genomes. Obviously, there is a strong inverse correlation between the increase in the number of wild-type genomes per cell and the selection pressure to retain the pacN-containing genome (Table 2).
FIG. 5.
Gene conversion under different selection pressures. A strain simultaneously containing genomes with the selD and the pacN gene at the selD locus was precultured under full selection pressure (2.5 μg of puromycin/ml) to minimize the number of selD-containing genomes. The preculture was used to inoculate four cultures, which were incubated under three different levels of selection (+++, 2.5 μg of puromycin/ml; ++, 0.6 μg of puromycin/ml; and +, 0.15 μg of puromycin/ml) and in the absence of selection (−). The four cultures were grown for ∼20 generations via consecutive dilution into fresh medium. After the first-, eighth-, and fourteenth-generation aliquots were withdrawn, the copy numbers of both types of genomes were determined by real-time PCR. Average values and SDs are shown. M, pacN-containing mutant genome; WT, selD-containing wild-type genome.
TABLE 2.
Effect of selection pressure on the number of wild-type genome copies
| Parameter | Puromycin concn (μg ml−1) |
|||
|---|---|---|---|---|
| 0 | 0.15 | 0.6 | 2.5 | |
| Selection pressure | − | + | ++ | +++ |
| No. of wild-type genomes | ||||
| After generation 1 | 2 | 1 | 1 | 1 |
| After generation 8 | 10 | 5 | 1 | 2 |
| After generation 14 | 21 | 17 | 9 | 3 |
| Avg increase per generation | 1.62 | 1.31 | 0.69 | 0.15 |
Taken together, gene conversion leads to a very fast replacement of the pacN gene with the native selD gene in the absence of selection pressure, and the velocity of gene replacement correlates directly with the strength of the selection pressure (Table 2).
DISCUSSION
Validation of the method for genome copy number determination.
The real-time PCR method for quantification of ploidy levels has been established only recently (5). For new methods it is very important to validate the results using independent methods, and this has been done very thoroughly with the real-time PCR method. The following approaches have been taken to validate the results. (i) In the first study, quantitative Southern blotting was used as an independent method and led to identical results for the analysis of two species of haloarchaea in various growth phases (5). (ii) For the first application of the method to bacteria, E. coli was deliberately chosen as the model, because a wealth of published knowledge on this organism exists. The numbers of origins and termini were quantified in both fast-growing (doubling time, 20 min) and slow-growing (doubling time, 103 min) cells. It could be shown that the number of origins is much higher than the number of termini in fast-growing cells, while this it not the case in slow-growing cells, and that the ploidy level is higher in fast-growing than in slow-growing cells. Both observations are thus in excellent agreement with previous studies using, e.g., fluorescence-activated cell sorting analyses and radioactive labeling. (29a). (iii) In the present study, spectroscopic quantification of DNA was used as an independent method to quantify the genome copy numbers of M. maripaludis and E. coli, and the results are in excellent agreement with the real-time PCR approach. (iv) Five different genomic regions were used for Halobacterium salinarum to prove that in this species with a doubling time of 4 h the results are independent of the region chosen for the analysis (S. Breuert and J. Soppa, unpublished results). (v) The fact that the method does not necessarily lead to high numbers but is useful to analyze monoploid species (Caulobacter crescentus, Wolinella succinogenes, and Corynebacterium glutamicum) can be taken as an indication that the method does not systematically overestimate the ploidy level. Taken together, the real-time PCR method has by now been used to quantify the genome copy number of more than 10 archaeal and bacterial species and has been validated against four different, independent methods.
Genome copy numbers in Euryarchaeota versus Crenarchaeota.
In the present study, the genome copy numbers of M. acetivorans and M. maripaludis were determined, doubling the number of species of methanogenic archaea with known ploidy level. Both of them turned out to be polyploid, and with more than 50 genome copies, M. maripaludis has the highest ploidy level found in the domain of archaea until now. Since polyploidy has been found in five of six investigated genera, it might be common in Euryarchaeota (5, 27). The only exception is M. thermoautotrophicus, which was described to contain two genome copies per cell (26). It should be noted that this species grows in filaments; therefore, each entity contains many genome copies. Of course, this might not be the reason for the low number of genomes per cell, and an alternative explanation is a variability of the ploidy level, as has also been observed in bacteria (29a). Nevertheless, none of the euryarchaeal species analyzed until now is monoploid.
