Abstract
Enhancing the knowledge on the genetic basis of germination and heterotrophic growth at extreme temperatures is of major importance for improving crop establishment. A quantitative trait loci (QTL) analysis was carried out at sub- and supra-optimal temperatures at these early stages in the model Legume Medicago truncatula. On the basis of an ecophysiological model framework, two populations of recombinant inbred lines were chosen for the contrasting behaviours of parental lines: LR5 at sub-optimal temperatures (5 or 10°C) and LR4 at a supra-optimal temperature (20°C). Seed masses were measured in all lines. For LR5, germination rates and hypocotyl growth were measured by hand, whereas for LR4, imbibition and germination rates as well as early embryonic axis growth were measured using an automated image capture and analysis device. QTLs were found for all traits. The phenotyping framework we defined for measuring variables, distinguished stages and enabled identification of distinct QTLs for seed mass (chromosomes 1, 5, 7 and 8), imbibition (chromosome 4), germination (chromosomes 3, 5, 7 and 8) and heterotrophic growth (chromosomes 1, 2, 3 and 8). The three QTL identified for hypocotyl length at sub-optimal temperature explained the largest part of the phenotypic variation (60% together). One digenic interaction was found for hypocotyl width at sub-optimal temperature and the loci involved were linked to additive QTLs for hypocotyl elongation at low temperature. Together with working on a model plant, this approach facilitated the identification of genes specific to each stage that could provide reliable markers for assisting selection and improving crop establishment. With this aim in view, an initial set of putative candidate genes was identified in the light of the role of abscissic acid/gibberellin balance in regulating germination at high temperatures (e.g. ABI4, ABI5), the molecular cascade in response to cold stress (e.g. CBF1, ICE1) and hypotheses on changes in cell elongation (e.g. GASA1, AtEXPA11) with changes in temperatures based on studies at the whole plant scale.
Introduction
Temperature is one of the most important environmental factors controlling seed germination and seedling heterotrophic growth and also limiting crop distribution. During crop establishment, extreme temperatures can decrease plant emergence and lead to drastic losses in crop yield and quality. Climate changes are leading to increasingly earlier spring sowings of crops in Western Europe (Qi and Jaggard 2008; Vocansson and Jeuffroy 2008) and low temperatures in early spring can slow down and decrease emergence. High temperatures can also have a negative impact on stand establishment, especially in the case of vegetables grown continuously throughout the year which may be sown during high temperature peak (Valdes et al. 1985; Cantliffe et al. 2000; Argyris et al. 2005). Breeding for improved germination and seedling growth at low and high temperatures can thus help to cope with these stresses during early crop establishment.
Germination and seedling growth are stages that are rarely a specific and primary target for breeders, even though cultivars with low seedling emergence may be rejected at the end of the breeding process. However, breeding for other traits can influence emergence traits. For instance, breeding for a smaller plant height in wheat has resulted in selecting dwarfing genes that reduce coleoptile length (Botwright et al. 2001; Rebetzke et al. 2007). Increasing knowledge on these stages—especially tolerance to extreme temperatures—could lead to possible genetic improvements. In recent years, quantitative trait loci (QTLs) associated with germination traits have been reported in different crops (Foolad et al. 1998, 1999; Bettey et al. 2000; Cui et al. 2002; Foolad et al. 2003; Al-Chaarani et al. 2005; Finch-Savage et al. 2005) and in Arabidopsis (Clerkx et al. 2004), some in relation with temperature response (Fujino et al. 2004; Argyris et al. 2005; Zhang et al. 2005; Hayashi et al. 2008). Many studies have focused on variations in germination and in early seedling growth after emergence under several abiotic stress conditions, but fewer have considered heterotrophic growth. To our knowledge, the genetic basis of heterotrophic seedling growth was only analyzed in Brassica oleracea by Bettey et al. (2000). No specific work has been reported for QTLs controlling the germination and heterotrophic growth of legumes. In Medicago truncatula (M. tr.), genetic diversity has been shown for germination and seedling heterotrophic growth at low temperatures and also at supra-optimal temperatures, i.e. 20°C and over (Brunel et al. 2009). Genetic and genomic tools, as well as extensive bioinformatics and database resources have been developed for M. tr. (Cannon et al. 2005). Its genome has been almost fully sequenced and several connected genetic maps created and anchored with the physical map (Huguet et al. 2007). These maps are helpful to access the genes positioned in the support intervals of QTLs. Medicago truncatula is phylogenetically close to several cultivated forage and grain legume species (Doyle and Luckow 2003; Choi et al. 2004b; Zhu et al. 2005). Macro and microsyntenies were observed between M. tr. and cultivated species such as lentil (Lens culinaris Medik., Phan et al. 2007), pea (Pisum sativum, Aubert et al. 2006), red clover (Trifolium pratense, Sato et al. 2005), alfalfa (Medicago sativa, Choi et al. 2004a) and soybean (Glycine max, Mudge et al. 2005; Li et al. 2008) and also in model plants such as Lotus japonicus (Cannon et al. 2006) and Arabidopsis thaliana (Mudge et al. 2005). Analyses of germination and seedling heterotrophic growth in M. tr. could thus help to improve the tolerance of other species to extreme temperatures.
The aim of the present study was to obtain data on the genetic basis of tolerance to adverse temperatures in order to improve early stages of crop establishment. The temperatures studied were chosen on the basis of previous characterization of cardinal temperatures in M. tr. (Brunel et al. 2009). Two genetic backgrounds of M. tr. were studied because the genotypes responsive to either low or high temperature were not the same during germination. We measured seed mass because both germination and heterotrophic growth may be correlated with this seed characteristic (Tekrony et al. 1987; Schneider 1998; Wang et al. 2004) and because seed mass is a major trait in crop breeding. Regarding phenotyping procedures, as far as possible we eliminated seed coat dormancy so it did not disturb germination, and we carefully separated the two stages leading to emergence in order to analyze their respective genetic bases. The automated device we used enabled identification of the imbibition rate and precise measurements of germination in a large number of lines. Seedling heterotrophic growth was studied with pots placed in the dark to measure growth before emergence under conditions close to sowing. Finally, integrating knowledge from the ecophysiological to the gene scale, M. tr. genome-wide sequence information and bioinformatics resources were used to identify a first set of genes possibly associated with the behaviours observed at the temperatures studied.
Materials and methods
Plant material
The genetic control of germination and hypocotyl heterotrophic growth was studied at sub-optimal temperatures using a population of 125 recombinant inbred lines (RILs), named LR5, which is derived from a cross between Jemalong A17 and a cold-sensitive line, F83005.5. At a supra-optimal temperature (20°C), we used a population of 178 recombinant inbred lines, named LR4, which is derived from a cross between Jemalong 6 and DZA315.16, the latter genotype germinating more slowly at 20°C and above. Both populations were F8 derived lines. In this paper Jemalong-6 and Jemalong-A17 are both denoted Jemalong as no genotypic or phenotypic differences were identified between them (Thoquet et al. 2002). Numerous genetic and genomic tools have been developed using Jemalong and the three parental lines belong to the M. tr. nested core collections (Ronfort et al. 2006).
The seeds of LR5 were produced in greenhouses by INP ENSAT-CNRS (Toulouse, France) and seeds of LR4 by the M.tr. Biological Resource Centre (Montpellier, France, http://www.montpellier.inra.fr/BRC-MTR/). For all the experiments, seeds were scarified with sand paper to break seed coat dormancy. Seeds were considered as germinated when the radicle protruded from the seed coat (Bewley and Black 1994). Hard seeds that escaped rubbing and remained unimbibed were excluded from the results. Seed mass (SM) was measured in order to test the possible correlations between this seed characteristic and genotypic differences in germination or seedling growth.
Sub-optimal temperature experiments
For the germination experiments, three replicates of 30 seeds were placed in plastic boxes (5.5 × 12 × 18 cm) on filter paper imbibed with 14 ml of deionised water and incubated in a climatic chamber at 5°C without light at the National Seed Testing Station (SNES, Angers France). Germination was recorded once a day for 3 weeks. For the seedling growth study, only final hypocotyl length (HL) and width (HW) were recorded as previous results had shown that there were no differences in radicle growth between the parental accessions (Brunel et al. 2009). Seedlings were grown at 10°C under dark conditions to mimic heterotrophic growth, and sampled 24 and 31 days after sowing to ensure that the seedling lengths had reached their plateau value. Seeds were sown in pots (height 10 cm, diameter 7.5 cm) filled with 500 g sand and watered with a nutrient solution (Saglio and Pradet 1980) to ensure that seedling growth conditions were not limiting. Pot water content was maintained constant at 0.20 g g−1. The experimental design was a randomized complete-block design with three blocks. All lines were present in one block, with one pot sown with seven seeds for each line for each sampling date (24 and 31 days). Measurements were thus made on three replicates of 14 seedlings for each genotype.
Supra-optimal temperature experiments
Two experiments were carried out at 20°C. For the first experiment performed at SNES in Angers (denoted SNES), three replicates of 25 seeds were used for each line. For image acquisition and analysis we used an automated set-up consisting of a Jacobsen table with three cameras positioned above the table (Ducournau et al. 2004, 2005). Seeds of 16 lines were positioned on filter paper under each camera, i.e. 48 lines for each sowing date. The time to reach full germination in a given line varied from 3 to 6 days. The experiment was designed as an incomplete block trial. Each block represented a given sowing date. Each of the three replicates of each line was sown at a different date. Thus, measurements of the three replicates of the 178 lines took 9 weeks. Each replicate was sown under a different camera, i.e. at a different place along the table, to ensure that differences in germination between lines could not be due to putative differences in thermal or water availability along the Jacobsen table. Temperature was recorded hourly with sensors placed on the table under each camera. Pictures were taken automatically every 2 h. An automatic process using Image J software (Rasband 1997) measured individual information on each seed (Demilly et al. 2007). We recorded the initial seed surface area (ISSA, mm2). The imbibition rate (IR, mm2 h−1) was estimated from the increase in seed surface area from 0 to 8 h after sowing, which had previously been shown to be correlated with seed water uptake (Demilly et al. 2007). Germination of each seed was determined from changes in the coordinates of its inertia centre. The germination times (Ti, number of hours to reach i% of germination, i < 90) were calculated from fittings to the germination time course on each replicate of 25 seeds. Final germination percentages (FGP) were recorded manually at the end of the experiment. The early embryo axis elongation rate (ELR, mm2 h−1) was determined by the difference in the embryo axis lengths measured at 4 and 10 h after germination, i.e. lengths of less than 5 mm.
Germination was also recorded during a second experiment (denoted LEG) carried out at INRA Dijon (Research Unit for Genetic and Ecophysiology of Grain Legumes). Here we took advantage of an experiment dedicated to phenotyping LR4 during vegetative growth. We measured seed mass (SM), Ti and FGP before the transfer of germinated seeds into pots for analysis of further growth. The seedlot used in the LEG experiment was different from the one used in the SNES experiment. Imbibed seeds, 50–90 per line, were vernalized for 3 days at 5°C before being transferred at 20°C. FGP was measured 10 days after sowing.
Data analysis
For each replicate, germination records were fitted to a Gompertz function (Richards 1959):
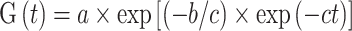 |
where G(t) is the cumulative germination rate at time t after sowing, a the maximum cumulative germination rate, and b and c are shape parameters. The fittings were used to calculate the time to reach 20, 50, 70 and 90% germination (T20, T50, T70, and T90).
All statistical analyses were performed with SAS 8.1 (SAS 2000). Data were subjected to analysis of variance using the GLM procedure. Regarding seedling elongation at low temperature (LR5), the absence of differences in HL measurements at 24 and 31 days was first checked and then all the data were combined. As a significant block effect was observed, we generated adjusted means by regression on the block effect by using the least squares method (LSmeans) in subsequent analyses. For the SNES experiment carried out at 20°C on LR4, heterogeneities could originate from the sowing position along the table (table position effect) and from variations over the 9 weeks (block effect). As a significant table position effect was observed, adjusted means were generated by regression on the table position effect by using the least squares method (LSmeans) for subsequent analyses. Simple Pearson correlation coefficients (r) were calculated between all the variables measured either on LR5 or on LR4.
QTL analysis of germination and heterotrophic growth
The genetic maps of LR5 and LR4 used for QTL mapping were based on 89 and 110 microsatellite markers, respectively (T. Huguet et al. unpublished). The markers are well distributed over the chromosomes and the framework maps span 665 and 608 cM, respectively, with an average distance between markers of 7.5 and 5.5 cM, respectively. These maps globally cover the entire genome of the species, the integrated genetic map of M. tr. covering 596 cM (http://www.medicago.org/). Before QTL analysis, the normality of distributions of all traits was tested by computing skewness and kurtosis indices. A trait was declared normally distributed when skewness and kurtosis values ranged between −2 and +2. Means or adjusted means (LSmeans) were used depending on the trait. QTL analysis was carried out with the PLABQTL software package (Utz and Melchinger 1996) using composite interval mapping (CIM). For each variable, a critical LOD score was determined using 1,000 permutations (Churchill and Doerge 1994) with a genome-wise error rate of α < 0.05. For CIM, PLABQTL uses the regression approach proposed by Haley and Knott (1992), using selected markers as cofactors. These cofactors, representing potential QTLs, were selected by forward stepwise regression with the statement cov SEL. The proportion of phenotypic variation accounted by each QTL (R²) was calculated by multiple regression in PLABQTL. When several QTLs were detected for one trait, an adjusted R 2 (R²adj), representing the proportion of the phenotypic variation explained by all QTLs, was calculated as described by Hospital et al. (1997). For each QTL, a support interval was automatically generated by the LOD fall of 1.0 method (Lander and Botstein 1989). We then searched for epistasis with the Epistat program (Chase et al. 1997). We used an automated search for both all-pairwise and conditional interactions with a log likelihood ratio threshold >6 as suggested by the authors. We subsequently used the Montecarlo simulation program associated with Epistat to analyze 106 resampled subgroups, as a way of estimating P values. To avoid false positive interactions, epistasis was only considered to be significant when P < 0.0005. The BioMercator software (Arcade et al. 2004) was used to generate a consensus genetic map of the two RIL populations. This was done by iterative projection of loci from the LR5 map onto the LR4 map. QTLs were projected onto this map. They were designated as trait abbreviations followed by the chromosome number (1–8) and the name of the genetic background (LR4 or LR5) from which they were identified. The genetic map and the QTLs were then drawn using MapChart, version 2.1 (Voorrips 2002).
Identification of candidate genes
First, all the genes present in the QTL support intervals were listed in silico. Then, some were selected in the light of specific a priori hypotheses we made at cellular and ecophysiological scales. The length of an organ (e.g. an internode, or a leaf, Tardieu and Granier 2000) at a given time L(t) can be described as:
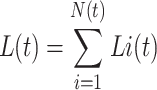 |
where N(t) is the number of cells at time t, and Li(t) the length of each cell i at this time. Based on previous studies (Gendreau et al. 1997; Barroco et al. 2005; Gimeno-Gilles et al. 2009), we hypothesized that germination and hypocotyl heterotrophic elongation were mainly related to cell elongation and not to an increase in the number of cells. Cell length which is related to changes in cell wall properties or in vacuole size can be altered not only by several environmental conditions, in particular temperature, but also by water availability and mechanical obstacles during heterotrophic growth. Such changes are strongly influenced by phytohormones (Hopkins and Huner 1999), especially abscissic acid (ABA) and gibberellins (GA), and depend on molecular cascades, including signal sensing and transduction, transcription control and stress responsive mechanisms that could be regulated differently in different genotypes. The list of Bacterial Artificial Chromosomes (BACs) included within the support interval for all QTLs was established from the positions of the upper and lower markers flanking the QTLs, by using the integrated M. tr. genetic map from the website http://www.medicago.org/genome/map.php. The chromosomal positions of the BACs on which given genes were annotated were confirmed by referring to the Oklahoma University website devoted to M. tr. (http://www.medicago.org/genome/index.php). We also used the website developed by the Kazusa DNA Research Institute for red clover (http://clovergarden.jp) to confirm certain BAC positions, because red clover is phylogenetically close to M. tr. (Choi et al. 2004b) and because the two species have a close syntenic relationship (Sato et al. 2005). The M. tr. gene Tentative Consensus (TCs) sequences were searched by keyword queries in the DCFI Medicago gene index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/Blast/index.cgi). Additionally, the M. tr. EST sequences obtained in the TIGR Medicago genome (IMGAG loci) browser were blasted between Arabidopsis (http://www.arabidopsis.org/Blast/index.jsp-BlastX) and the red clover EST bank (http://est.kazusa.or.jp/en/plant/red_clover/EST/).
Results
Phenotypic variation
Table 1 gives mean values for seed mass, germination and seedling growth characteristics measured on LR5 at low temperatures, on LR4 at 20°C, and on their parental lines. It also includes skewness and kurtosis values that gave information on trait distribution shapes. Analysis of variance indicated highly significant differences between the RILs for seed mass and all the variables measured for germination and heterotrophic growth in LR5 and LR4 (SNES experiment only, as no replicate was made for the LEG experiment).
Table 1.