In stark contrast, seven species of four crenarchaeal genera were found to be monoploid (3, 24). All of them were shown to have a short G1 phase and a long G2 phase and thus contain two copies of the chromosome for a major part of the cell cycle. Table 3 summarizes the results of all studies in which genome copy numbers of archaeal species were analyzed. Growth temperatures and doubling times are also listed to possibly reveal whether a correlation with the ploidy levels might exist. However, no such systematic correlation to either of the two factors could be found. In addition, the ploidy level is independent from the growth rate for H. salinarum (5), while, in contrast, the growth rate has a considerable effect on the number of genome copies in M. acetivorans (the present study). In the majority of euryarchaeal species the ploidy level is influenced by the growth phase, i.e., the number of copies decreases when the cells enter stationary phase.
TABLE 3.
Archaeal species, selected features, and genome copy numbers in exponential and stationary growth phases
| Species | Growth temp (°C) | Doubling time (h) | Genome copy no. |
Ploidy | Source or reference | |
|---|---|---|---|---|---|---|
| Exponential phase | Stationary phase | |||||
| Euryarchaeota | ||||||
| Halobacterium cutirubrum | 6.7 | 10.6 | Polyploid | 6 | ||
| Halobacterium cutirubrum | 13.3 | 6.3 | Polyploid | 6 | ||
| Halobacterium salinarum | 42 | 4 | 25 | 1 | Polyploid | 5 |
| Halobacterium salinarum | 30 | 8 | 25 | 15 | Polyploid | 5 |
| Halobacterium salinarum (anaerobic) | 42 | 8 | 25 | 15 | Polyploid | 5 |
| Haloferax volcanii | 42 | 4 | 17 | 10 | Polyploid | 5 |
| Methanocaldococcus jannaschii | 85 | 0.5 | 10-15 | 1-5 | Polyploid | 25 |
| Methanococcus maripaludis | 37 | 2 | 55 | 30 | Polyploid | This study |
| Methanosarcina acetivorans | 37 | 6 | 18 | 16 | Polyploid | This study |
| Methanosarcina acetivorans | 37 | 49 | 3 | 5 | Oligoploid | This study |
| Methanothermobacter thermoautotrophicus | 65 | 2a | 1-2a | Diploid | 24 | |
| Crenarchaeota | ||||||
| Acidianus hospitalis | 80 | 1-2 | 2 | Monoploid | 22 | |
| Aeropyrum pernix | 95 | 3.3 | 1-2 | 1 | Monoploid | 22 |
| Pyrobaculum aerophilum | 100 | 1-2 | 2 | Monoploid | 22 | |
| Pyrobaculum calidifontis | 90 | 3.4 | 1-2 | 2 | Monoploid | 22 |
| Sulfolobus acidocaldarius | 79 | 3.5 | 1-2 | 2 | Monoploid | 3 |
| Sulfolobus tokodai | 80 | 8 | 1-2 | 2 | Monoploid | 22 |
| Sulfolobus solfataricus | 79 | 7 | 1-2 | 2 | Monoploid | 3 |
aThe cells grow in filaments; the numbers given are thus genome copies per cell, not per filament.
Possible evolutionary advantages of polyploidy in euryarchaea.
There are several possible evolutionary advantages of polyploidy for prokaryotic species, and their relevance for euryarchaeal species are discussed briefly below.
(i) The rate of spontaneous mutations can be reduced in comparison to monoploid species, as has been described for H. volcanii (25).
(ii) The resistance against DNA-damaging conditions, especially conditions causing double-strand breaks (DSBs), can be enhanced. In the laboratory, radiation with X-rays is often used to induce DSBs; in terrestrial environments the evolutionary advantage is most likely the resistance against desiccation, which also induces DSBs. M. maripaludis S2 was isolated from an intertidal estuarine sediment and is thus probably subject to desiccation and drastic changes in the salinity of its surroundings (48). For the bacterium D. radiodurans, which contains five to eight genome copies, it was shown that the same functions are involved in strong resistance against both radiation and desiccation (28). It has long been known that D. radiodurans can restore intact chromosomes from heavily fragmented chromosomes, and recently it was shown that this is a two-stage mechanism involving a high induction of DNA repair synthesis, followed by recombination (40, 50). H. salinarum is also very resistant to ionizing radiation and to desiccation, and it has been shown that it is also able to restore intact chromosomes from fragmented chromosomes (20). Mutants of H. salinarum could be isolated that show a considerably higher radiation resistance than D. radiodurans (9). Several species of methanogenic archaea have been shown to be very resistant against desiccation (17, 23).