Seed characteristics, germination and heterotrophic growth (a) at sub-optimal temperature in the LR5 population (Jemalong × F83005.5) and (b) at supra-optimal temperature in the LR4 population (Jemalong × DZA315.16; two experiments SNES and LEG)
| Stage | Trait | Temperature | Parental lines† | Recombinant inbred lines | ||||
|---|---|---|---|---|---|---|---|---|
| Jemalong | F83005.5 | Mean†† | Range | Skewness | Kurtosis | |||
| (a) | ||||||||
| Dry seed | SM (mg) | 4.1 | 3.9 | 3.7 | 2.3–5.0 | 0.0 | 0.2 | |
| Germination | T20 (h) | 5°C | 89 | 109 | 92 | 60–144 | 1.3 | 2.0 |
| T50 (h) | 113 | 144 | 114 | 75–202 | 1.3 | 1.6 | ||
| T70 (h) | 139 | 181 | 134 | 85–262 | 1.3 | 1.3 | ||
| T90 (h) | 140 | 212 | 156 | 105–303 | 0.8 | 0.3 | ||
| FGP (%) | 94 | 94 | 93 | 55–100 | −1.5 | 2.0 | ||
| Heterotrophic growth | HL (mm) | 10°C | 67.2 | 49.9 | 52.9 | 38–67 | 0.4 | 0.2 |
| HW (mm) | 1.1 | 1.0 | 1.0 | 0.8–1.2 | 0.3 | 0.6 | ||
| Stage | Trait | Temperature | Parental lines††† | Recombinant inbred lines | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jemalong | DZA315.16 | Mean | Range | Skewness | Kurtosis | |||||||
| SNES†† | LEG | SNES | LEG | SNES | LEG | SNES | LEG | |||||
| (b) | ||||||||||||
| Dry seed | SM (mg) | 20°C | 4.4 | 4.3 | 4.1 | 5.8 | 2.4–5.4 | 4.4–7.0 | 0.1 | 0.0 | 0.0 | 0.3 |
| ISSA (mm2) | 8.3 | 8.5 | 6.9 | – | 4.4–9.5 | – | 0.2 | – | 0.0 | – | ||
| Imbibition | IR (mm2 h−1) | 32 | 27 | 52.5 | – | 19–89 | – | 0.2 | – | −0.4 | – | |
| Germination | T20 (h) | 16 | 18 | 17 | 73 | 14–20 | 63–82 | 0.2 | −0.1 | −0.2 | 1.9 | |
| T50 (h) | 19 | 24 | 21 | 78 | 16–26 | 69–88 | 0.6 | 0.3 | 0.8 | 0.3 | ||
| T70 (h) | 21 | 27 | 24 | 82 | 19–35 | 72–96 | 1.0 | 0.4 | 1.5 | 0.4 | ||
| T90 (h) | 26 | 37 | 31 | 90 | 23–52 | 79–102 | 2.6 | −0.2 | 1.3 | −0.8 | ||
| FGP (%) | 100 | 99 | 99 | 92 | 94–100 | 64–100 | −1.8 | −5.6 | 3.8 | 6.4 | ||
| Heterotrophic growth | ERL (mm h−1 10−2) | 53 | 53 | 39 | 36 | 21–58 | 10–59 | 0.0 | −0.2 | 0.1 | 0.0 | |
SM seed mass. T20, T50, T70 and T90: times to reach 20, 50, 70 and 90% of germination. FGP final germination percentage. HL, HW final length and width of hypocotyls. ISSA initial seed surface area. IR imbibition rate. ERL elongation rate
†Parental means were obtained from four previous experiments
††Means or LS means depending on the trait. Differences between genotypes were significant for all traits at P < 0.001, except for T20 at 20°C (P < 0.01)
†††Parental means were obtained from the SNES experiment
Seed characteristics
Ranges of variation in seed mass (SM) in LR5 and LR4 (SNES experiment) seed lots were close, from 2.3 to 5.0 mg (3.7 mg in average) and from 2.4 to 5.4 mg (4.1 mg in average), respectively. In the LEG experiment (LR4), which was carried out with a different seed lot, SM was higher (Table 1b). SM and initial seed surface area (ISSA) followed a normal distribution with a range of variation reaching 50% of the maximum value.
Germination and heterotrophic growth at sub-optimal temperatures
The range of variation in germination times (Ti) across LR5 was wide, about 60% of the maximum values (Table 1a). For instance, T50 varied from 75 to 202 h. Transgressive phenotypes were found, with lines taking more time to germinate, i.e. more sensitive to cold. FPG reached a high average value of 93%, but its range of variation was wide (45% of the maximal value), with some lines showing an FPG lower than 60% (Table 1a). Hypocotyl length (HL) at 10°C averaged 53 mm with variation among the RILs corresponding to 43% of the maximal value, while hypocotyl width (HW) varied from 0.8 to 1.2 mm.
Germination and early growth at the supra optimal temperature
All the variables varied widely across the LR4 population of lines in the SNES experiment. They showed a normal distribution, except for FPG whose values exceeded 95%, and for which many lines reached 100% (Table 1b). The ranges of variation of imbibition and early elongation rates (IR and ELR) represented 80 and 50% of the maximum values, respectively, while a variation of 30% was observed in germination times (Table 1b). The seeds used in the LEG experiment germinated more slowly and had a lower FPG than those used in the SNES experiment.
Correlations between traits
In both experiments, all germination times (T20 to T90) were positively correlated (r > 0.5***). At 5°C (LR5), the final percentage of germination (FPG) varied and was also negatively correlated with germination times (−0.74*** < r < −0.6***). This indicated that a RIL that began germinating early also germinated rapidly and reached a high final percentage. Germination times and FPG were thus all indicators of the same trend in the germination curve of a RIL. Seed mass was not correlated with germination time courses, FPG or HL. It was only positively correlated with HW (r = 0.59***), whereas HL and HW were not correlated. In the LR4 SNES experiment, no variables were correlated with SM except ISSA (r = 0.49***). The negative correlation observed between ISSA and IR (r = −0.65***) indicated a higher imbibition rate for small seeds. Weak but significant negative correlations (r < −0.25**) appeared between germination times and ELR, even though measurements were made for each seed so as to carefully separate these two variables.
QTL analysis at sub-optimal temperatures on LR5
Regarding germination, one QTL was found for FPG on chromosome 3 and one for T90 on chromosome 8. QTLs for other Ti were not significant but co-localized with the QTL for T90 (Table 2a; Fig. 1). The latter QTL explained 15% of phenotypic variation and showed a high additive effect, the F83005.5 alleles decreasing the trait value (Table 2a).
Table 2.
QTLs identified for seed mass, germination and heterotrophic growth, (a) at sub-optimal temperatures in the LR5 population (Jemalong × F83005.5), (b) at 20°C, a supra-optimal temperature in the LR4 population (Jemalong × DZA315.16; two experiments SNES and LEG)
| Trait | QTLa | Chrb | QTL position (cM) | Support interval (cM) | Closest marker | Peak LOD score | R 2%c | Additive allele effecte |
|---|---|---|---|---|---|---|---|---|
| (a) | ||||||||
| SM (mg) | SM5-LR5 | 5 | 38 | 30–46 | MTE33 | 3.4 | 11.7 | −0.21 |
| SM8-LR5 | 8 | 80 | 78–82 | Mtic1194 | 3.1 | 10.9 | 0.26 | |
| R 2adjd = 21.2 | ||||||||
| T90 (h) | T90.8-LR5 | 8 | 8 | 4–10 | MTE118 | 3.0 | 15.0 | −24.20 |
| FPG (%) | FPG3-LR5 | 3 | 26 | 20–36 | MTE19 | 3.1 | 12.5 | −2.97 |
| HL (mm) | HL1-LR5 | 1 | 4 | 0–12 | MTE75 | 5.1 | 17.7 | −1.82 |
| HL2-LR5 | 2 | 6 | 2–12 | MTE10 | 6.7 | 22.0 | −2.10 | |
| HL8-LR5 | 8 | 66 | 62–70 | MTE136 | 8.0 | 25.5 | −2.00 | |
| R 2adjd = 59.4 | ||||||||
| HW (mm) | HW2-LR5 | 2 | 70 | 64–70 | MTE80 | 3.4 | 12.1 | 0.02 |
| Trait | QTLa | Chrb | QTL position (cM) | Support interval (cM) | Closest marker | Peak LOD score | R2%c | Additive allele effecte | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (b) | |||||||||||||||
| Experiment | SNES | LEG | SNES | LEG | SNES | LEG | SNES | LEG | SNES | LEG | SNES | LEG | SNES | LEG | |
| SM (mg) | SM1-LR4 | 1 | 1 | 52 | 56 | 50–60 | 50–60 | MTE62 | MTE62 | 3.4 | 2.8 | 8.5 | 11.4 | 0.11 | 0.15 |
| SM5-LR4 | 5 | 2 | 0–8 | MTE30 | 3.0 | 8.0 | 0.11 | ||||||||
| SM7-LR4 | 7 | 7 | 52 | 48 | 48–58 | 42–52 | Mtic432 | Mtic432 | 7.6 | 2.7 | 18.5 | 10.8 | −0.18 | −0.15 | |
| SM8-LR4 | 8 | 70 | 66–76 | MTE56 | 3.4 | 8.6 | 0.11 | ||||||||
| R 2adjd = 35.4 | R 2adjd = 20.2 | ||||||||||||||
| IR (mm2/h) | IR4-LR4 | 4 | 44 | 34–48 | MTE29 | 3.1 | 7.9 | −6.31 | |||||||
| T20 (h) | T20.5-LR4 | 5 | 8 | 0–12 | MTE31 | 4.9 | 12.3 | 0.44 | |||||||
| T50 (h) | T50.5-LR4 | 5 | 8 | 0–14 | MTE31 | 4.6 | 11.6 | 0.59 | |||||||
| T70 (h) | T70.5-LR4 | 5 | 48 | 36–52 | MTE35 | 2.7 | 6.9 | 0.70 | |||||||
| T90 (h) | T90.5-LR4 | 5 | 82 | 74–86 | MTE37 | 2.9 | 19.9 | 2.81 | |||||||
| T90.7-LR4 | 7 | 62 | 58–66 | MTE125 | 2.9 | 20.6 | −3.04 | ||||||||
| R 2adjd = 38.7 | |||||||||||||||
| ELR (mm/h) | ELR3-LR4 | 3 | 54 | 48–58 | MTE20 | 3.3 | 8.5 | 0.03 | |||||||
SM seed mass, T20–T90 times to reach 20–90% of germination, FPG final percentage of germination, IR imbibition rate, HL final hypocotyl length, HW final hypocotyl width, ELR early embryonic axis elongation rate
aQTL designation
bChromosome
cProportion of the phenotypic variation accounted for each QTL
dProportion of the phenotypic variation explained by all QTLs
eAdditive allele effect is positive when the F83005.5 alleles or the DZA315.16 alleles increase the trait value in the LR5 or LR4 genetic backgrounds, respectively
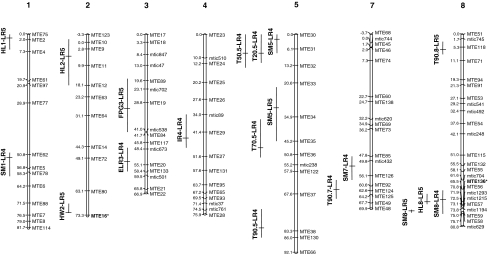
Fig. 1.
Consensus linkage projection from LR5 map onto LR4 map with locations of QTLs. Chromosome 6, without QTLs was omitted. The markers’ names are indicated on the right of the chromosomes and their positions on the left. To the left of the chromosomes, vertical bars indicate the confidence intervals of QTLs. Horizontal bars indicate the position of the peaks of QTLs. Markers followed by an asterisk are involved in an epistatic effect for HW at sub-optimal temperature (10°C). SM seed mass. T20, T50, T70 and T90 times to reach 20, 50, 70 and 90% of germination. FGP final germination percentage; HL, HW final length and width of hypocotyls; IR imbibition rate; ERL elongation rate
Regarding heterotrophic growth, QTLs were identified for both HL and HW. The three HL QTLs each contributed to a high percentage of variation (>17%). They were located on chromosomes 1, 2 and 8 (Table 2a; Fig. 1). For all QTLs associated with variations in HL, the F83005.5 alleles decreased the trait value. The HW QTL was localized on chromosome 2, the F83005.5 alleles increasing HW but with a low additive effect. For this trait, a significant epistatic interaction (P < 0.0002) was found between MTE16 (chromosome 2) and MTE136 (chromosome 3). This interaction implicated additive QTL-linked markers associated with hypocotyl elongation at low temperature (QTL HW2-LR5 nearby MTE16 and QTL HL8-LR5 nearby MTE136). When combined, F83005.5 alleles increased HW. QTLs identified for the different variables were not co-localized. In particular the QTLs for germination and heterotrophic growth were distinct.
QTLs identified at the supra-optimal temperature in LR4
In the SNES experiment, QTLs were found for SM, IR, Ti (i < 70%) and for ELR and one to four QTLs were identified for each trait (Table 2b; Fig. 1). QTLs for the different traits all mapped into distinct genomic positions. Considering the germination times, two QTLs were identified on chromosome 5, one at position 8 (T20 and T50.5-LR4) and the other at position 48 (T70.5-LR4). The LOD value of the last QTL was lower. The DZA315.16 alleles increased the Ti values for both QTLs, i.e. they decreased the germination rate. Concerning heterotrophic growth, one QTL was mapped for early ELR on chromosome 3 in a distinct chromosomal region from the QTLs for SM and for traits characterizing germination. In the LEG experiment, two QTLs involved in the variation in T90 were detected on chromosomes 5 and 7. These QTLs were distinct from those identified with SNES data.
QTLs of seed mass
SM QTLs identified in LR5 were located on chromosomes 5 and 8 (Table 2a; Fig. 1) each explaining more than 10% of SM variation. In LR4 (SNES), four QTLs were identified on chromosomes 1, 5, 7 and 8 (Table 2b; Fig. 1). The QTL on chromosome 7 had the highest LOD value (7.6) and the highest explained variance (18.5%). The genetic effect at this QTL was negative, i.e. the DZA315.16 alleles decreased SM values. SM QTLs identified with the LEG seed lot on chromosomes 1 and 7 were a subset of those found with the SNES seed lot and could therefore be considered as stable QTLs in the LR4 genetic background. The support intervals of the QTLs identified on chromosome 8 for both populations overlapped but their peaks were not in the same marker intervals, suggesting they may be different.
Whatever the RIL population used, SM QTLs were not co-localized with any QTL of other variables in spite of the correlations observed between SM and other traits (HW, ISSA). The QTLs detected for SM and Ti (i < 50%) on chromosome 5 in the LR4 SNES experiment overlapped, suggesting a pleiotropic effect or tight linkage between them. If so, alleles that resulted in a high SM at these QTLs would result in a longer germination time (lower germination rate). However, these QTLs were probably distinct as the two variables were not correlated and the peaks of both QTLs were not in the same marker intervals.
Candidate genes identified by data mining
Depending on the size of the QTL support interval (ranging from 4 to 16 cM), 10 to 34 BACs and 20 to 40 BACs were analyzed per QTL in LR5 and LR4, respectively. A total of 204 BACs were analyzed in LR5 and 151 in LR4. The number of genes analysed in support intervals was 700 in average. Up to now, about 50% of M. tr. genes have unknown function (http://www.medicago.org/). Table 3 lists the candidate genes selected for SM and for the other traits, from imbibition to elongation, based on a priori hypotheses we had made at cellular and ecophysiological scales.
Table 3.