(iii) Polyploidy offers a mechanism of global regulation of gene expression via regulation of the genome copy number, e.g., in response to environmental changes that influence the growth rate. Although this has not yet been addressed experimentally, the observed ploidy levels indicate that Euryarchaeota make use of this possibility; in M. acetivorans the genome copy number is reduced 6-fold with a lower growth rate (the present study), and in M. jannaschii it decreases from 10 to 15 to increases from 1 to 5 upon entering stationary phase (27).
(iv) Another possible advantage is gene redundancy, including the possibility to mutate the genome under unfavorable conditions while keeping the wild-type information in other copies. We have shown here that M. maripaludis can indeed harbor different genomes simultaneously when that offers a selective advantage, and the same could be shown for H. volcanii (C. Lange, S. Breuert, and J. Soppa, unpublished data).
Taken together, polyploidy offers several possible evolutionary advantages for euryarchaeal species. In species that evolved polyploidy independently from one another a different advantage or a different combination of advantages might have been the driving force to raise the genome copy number. If so many evolutionary advantages of polyploidy exist, the question arises as to why Crenarchaeota are still monoploid. Part of the answer might be found in their cell cycle that is characterized by a short G1 phase and a long G2 phase (for a review, see reference 24). Therefore, most of the time they contain two copies of the chromosome. In addition, they may have evolved alternative mechanisms that allow making use of multiple genomes for the repair of DNA damage. It has been shown that Sulfolobus solfataricus forms large aggregates after UV irradiation, as well as after exposure to chemicals that induce DNA DSBs (11). Although not yet experimentally investigated, it can be speculated that DNA transfer between cells in the aggregates might take place, thus enhancing the ability for DNA repair via homologous recombination in the population.
Growth rate and cell sizes.
It was observed decades ago that the cell size of Salmonella enterica serovar Typhimurium is not constant but is influenced by temperature and medium (37). Recently, a molecular sensor was identified in Bacillus subtilis that is responsible for cell size regulation (46). However, differential cell size regulation has only been described for a few bacterial species, and it is not clear whether this is a general phenomenon. Several features that were characterized using only one or a few model species such as E. coli subsequently turned out not to be extendable to other species. For example, it is now clear that the ploidy level of E. coli cannot be generalized, and the textbook view of “run-and-tumble” for “bacterial” chemotaxis is true for E. coli but not for polarly flagellated bacteria and archaea. Therefore, it will be of interest to quantify the influence of different parameters on cell size for a variety of prokaryotic species. This kind of information is particularly sparse for archaeal species. The fact that the sizes of fast- and slow-growing Methanosarcina cells are rather different has been qualitatively observed by several groups working with this species, but to our knowledge this cell growth regulation has never been published before.
For stationary-phase cells of M. maripaludis S2, which had been grown in complex medium, a cell diameter of 1.1 μm (SD = 0.2 μm) has been described (18), which is somewhat smaller but similar to the value of 1.5 μm (SD = 0.2 μm) measured by us for exponentially growing cells in formate medium.
Genome copies are unified by gene conversion, and the velocity can be influenced by the selection pressure.
“Gene conversion” is defined as the nonreciprocal transfer of information between homologous sequences (36) and is the basis for antigenic variation in many pathogenic bacteria and eukaryotes or mating type switching in yeast. It includes homologous recombination between two DNA molecules (or two different sites of one molecule), and several possible molecular mechanisms have been discussed (36). Gene conversion is mostly studied in eukaryotes; a recent literature survey found more than 2,000 publications with eukaryotes, 137 with bacteria, and a single publication with archaea (21), the last of which turned out to be a bioinformatic and not an experimental analysis (2). Apart from one study that had a focus on DNA repair (8), we therefore present here the first experimental study of gene conversion in archaea. Most studies of gene conversion in bacteria either aim to characterize concerted evolution of gene families (e.g., rRNA operons) or its role in antigenic variation and phase variation (36). In contrast, we have addressed its role in the equalization of genomes of polyploid organisms. For several reasons, we predicted that a mechanism for genome equalization has to exist. One reason is that gene conversion allows escaping from “Muller's ratchet,” which is the accumulation of deleterious mutations in organism without sexual reproduction until no intact chromosome is present in the cell (22). Another reason is the ease with which chromosomal mutants can be constructed in a targeted or random fashion in polyploid halophilic and methanogenic archaea (1, 14, 32, 33, 41, 42).