Genes identified in the support intervals of QTLs, by data mining and BLAST results for similarity with A. thaliana genes (a) for QTLs identified at sub-optimal temperatures in the LR5 population and (b) for QTLs identified at a supra-optimal temperature in the LR4 population
| Trait | QTL | Genea | Proteinb | GO biological processc | Gene position on UMN mapd | Gene position on consensus mape | M. truncatula BAC accession number | A. thaliana homolog locus | E valuef | M. truncatula TC number |
|---|---|---|---|---|---|---|---|---|---|---|
| (a) | ||||||||||
| Dry seed | SM5 | KING1 | SNF1-related protein kinase regulatory subunit gamma 1 | Unknown | 20.9 | 21.6 | AC136286_33 | At3g48530 | 6.00E-50 | NP7273625 |
| Germination | T90.8 | AQP1 | Aquaporin TIP2-1 | Transport | 4 | 2.6 | AC144721_16 | At3g16240 | 2.00E-69 | TC113742 |
| FPG3 | OBF4 | bZIP transcription factor | Regulation of transcription | 23.9 | 34.9 | AC150247_11 | At5g10030 | 5.00E-23 | AW685623 | |
| Heterotrophic growth | HL1 | K21H1.15 | DREB1 transcription factor, ERF | Regulation of transcription | 1 | 4.4 | AC148486_29 | At5g67190 | 2.00-34 | TC129893 |
| HL8 | CBF1/DREB1B | C-repeat/DRE binding factor | 50.9 | 70.8 | AC119408_11 | At4g25490 | 2.00E-21 | TC132828 | ||
| HL1 | RD22 | Dehydration responsive protein | Response to desiccation, salt stress and ABA | 0 | 3.3 | AC127429_30 | At5g25610 | 5.00E-64 | TC127406 | |
| HL1 | ZAT10 | Salt tolerance zinc finger protein | Response to stress and ABA | 2.2 | 5.0 | AC140548__9 | At1g27730 | 6.00E-37 | TC113919 | |
| HL8 | CSDP1 | Glycine rich protein, cold shock domain protein 1 | Response to cold, DNA duplex and RNA secondary structure unwinding | 49.5 | 70.4 | AC160096_24 | At4g36020 | 3.00E-37 | TC121448 | |
| HL2 | HSP70B | Heat shock protein 70kD | Protein folding, response to stress | 3 | 6.2 | AC140547_7 | At1g16030 | 1.00E-178 | NP7266868 | |
| HL8 | HSP70T-2 | Similar to heat shock protein 70kD, ATP binding | 56.1 | 71 | AC122726_8 | At2g32120 | 1.00E-124 | TC114123 | ||
| HL8 | GASA1 | Gibberellin acid-regulated protein | Cell growth, response to gibberellin stimulus | 49.5 | 70.4 | AC138453_32 | At1g75750 | 2.00E-21 | NP7275579 | |
| HL1 | AtEXPA11 | Expansin A11 | Cell organization and biogenesis | 6 | 6.5 | AC146790_52 | At1g20190 | 2.00E-54 | TC124171 | |
| HL2 | At5g44310.1 | LEA domain-containing protein | Embryonic development ending in seed dormancy | 1.5 | 1.7 | AC146855_27 | At5g44310 | 5.00E-20 | TC119095 | |
| HW2 | ICE1 | Inducer of CBF expression 1 | Regulation of transcription, response to stress | 54.7 | 69.2 | AC126013_9 | At3g26744 | 4.00E-16 | TC114412 | |
| HW2 | CBF1/DREB1B | C-repeat/DRE binding factor 1 | Regulation of transcription, response to stress, cold acclimation | 59.9 | 72 | AC138130_21 | At4g25490 | 3.00E-23 | NP7267052 | |
| HW2 | HSP70B | Heat shock protein 70kD | Protein folding, response to stress | 59.9 | 72 | AC138130_37 | At3g12580 | 1.00E-169 | TC131453 | |
| HW2 | XTH9 | Xyloglucan endo-transglucosylase | Carbohydrate or cellular glucan metabolic process | 58.6 | 71.3 | AC145218_27 | At4g03210 | 3.00E-86 | TC113887 | |
| HW2 | At3g54580.1 | Proline-rich extensin-like family protein | Cell organization and biogenesis | 59.6 | 71 | AC161406_29 | At3g54580 | 2.00E-27 | TC138188 | |
| Stage | QTL | Genea | Proteinb | GO biological processc | Gened position on UMN map | Genee position on consensus map | M. truncatula BAC accession number | A. thaliana homolog locus | E valuef | M. truncatula TC number |
|---|---|---|---|---|---|---|---|---|---|---|
| (b) | ||||||||||
| Dry seed | SM7 | F8J2.210 | LEA protein-like | Embryonic development ending in seed dormancy | 49.1 | 58 | AC124218_29 | At3g53040 | 3.00E-05 | TC124162 |
| SM8 | At5g44310.1 | LEA protein-like | 65.3 | 71.2 | AC146862_4 | At5g44310 | 5.00E-20 | TC115299 | ||
| SM5 | F24B9.13 | Cupin family protein, seed storage protein | Unknown | 6.7 | 6.3 | AC123571_46 | At1g07750 | 4.00E-69 | TC132176 | |
| Imbibition | IR4 | AtTT12 | Transparence testa 12 protein | Biosynthetic process, maintenance of seed dormancy | 33.2 | 34.7 | AC124969_55 | At3g59030.1 | 7.00E-13 | TC131970 |
| Germination | T20.5, T50.5 | AtLPP2 | Phosphatidic acid phosphatase, ABI4 regulator | ABA mediating signalling pathway | 0 | 1 | CR931729_1 | At1g15080 | 2.00E-39 | TC121991 |
| T90.5 | ABI4 | Pathogenesis-related transcriptional factor, ERF | Regulation of transcription, ABA mediating signalling pathway | 72.2 | 72.7 | AC144729_8 | At2g40220 | 7.00E-31 | NP7260064 | |
| T90.7 | ABI5 | ABA insensitive-protein | 53.3 | 61 | AC149134_18 | At2g36270 | 4.00E-16 | TC129233 | ||
| T20.5, T50.5 | HSP70T-2 | Heat shock protein 70kD | Protein folding, response to stress | 0 | 1 | AC145222_40 | At2g32120 | 0.00E + 00 | TC114123 | |
| T90.7 | F28J15.2 | DNAJ heat shock N-terminal domain-containing protein | Protein folding | 53.3 | 61 | AC155881_12 | At3g12170.1 | 2.00E-23 | TC113877 | |
| T20.5, T50.5 | F16J14.9 | Similar to HSP20-like chaperone | Unknown | 0 | 1 | AC135320_14 | At3g22530 | 8.00E-33 | TC118335 | |
| T70.5 | GLP2A | Germin-like protein | 33.6 | 39.6 | AC140031_7 | At3g05950 | 3.00E-64 | TC122225 | ||
| T90.7 | FAD2 | Omega-6 fatty acid desaturase | Fatty acid biosynthetic process | 57.7 | 64.9 | AC186676_34 | At3g12120 | 1.00E-118 | TC114302 | |
| T20.5, T50.5 | AtEXLB1 | Expansin-like B1 | Cell organization and biogenesis, other biological processes | 0 | 1 | CR931735_16 | At4g17030 | 5.00E-35 | TC118172 | |
| Early elongation rate | ELR3 | AtEXPA13 | Expansin A13 | Cell organization and biogenesis, other biological processes | 49.3 | 50.7 | AC151822_28 | At3g03220 | 3.00E-41 | CX522867 |
| KOR1 | Endoglucanase 25 | 62.2 | 57.4 | AC144618_29 | At5g49720 | 6.00E-78 | NP7251674 | |||
SM seed mass, T20–T90 times to reach 20–90% of germination, FPG final percentage of germination, IR imbibition rate, HL final hypocotyl length, HW final hypocotyl width, ELR early embryonic axis elongation rate
a, b, cGene name, protein name and GO gene onthology biological process verified using UniProtKB website SwissProt module
dUMN map: integrated genetic map of Medicago truncatula (http://medicago.org/)
eConsensus map: projection of LR5 map onto LR4 map
fClosest Arabidopsis homolog identified using Tair BLAST. AGI protein database was queried with Medicago truncatula nucleotide sequences using BLASTX
Seed mass, seed imbibition and germination
Regarding SM QTLs, several genes found in the support intervals encode proteins accumulated at the end of seed development (Table 3): late embryogenesis abundant protein LEAs (Dure 1985) and the SNF1-related (SnRK1 gamma subunit). SnRK1 gamma subunit influences seed longevity in Medicago truncatula (Buitink et al. 2004; Rosnoblet et al. 2007). Co-localized with the QTL for imbibition rate, we found the ortholog of AtTT12, involved in proanthocyanidin biosynthesis (Debeaujon et al. 2001). It has been suggested that flavonoids such as proanthocyanidins present in the seed coat could slow down imbibition (Kantar et al. 1996; Debeaujon et al. 2000; Debeaujon et al. 2001).
Two genes were selected in the support intervals of QTLs identified for germination at 5°C (Table 3a). One encoded a bZIP transcription factor type (OBF4) known to be involved in responses to cold and biotic stresses (Zhang et al. 1993; Johnson et al. 2001; Singh et al. 2002; Bae et al. 2003). The second gene corresponded to the human gene AQP1 that encodes a δ-TIP-aquaporin. Such aquaporins are involved in water channel activity in the tonoplast (Sarda et al. 1999; Maurel and Chrispeels 2001). At supra-optimal temperatures, ABA biosynthesis plays a key role in the regulation of germination (Larkindale et al. 2005; Tamura et al. 2006; Toh et al. 2008). Argyris et al. (2008) found contrasting seed thermoinhibition in two lettuce genotypes with different sensitivity to ABA. They found a QTL associated with thermotolerance that was co-localized with a QTL for ABA sensitivity, and with a gene encoding an enzyme of the ABA biosynthetic pathway. In our study several genes related to ABA signalling were positioned under QTLs associated with germination at supra-optimal temperature: orthologs of AtLPP2, ABI4 and ABI5, respectively (Table 3b). The latter genes encode transcription factors with an APETALA2 and a basic leucine zipper (bzip) domain, respectively. ABA-insensitive abi4 mutants show decreased sensitivity to ABA inhibition of germination (Finkelstein et al. 1998) and AtLPP2 encodes a lipid phosphate phosphatase which functions as a negative regulator upstream of ABI4 (Katagiri et al. 2005). We also found heat-shock protein (HSP) genes co-localized with QTLs for Ti. HSPs have chaperone-like functions and implications for signal transduction, and could help maintain metabolic process integrity affected by heat stress (Feder and Hoffman 1999; Wahid et al. 2007). Changes in membrane fluidity are one of the immediate reactions of plants subjected to temperature stress (Sung et al. 2003). Fluidity increases with membrane lipid unsaturation. In photosynthesising plants, for example in soybean, mutants deficient in fatty acid unsaturation rendered the plant more tolerant to high temperatures (Alfonso et al. 2001). We found an omega 6 fatty acid desaturase (FAD2) gene in the T90.7-LR4 support interval. Finally, cell expansion is among the events necessary for radicle protrusion and we found a gene encoding an expansin-like B1 co-mapped with T20.5- and T50.5-LR4.
Heterotrophic growth
Concerning heterotrophic growth at 10°C, we selected several genes associated with cell elongation in the support intervals of QTLs (Table 3a): AtEXPA11, a gene encoding an α-expansin protein which causes cell-wall loosening in growing cells (Sampedro and Cosgrove 2005); orthologs of At3g54580 and XTH9 that encode, respectively, an extensin, which plays a role in the strength and rigidity of the cell wall (Weiser et al. 1990), and a xyloglucan endotransglucosylase/hydrolase, which is involved in cell wall loosening and cell elongation (Hyodo et al. 2003); a gibberellin responsive gene GASA1 (Herzog et al. 1995; Vriezen et al. 2004) that might be involved in cell elongation. At the supra-optimal temperature tested, several genes related to cell elongation or to the synthesis of cell wall compounds were also found: AtEXPA13, which encodes an α-expansin protein, and AtGH9A1, also called KOR1, which encodes a glycoside transferase, a membrane bound endo-1, 4-β-d-glucanase involved in cellulose biosynthetic process (Nicol et al. 1998). In Arabidopsis, dark-grown kor1 mutants had extreme dwarf phenotypes with pronounced architectural alterations in the primary cell wall (Nicol et al. 1998; His et al. 2001).
We found seven genes encoding transcription factors involved in the stress-response cascade linked to or independent of the CBF/DREB mediated pathway (C-repeat Binding Factor/Dehydration Responsive Element Binding; see for instance reviews by Sung et al. 2003, and Yamaguchi-Shinozaki and Shinozaki 2006) within the support interval of QTLs identified at 10°C (Table 3a): three CBFs, i.e. ICE1, a constitutive transcription factor involved in the induction of CBF genes under cold or freezing conditions (Chinnusamy et al. 2003; Lee et al. 2005); RD22, involved in tolerance to dehydration (Abe et al. 1997); ZAT10, a C2H2 zinc finger type protein, which might down-regulate the expression of cold-responsive (COR) genes and which is responsive to dehydration (Lee et al. 2002); and CSDP1, a zinc finger CCHC type protein (Kim et al. 2007). We also found genes involved in cellular protection mechanisms: genes encoding HSP70s, chaperone proteins playing a role in membrane stabilization and protein refolding under stress conditions (Wang et al. 2003) and one gene encoding a LEA protein, such protein being assumed to act as water-binding molecules in ion sequestration and in membrane stabilization (Wang et al. 2003; Tunnacliffe and Wise 2007; Hundertmark and Hincha 2008).
Discussion
The present study is an initial step towards combining ecophysiological knowledge on crop emergence and genomic information, for QTL analysis and for the identification of genes that are important for germination and heterotrophic growth in sowing conditions. Such studies combining several scales of knowledge are still rare but necessary. The segregating populations we analysed were chosen because when described with an agro-ecophysiological framework of emergence in field conditions, their parental lines displayed contrasting traits (Brunel et al. 2009). QTL mapping was enhanced because several variables associated with emergence were measured and defined on the basis of an ecophysiological model. The two stages studied, germination and early seedling growth, depend on different physiological processes, and the phenotyping procedures were carefully defined to distinguish these two stages. This led to the detection of distinct QTLs for these two phases and all traits, whatever the temperature tested or genetic background, supporting the existence of different genetic controls. Bettey et al. (2000) also found distinct QTLs for germination and seedling heterotrophic growth in Brassica oleracera. In contrast, Argyris et al. (2005) found co-localized QTLs for such traits in Lactuca but their measurements did not de-correlate values for the two stages. This demonstrated the need to separate the different stages and to perform multiple trait analyses, considering the complexity of analyzing the genetic control of the emergence process. As seed mass has often been shown to influence imbibition and germination rates, and also hypocotyl width (Sinha and Ghildyal 1979; Calero et al. 1981; Rao 1981), this trait was also measured and except with hypocotyl width no correlations were found with other traits. Moreover, QTLs for seed mass were not co-localized with QTLs associated with germination characteristics and hypocotyl length and width, as also observed by Bettey et al. (2000). Three SM QTLs (SM1-LR4, SM5-LR4 and SM7-LR4) coincided with stable QTLs identified for flowering date (Pierre et al. 2008), suggesting an influence of flowering date on SM in LR4. Concerning epistatic interactions, we chose to use a high statistical threshold to avoid false-positive interactions (Boer et al. 2002). Epistasis has already been tested in some QTL studies focused on germination and seedling growth (e.g. Foolad et al. 1998; Clerkx et al. 2004; Argyris et al. 2005; Zhang et al. 2005; Hayashi et al. 2008) and significant interactions between loci were reported for seed dormancy (Alonso-Blanco et al. 2003), for germination on ABA and at high temperatures (Clerkx et al. 2004; Argyris et al. 2005), and for seedling growth after emergence under cold conditions (Zhang et al. 2005). In the present study, we found only one significant interaction at the chosen threshold concerning hypocotyl width for seedlings grown under sub-optimal temperature.
Studies on the effects of several abiotic stresses during germination have reported shared QTLs (Finch-Savage et al. 2005; Foolad et al. 2007). Many possible cross-talk mechanisms exist at molecular level between different stresses (Bae et al. 2003; Kaplan et al. 2004; Lee et al. 2005; Fujita et al. 2006), and responses to abiotic stresses, including temperature-induced stresses, range from general to specific (Kreps et al. 2002). In the present study, we used the loci of the consensus genetic-physical map developed by Huguet et al. (2007) to connect the genetic maps, to enable comparison of QTLs identified for low and high temperatures in different genetic backgrounds and we did not identify any shared QTLs. But we cannot conclude definitively on a different genetic basis as two genetic backgrounds were analysed. As phenotyping germination and heterotrophic growth of a large number of lines is time-consuming, we decided to take advantage of two experiments being conducted on LR4 at supra-optimal temperature. When comparing the results of the two experiments, we observed not only the lower germination rates in the LEG experiment, but also that the QTLs identified in the two experiments were different. This could be linked to differences in seed preparation and vernalization before germination at 20°C, and, as different seedlots were used, it could also be due to different seedlot characteristics. This confirms the need to pay careful attention to phenotyping procedures to identify stable QTLs, that are also free from the influence of seedlot characteristics, before designing markers to assist selection or cloning QTLs.
One advantage of conducting QTL analyses on a model species is that it facilitates the search for candidate genes underlying these QTLs. The present QTL analysis should enable identification of key genes associated with variations in the different variables which characterise each stage. Genetic maps of M. tr. are anchored in the physical map (Huguet et al. 2007), and genetic markers thus constitute reliable boundaries for in silico searches for candidate genes. Data mining for candidate genes depends to a great extent on the state of knowledge, and there is a great need to combine several scales to improve our knowledge of plant functioning in their environment. Because of ecophysiological hypotheses, we focused on genes involved in the signalling of and response to extreme temperature stress and linked to modifications in the membrane and in the cell wall. Due to results showing the absence of complete cell division until radicle protrusion and during hypocotyl growth in the dark (Gendreau et al. 1997) careful attention was paid to the role of cell elongation. Cell wall modifications are involved in hypocotyl growth regardless of temperature (Refregier et al. 2004; Derbyshire et al. 2007) and the balance between cell wall rigidity and loosening is controlled by temperature. Changes in membrane fluidity have been hypothesized to be the primary temperature sensing event (Murata and Los 1997; Penfield 2008). These main hypotheses enabled us to identify an initial set of candidate genes including expansin, extensin, xyloglucan endotransglucosylase and gibberellin responsive genes. Candidate genes encoding proteins involved in membrane protection and repair in the case of abiotic stress (LEA like proteins, HSP70 proteins) were also found, as well as genes that encode transcription factors (RD22, ZAT10, ICE1, CBF1, CSDP1, OBF4) involved in cold and drought stress signalling cascades (Thomashow 2001; Singh et al. 2002; Xiong et al. 2002; Wang et al. 2003; Kacperska 2004; Van Buskirk and Thomashow 2006). Most molecular studies have focused on freezing acclimation at later stages in photosynthetic seedlings and our results suggest that this regulation cascade during cold stress could be more general. At supra-optimal temperature, the QTLs controlling germination times were co-localized with several genes related to response to and protection against stress damage, to ABA signalling or to membrane fluidity. In particular, some genes found under QTLs were related to sensitivity to ABA (ABI4 and ABI5, AtLPP2). These genes could be potential candidates, as the ABA–GA balance is pivotal in controlling germination (Cadman et al. 2006), and endogenous production and sensitivity to ABA play a significant role in regulating seed germination at high temperatures in Arabidopsis (Tamura et al. 2006), lettuce (Argyris et al. 2008), and wheat (Walker-Simmons 1988).