Starting from a culture containing, on average, about two wild-type copies of the genome and more than 20 pacN-containing copies, we could indeed show that in the absence of selection pressure gene conversion leads to an accumulation of genomes with the native selD allele at the selD site in a very fast and efficient way. Within only 14 generations after the removal of selection, which is a very short time for processes of genome evolution, the situation was totally reversed. Furthermore, the strength of the selection pressure correlates with the velocity of gene conversion and the proportions of the two genomes. The average increase of wild-type genomes after 14 generations is much higher in the absence of selection than in the presence of the full selection pressure, where it is negligible (Table 2). The selD gene was ideally suited for the approach because (i) it is an essential gene that cannot be deleted in M. maripaludis S2 during growth on formate and (ii) it has a very low expression level, allowing the cells to grow with only one or two selD-containing genomes with the same growth rate as the wild-type that is homozygous for selD (44). The experiment shows that genome redundancy can under specific selection conditions allow heterozygoty at a locus within one cell, enabling its survival and growth. This would be impossible for a monoploid cell, which underscores the notion that one selective advantage of polyploidy is the possibility to mutate the chromosome while simultaneously retaining the wild-type information.
It should be noted that alternative, less likely, mechanisms, other than gene conversion, could underlie the experimental observations. One is that integration of the pacN gene at the selD locus may compromise the initiation of replication. Thus, in the absence of selection pressure, wild-type genomes would be replicated with greater efficiency (i.e., faster) than pacN-containing genomes, leading to their accumulation. There is no experimental evidence about the site of replication initiation in M. maripaludis. Four genes are annotated to encode a replication initiator protein, all of which are more than 100,000 bp away from selD.
Another possible explanation is that upon cell division, daughter cells with various numbers of wild-type alleles are generated and that cells with a higher number of wild-type copies are positively selected. Consequently, if a positive selection for a high number of wild-type copies existed, the average increase of wild-type genomes in the population would have to be accompanied by an increase in growth rate in order for the positive selection to take place. However, this was not observed. The growth rates of the culture at the beginning of the experiment, containing mostly pacN-containing genomes, throughout the experiment, and at the end of the experiment, containing nearly exclusively wild-type genomes, were the same (data not shown). It should be noted that the cultures were deliberately grown on H2/CO2 for this experiment to avoid a possible selective advantage of cells with more wild-type genomes, which could be envisaged during growth on formate, when a large amount of formate dehydrogenase is needed. In addition, even if a small selective advantage of cells containing a very small number of wild-type genomes is assumed, this must readily be lost as soon as a few additional copies are acquired, and thus positive selection cannot explain the observed outcome, i.e., cells containing nearly exclusively wild-type genomes.
A similar alternative explanation would be a counterselection against pacN in the absence of puromycin. However, again, a prerequisite for the selection would be that cells with fewer pacN alleles grow faster than the cells with a higher number of pacN alleles, but the growth rates of cells were the same regardless of the numbers of pacN alleles (from more than 20 to less than 2).
The possibility that the heterozygous SKoD4 strain (44) is not pure but consists of two clones, which depend on each other, one (carrying only the wild-type allele) “feeding” selenomonophosphate to the other (carrying only the pacN allele) which in turn detoxifies puromycin, is highly unlikely. First, selenophosphate is highly reactive and thus unstable (13). Second, if the pacN-carrying “population” excreted puromycin acetyltransferase, which would inactivate the puromycin in the medium, the absence of selection would lead to an accumulation of cells with wild-type genomes due to positive selection. However, also in this scenario the growth rate of the population would have to change to allow positive selection to take place. This was not observed. Therefore, we think that gene conversion is the most likely explanation for the observed results.
An experiment addressing gene conversion has also been performed with halophilic archaea (Lange et al., unpublished). Heterozygoty was enforced by suitable selective conditions in Haloferax volcanii. In this case the equalization of genomes by gene conversion was possible and could indeed be observed, in both directions, depending on the absence or presence of selective forces. Therefore, it seems that the equalization of genomes by gene conversion is generally present in polyploid Euryarchaeota. A future challenge will be to unravel the underlying molecular mechanisms and whether the cell senses in which direction gene conversion has to occur.
Acknowledgments
The project was supported by the Deutsche Forschungsgemeinschaft through grants So264/16-1 to J.S. and SFB 579 to M.R.
T.S. and M.R. thank V. Müller, Frankfurt, Germany, for his support. We thank three anonymous reviewers for the critical reading of the manuscript and their many constructive suggestions.