Finally, we focused on candidate genes that appeared to be relevant considering our hypotheses on the main physiological pathways that are possibly involved in response to temperature stresses. We found several valuable candidate genes in the same chromosomal area. Some studies on cereals have reported clusters of relevant candidate genes that co-mapped with one abiotic stress tolerance QTL (for instance Teulat et al. 2003; Tondelli et al. 2006). But many genes in the QTL support intervals had no known functions. Moreover, many publications on the effects of temperature at molecular level are not specific to these stages and often concern later stages in photosynthesizing plants. Specific metabolic pathways may exist for early heterotrophic stages when no photosynthesis resources are available and seedlings live on seed resources. Transcriptome profiling during heterotrophic hypocotyl growth will help to identify genes differentially expressed at extreme temperatures in different genotypes. These sets of genes could be then compared with the candidate genes we selected, and more generally to all genes positioned in the QTL support intervals. Combining ecophysiological, genetic and molecular analysis should provide reliable markers to assist selection with a view to improve crop stand establishment. Pea and other legumes should be the first species concerned, although the results could be generalized to other species.
Acknowledgments
This research was funded by Region Pays de la Loire, INRA and GEVES. We are grateful to Lydie Ledroit and the technical staff of the Seed National Testing Laboratory, Delphine Moreau and the technical staff of UMR LEG in Dijon for sharing their experiments on LR4. We also thank Jean-Marie Prosperi, of the Medicago truncatula Biological Resource Centre in Montpellier (UMR 1097 France) for providing seeds from the LR4 population.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chaarani GR, Gentzbittel L, Wedzony M, Sarrafi A. Identification of QTLs for germination and seedling development in sunflower (Helianthus annuus L.) Plant Sci. 2005;169:221–227. doi: 10.1016/j.plantsci.2005.03.016. [DOI] [Google Scholar]
- Alfonso M, Yruela I, Almárcegui S, Torrado E, Pérez MA, Picorel R. Unusual tolerance to high temperatures in a new herbicide-resistant D1 mutant from Glycine max (L.) Merr. cell cultures deficient in fatty acid desaturation. Planta. 2001;212:573–582. doi: 10.1007/s004250000421. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Vries HBE, Koornneef M. Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics. 2003;164:711–729. doi: 10.1093/genetics/164.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J. BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics. 2004;20:2324–2326. doi: 10.1093/bioinformatics/bth230. [DOI] [PubMed] [Google Scholar]
- Argyris J, Truco MJ, Ochoa O, Knapp SJ, Still DW, Lenssen GM, Schut JW, Micelmore RW, Bradford KJ. Quantitative trait loci associated with seed and seedling traits in Lactuca. Theor Appl Genet. 2005;111:1365–1376. doi: 10.1007/s00122-005-0066-4. [DOI] [PubMed] [Google Scholar]
- Argyris J, Dahal P, Hayashi E, Still DW, Bradford KJ. Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism and response genes. Plant Physiol. 2008;148:926–947. doi: 10.1104/pp.108.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert G, Morin J, Jacquin F, Loridon K, Quillet MC, Petit A, Rameau C, Lejeune-Hénaut I, Huguet T, Burstin J. Functional mapping in pea, as an aid to the candidate gene selection and for investigating synteny with the model legume Medicago truncatula. Theor Appl Genet. 2006;112:1024–1041. doi: 10.1007/s00122-005-0205-y. [DOI] [PubMed] [Google Scholar]
- Bae MS, Cho EJ, Choi E-Y, Park OK. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003;36:652–663. doi: 10.1046/j.1365-313X.2003.01907.x. [DOI] [PubMed] [Google Scholar]
- Barroco RM, Van Poucke K, Bergervoet JHW, De Veylder L, Groot SPC, Inze D, Engler G. The role of the cell cycle machinery in resumption of postembryonic development. Plant Physiol. 2005;137:127–140. doi: 10.1104/pp.104.049361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettey M, Finch-Savage WE, King GJ, Lynn JR. Quantitative genetic analysis of seed vigour and pre-emergence seedling growth traits in Brassica oleracea. New Phytol. 2000;148:277–286. doi: 10.1046/j.1469-8137.2000.00760.x. [DOI] [Google Scholar]
- Bewley JD, Black M. Seeds: physiology of development and germination. 2. New York: Plenum Press; 1994. [Google Scholar]
- Boer MP, ter Braak CJF, Jansen RC. A penalized likelihood method for mapping epistatic quantitative trait loci with one-dimensional genome searches. Genetics. 2002;162:951–960. doi: 10.1093/genetics/162.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botwright TL, Rebetze G, Condon T, Richards R. The effect of rht genotype and temperature on coleoptile growth and dry matter partitionning in young wheat seedlings. Aust J Plant Physiol. 2001;28:417–423. [Google Scholar]
- Brunel S, Teulat-Merah B, Wagner M-H, Huguet T, Prosperi JM, Durr C. Using a model-based framework for analysing genetic diversity during germination and heterotrophic growth of Medicago truncatula. Ann Bot. 2009;103:1103–1117. doi: 10.1093/aob/mcp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitink J, Thomas M, Gissot L, Leprince O. Starvation, osmotic stress and desiccation tolerance lead to expression of different genes of the regulatory beta and gamma subunits of the SnRK1 complex in germinating seeds of Medicago truncatula. Plant Cell Environ. 2004;27:55–67. doi: 10.1046/j.0016-8025.2003.01125.x. [DOI] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- Calero E, West SH, Hinson K. Water absorption of soybean seeds and associated causal factors. Crop Sci. 1981;21:926–933. doi: 10.2135/cropsci1981.0011183X002100060030x. [DOI] [Google Scholar]
- Cannon SB, Crow JA, Heuer ML, Wang X, Cannon EKS, Dwan C, Lamblin AF, Vasdewani J, Mudge J, Cook A, Gish J, Cheung F, Kenton S, Kunau TM, Brown D, May GD, Kim D, Cook DR, Roe BA, Town CD, Young ND, Retzel EF. Databases and Information Integration for the Medicago truncatula genome and transcriptome. Plant Physiol. 2005;138:38–46. doi: 10.1104/pp.104.059204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Schiex T, Spannagl M, Monaghan E, Nicholson C, Humphray SJ, Schoof H, Mayer KFX, Rogers J, Quetier F, Oldroyd GE, Debelle F, Cook DR, Retzel EF, Roe BA, Town CD, Tabata S, Van de Peer Y, Young ND. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc Natl Acad Sci USA. 2006;103:14959–14964. doi: 10.1073/pnas.0603228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantliffe D, Schuler KD, Guedes AC. Overcoming seed thermodormancy in a heat- sensitive romaine lettuce by seed priming. HortScience. 2000;16:196–198. [Google Scholar]
- Chase K, Adler FR, Lark KG. Epistat: a computer program for identifying and testing interactions between pairs of quantitative trait loci. Theor Appl Genet. 1997;94:724–730. doi: 10.1007/s001220050471. [DOI] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Kim D, Uhm T, Limpens E, Lim H, Mun JH, Kalo P, Penmetsa RV, Seres A, Kulikova O, Roe BA, Bisseling T, Kiss GB, Cook DR. A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa. Genetics. 2004;166:1463–1502. doi: 10.1534/genetics.166.3.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, Young ND, Cook DR. Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA. 2004;101:15289–15294. doi: 10.1073/pnas.0402251101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx EJM, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-de Vries H, Groot SPC, Vreughenhil D, Koorneef M. Analysis of natural variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 2004;135:432–443. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Peng B, Xing Z, Xu G, Yu B, Zhang Q. Molecular dissection of seedling-vigor and associated physiological traits in rice. Theor Appl Genet. 2002;105:745–753. doi: 10.1007/s00122-002-0908-2. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination and longevity in Arabidopsis. Plant Physiol. 2000;122:403–413. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 2001;13:853–871. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demilly D, Wagner MH, Brunel S, Dürr C (2007) Computer vision for helping analysis of relationship between seed physical characteristics and germination and root elongation on Medicago truncatula. In: 28th ISTA Seed Symposium, May 2007, Iguassu Falls, Brazil
- Derbyshire P, Findlay K, McCann MC, Roberts K. Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J Exp Bot. 2007;58:2079–2089. doi: 10.1093/jxb/erm074. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Luckow MA. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 2003;131:900–910. doi: 10.1104/pp.102.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducournau S, Feutry A, Plainchault P, Revollon P, Vigouroux B, Wagner MH. An image acquisition system for automated monitoring of the germination rate of sunflower seeds. Comput Electron Agric. 2004;44:189–202. doi: 10.1016/j.compag.2004.04.005. [DOI] [Google Scholar]