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Allers, T., and M. Mevarech. 2005. Archaeal genetics: the third way. Nat. Rev. Genet. 6:58-73. [DOI] [PubMed] [Google Scholar]
- 2.Archibald, J. M., and A. J. Roger. 2002. Gene conversion and the evolution of euryarchaeal chaperonins: a maximum likelihood-based method for detecting conflicting phylogenetic signals. J. Mol. Evol. 55:232-245. [DOI] [PubMed] [Google Scholar]
- 3.Bernander, R., and A. Poplawski. 1997. Cell cycle characteristics of thermophilic archaea. J. Bacteriol. 179:4963-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell growth rate, p. 1553-1569. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 5.Breuert, S., T. Allers, G. Spohn, and J. Soppa. 2006. Regulated polyploidy in halophilic archaea. PLoS One 1:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chant, J., I. Hui, D. De Jong-Wong, L. Shimmin, and P. Dennis. 1986. The protein synthesizing machinery of archaebacterium Halobacterium cutirubrum: molecular characterization. Syst. Appl. Microbiol. 7:106-114. [Google Scholar]
- 7.Comai, L. 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6:836-846. [DOI] [PubMed] [Google Scholar]
- 8.Delmas, S., L. Shunburne, H. P. Ngo, and T. Allers. 2009. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet. 5:e1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeVeaux, L. C., et al. 2007. Extremely radiation-resistant mutants of a halophilic archaeon with increased single-stranded DNA-binding protein (RPA) gene expression. Radiat. Res. 168:507-514. [DOI] [PubMed] [Google Scholar]
- 10.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K.-U. Ludtke, and H. Sahm. 1994. Nucleotide sequence, expression, and analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 11.Frols, S., et al. 2008. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 70:938-952. [DOI] [PubMed] [Google Scholar]
- 12.Gernhardt, P., O. Possot, M. Foglino, and A. Klein. 1990. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol. Gen. Genet. 221:273-279. [DOI] [PubMed] [Google Scholar]
- 13.Glass, R. S., W. P. Singh, W. Jung, Z. Veres, T. D. Scholz, and T. C. Stadtman. 1993. Biochemistry 32:12555-12559. [DOI] [PubMed] [Google Scholar]
- 14.Hammelmann, M., and J. Soppa. 2008. Optimized generation of vectors for the construction of Haloferax volcanii deletion mutants. J. Microbiol. Methods 75:201-204. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, M. T. 1978. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J. Bacteriol. 134:71-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegarty, M. J., and S. J. Hiscock. 2008. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 18:R435-R444. [DOI] [PubMed] [Google Scholar]
- 17.Kendrick, M. G., and T. A. Kral. 2006. Survival of methanogens during desiccation: implications for life on Mars. Astrobiology 6:546-551. [DOI] [PubMed] [Google Scholar]
- 18.Keswani, J., et al. 1996. Phylogeny and taxonomy of mesophilic Methanococcus spp. and comparison of rRNA, DNA hybridization, and phenotypic methods. Int. J. Syst. Bacteriol. 46:727-735. [DOI] [PubMed] [Google Scholar]
- 19.Kitten, T., and A. G. Barbour. 1992. The relapsing fever agent Borrelia hermsii has multiple copies of its chromosome and linear plasmids. Genetics 132:311-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kottemann, M., A. Kish, C. Iloanusi, S. Bjork, and J. DiRuggiero. 2005. Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation. Extremophiles 9:219-227. [DOI] [PubMed] [Google Scholar]
- 21.Lawson, M. J., J. Jiao, W. Fan, and L. Zhang. 2009. A pattern analysis of gene conversion literature. Comp. Funct. Genomics 2009:761512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehman, N. 2003. A case for the extreme antiquity of recombination. J. Mol. Evol. 56:770-777. [DOI] [PubMed] [Google Scholar]
- 23.Liu, C. T., T. Miyaki, T. Aono, and H. Oyalzu. 2008. Evaluation of methanogenic strains and their ability to endure aeration and water stress. Curr. Microbiol. 56:214-218. [DOI] [PubMed] [Google Scholar]
- 24.Lundgren, M., L. Malandrin, S. Eriksson, H. Huber, and R. Bernander. 2008. Cell cycle characteristics of crenarchaeota: unity among diversity. J. Bacteriol. 190:5362-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackwan, R. R., G. T. Carver, J. W. Drake, and D. W. Grogan. 2007. An unusual pattern of spontaneous mutations recovered in the halophilic archaeon Haloferax volcanii. Genetics 176:697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majernik, A. I., M. Lundgren, P. McDermott, R. Bernander, and J. P. Chong. 2005. DNA content and nucleoid distribution in Methanothermobacter thermautotrophicus. J. Bacteriol. 187:1856-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malandrin, L., H. Huber, and R. Bernander. 1999. Nucleoid structure and partition in Methanococcus jannaschii: an archaeon with multiple copies of the chromosome. Genetics 152:1315-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalf, W. W., J. K. Zhang, X. Shi, and R. S. Wolfe. 1996. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J. Bacteriol. 178:5797-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Pecararo, V., K. Zerulla, C. Lange, and J. Soppa. Quantification of ploidy in proteobacteria revealed the existence of monoploid, (mero-)oligoploid, and polyploid species. PLoS One, in press. [DOI] [PMC free article] [PubMed]
- 30.Postgate, J. R., H. M. Kent, R. L. Robson, and J. A. Chesshyre. 1984. The genomes of Desulfovibrio gigas and D. vulgaris. J. Gen. Microbiol. 130:1597-1601. [DOI] [PubMed] [Google Scholar]
- 31.Riddle, N. C., H. Jiang, L. An, R. W. Doerge, and J. A. Birchler. 2010. Gene expression analysis at the intersection of ploidy and hybridity in maize. Theor. Appl. Genet. 120:341-353. [DOI] [PubMed] [Google Scholar]
- 32.Rosenshine, I., and M. Mevarech. 1991. The kinetic of the genetic exchange process in Halobacterium volcanii mating. Gen. Appl. Aspects Halophilic Microorg. (NATO ADV SCI I A-LIF) 201:265-270. [Google Scholar]
- 33.Rother, M., and W. W. Metcalf. 2005. Genetic technologies for archaea. Curr. Opin. Microbiol. 8:745-751. [DOI] [PubMed] [Google Scholar]
- 34.Rother, M., I. Mathes, F. Lottspeich, and A. Bock. 2003. Inactivation of the selB gene in Methanococcus maripaludis: effect on synthesis of selenoproteins and their sulfur-containing homologs. J. Bacteriol. 185:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Santoyo, G., and D. Romero. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169-183. [DOI] [PubMed] [Google Scholar]
- 37.Schaechter, M., O. Maaloe, and N. O. Kjeldgaard. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19:592-606. [DOI] [PubMed] [Google Scholar]
- 38.Semon, M., and K. H. Wolfe. 2007. Consequences of genome duplication. Curr. Opin. Genet. Dev. 17:505-512. [DOI] [PubMed] [Google Scholar]
- 39.Skarstad, K., E. Boye, and H. B. Steen. 1986. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 5:1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slade, D., A. B. Lindner, G. Paul, and M. Radman. 2009. Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136:1044-1055. [DOI] [PubMed] [Google Scholar]
- 41.Soppa, J. 1998. Optimization of the 5-bromo-2′-deoxyuridine selection and its application for the isolation of nitrate respiration-deficient mutants of Haloferax volcanii. J. Microbiol. Methods 34:41-48. [Google Scholar]
- 42.Soppa, J., and D. Oesterhelt. 1989. Bacteriorhodopsin mutants of Halobacterium sp. GRB. I. The 5-bromo-2′-deoxyuridine selection as a method to isolate point mutants in halobacteria. J. Biol. Chem. 264:13043-13048. [PubMed] [Google Scholar]
- 43.Sowers, K. R., J. G. Baron, and S. F. Ferry. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stock, T., M. Selzer, and M. Rother. 2010. In vivo requirement of selenophosphate for selenoprotein synthesis in archaea. Mol. Microbiol. 75:149-160. [DOI] [PubMed] [Google Scholar]
- 45.Tumbula, D. L., T. L. Bowen, and W. B. Whitman. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 121:309-314. [Google Scholar]
- 46.Weart, R. B., A. H. Lee, A. C. Chien, D. P. Haeusser, N. S. Hill, and P. A. Levin. 2007. A metabolic sensor governing cell size in bacteria. Cell 130:335-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webb, C. D., et al. 1998. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol. 28:883-892. [DOI] [PubMed] [Google Scholar]
- 48.Whitman, W. B., J. Shieh, S. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterisation of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]
- 49.Wolfe, K. H. 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2:333-341. [DOI] [PubMed] [Google Scholar]
- 50.Zahradka, K., et al. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569-573. [DOI] [PubMed] [Google Scholar]