- Ducournau S, Feutry A, Plainchault P, Revollon P, Vigouroux B, Wagner M-H. Using computer vision to monitor germination time course of sunflower (Helianthus annuus L.) seeds. Seed Sci Tech. 2005;33:329–340. [Google Scholar]
- Dure L. Embryogenesis and gene expression during seed formation. Oxf Surv Plant Mol Cell Biol. 1985;2:179–197. [Google Scholar]
- Feder ME, Hoffman GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Ann Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Côme D, Lynn JR, Corbineau F. Sensitivity of Brassica oleracea seed germination to hypoxia: a QTL analysis. Plant Sci. 2005;169:753–759. doi: 10.1016/j.plantsci.2005.05.026. [DOI] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foolad MR, Chen FQ, Lin GY. RFLP mapping of QTLs conferring cold tolerance during seed germination in an interspecific cross of tomato. Mol Breed. 1998;4:519–529. doi: 10.1023/A:1009648112491. [DOI] [Google Scholar]
- Foolad MR, Lin GY, Chen FQ. Comparison of QTLs for seed germination under non-stress, cold stress and salt stress in tomato. Plant Breed. 1999;118:167–173. doi: 10.1046/j.1439-0523.1999.118002167.x. [DOI] [Google Scholar]
- Foolad MR, Zhang LP, Subbiah P. Genetics of drought tolerance during seed germination in tomato: inheritance and QTL mapping. Genome. 2003;46:536–545. doi: 10.1139/g03-035. [DOI] [PubMed] [Google Scholar]
- Foolad MR, Subbiah P, Zhang LP (2007) Common QTL affect the rate of tomato seed germination under different stress and non stress conditions. Int J Plant Genomics. doi:10.1155/2007/97386 [DOI] [PMC free article] [PubMed]
- Fujino K, Sekiguchi H, Sato T, Kiuchi H, Nonoue Y, Takeuchi Y, Ando T, Lin SY, Yano M. Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.) Theor Appl Genet. 2004;108:794–799. doi: 10.1007/s00122-003-1509-4. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opi Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H. Cellular Basis of Hypocotyl Growth in Arabidopsis thaliana. Plant Physiol. 1997;114:295–305. doi: 10.1104/pp.114.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno-Gilles C, Lelièvre E, Viau L, Malik-Ghulam M, Ricoult C, Niebel A, Leduc N, Limami AM. ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture : modifying enzymes and structural proteins in Medicago truncatula embryo axis. Mol Plant. 2009;2:108–119. doi: 10.1093/mp/ssn092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hayashi E, Aoyama N, Still DW. Quantitative trait loci associated with lettuce seed germination under different temperature and light environments. Genome. 2008;51:928–947. doi: 10.1139/G08-077. [DOI] [PubMed] [Google Scholar]
- Herzog M, Dorne A-M, Grellet F. GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol Biol. 1995;27:743–752. doi: 10.1007/BF00020227. [DOI] [PubMed] [Google Scholar]
- His I, Driouich A, Nicol F, Jauneau A, Hofte H. Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1, 4-beta-glucanase. Planta. 2001;212:348–358. doi: 10.1007/s004250000437. [DOI] [PubMed] [Google Scholar]
- Hopkins WG, Huner NPA (1999) Introduction to plant physiology. 3rd edn. Wiley, New York, 576 p
- Hospital F, Moreau L, Lacoudre F, Charcosset A, Gallais A. More on the efficiency of marker-assisted selection. Theor Appl Genet. 1997;95:1181–1189. doi: 10.1007/s001220050679. [DOI] [Google Scholar]
- Huguet T, Ghérardi M, Chardon F, Sartorel EJP (2007) Creation of consensus genetic-physical map (CGPM) for the identification of Medicago truncatula genes involved in natural populations. Abstracts of the Model Legume Congress, Tunis, Tunisia, 24–28 March
- Hundertmark M, Hincha DK (2008) LEA (Late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118 [DOI] [PMC free article] [PubMed]
- Hyodo H, Yamakawa S, Takeda Y, Tsuduki M, Yokota A, Nishitani K, Kohchi T. Active gene expression of a xyloglucan endotransglucosylase/hydrolase gene, XTH9, in inflorescence apices is related to cell elongation in Arabidopsis thaliana. Plant Mol Biol. 2003;52:473–482. doi: 10.1023/A:1023904217641. [DOI] [PubMed] [Google Scholar]
- Johnson C, Boden E, Desai M, Pascuzzi P, Arias J. In vivo target promoter-binding activities of a xenobiotic stress-activated TGA factor. Plant J. 2001;28:237–243. doi: 10.1046/j.1365-313X.2001.01147.x. [DOI] [PubMed] [Google Scholar]
- Kacperska A. Sensor types in signal transduction pathways in plant cells responding to abiotic stressors: do they depend on stress intensity? Physiol Plantarum. 2004;122:159–168. doi: 10.1111/j.0031-9317.2004.00388.x. [DOI] [Google Scholar]
- Kantar F, Pilbeam CJ, Hebblethwaite PD. Effect of tannin content of Faba bean (Vicia faba) seed on seed vigour, germination and field emergence. Ann Appl Biol. 1996;128:85–93. doi: 10.1111/j.1744-7348.1996.tb07092.x. [DOI] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Ishiyama K, Kato T, Tabata S, Kobayashi M, Shinozaki K. An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. Plant J. 2005;43:107–117. doi: 10.1111/j.1365-313X.2005.02431.x. [DOI] [PubMed] [Google Scholar]
- Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung CH, Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 2007;35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121(1):185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Hall JD, Knight MR, Vierling E. Heat stress phenotypes of Arabidopsis mutants implicate multiple signaling pathways in the acquisition of thermotolerance. Plant Physiol. 2005;138:882–897. doi: 10.1104/pp.105.062257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong LM, Stevenson B, Zhu JK. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 2002;21:2692–2702. doi: 10.1093/emboj/21.11.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, He H, Zhang J, Wang X, Bai S, Stolc V, Tongprasit W, Young N, Yu O, Deng XW. Transcriptional analysis of highly syntenic regions between Medicago truncatula and Glycine max using tiling microarrays. Genome Biol. 2008;9:R57. doi: 10.1186/gb-2008-9-3-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Chrispeels MJ. Aquaporins. A molecular entry into plant water relations. Plant Physiol. 2001;125:135–138. doi: 10.1104/pp.125.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge J, Cannon S, Kalo P, Oldroyd G, Roe B, Town C, Young N. Highly syntenic regions in the genomes of soybean, Medicago truncatula, and Arabidopsis thaliana. BMC Plant Biol. 2005;5:15. doi: 10.1186/1471-2229-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N, Los DA. Membrane fluidity and temperature perception. Plant Physiol. 1997;115:875–879. doi: 10.1104/pp.115.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Hofte H. A plasma membrane-bound putative endo-1, 4-beta-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. Temperature perception and signal transduction in plants. New Phytol. 2008;179:615–628. doi: 10.1111/j.1469-8137.2008.02478.x. [DOI] [PubMed] [Google Scholar]
- Phan H, Ellwood S, Hane J, Ford R, Materne M, Oliver R. Extensive macrosynteny between Medicago truncatula and Lens culinaris ssp. culinaris. Theor Appl Genet. 2007;114:549–558. doi: 10.1007/s00122-006-0455-3. [DOI] [PubMed] [Google Scholar]
- Pierre JB, Huguet T, Barre P, Huyghe C, Julier B. Detection of QTLs for flowering date in three mapping populations of the model legume species Medicago truncatula. Theor Appl Genet. 2008;117:609–620. doi: 10.1007/s00122-008-0805-4. [DOI] [PubMed] [Google Scholar]
- Qi AM, Jaggard KW. Assessing the impact of future climate change on the UK sugar beet production. Aspects Appl Biol. 2008;88:153–159. [Google Scholar]
- Rao SK. Influence of seed size on field germination, seedling vigour, yield and quality in self pollinated crops—a review. Agric Rev. 1981;2:95–101. [Google Scholar]
- Rasband WS (1997) ImageJ—a software for image analysis. US National Institutes of Health
- Rebetzke GJ, Richards RA, Fettell NA, Long M, Condon AG, Forrester RI, Botwright TL. Genotypic increases in coleoptile length improves stand establishment, vigour and grain yield of deep-sown wheat. Field Crop Res. 2007;100:10–23. doi: 10.1016/j.fcr.2006.05.001. [DOI] [Google Scholar]
- Refregier G, Pelletier S, Jaillard D, Hofte H. Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol. 2004;135:959–968. doi: 10.1104/pp.104.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards FJ. A flexible growth function for empirical use. J Exp Bot. 1959;10:290–300. doi: 10.1093/jxb/10.2.290. [DOI] [Google Scholar]
- Ronfort J, Bataillon T, Santoni S, Delalande M, David J, Prosperi JM. Microsatellite diversity and broad scale geographic structure in a model legume: building a set of nested core collection for studying naturally occurring variation in Medicago truncatula. BMC Plant Biol. 2006;6:28. doi: 10.1186/1471-2229-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnoblet C, Aubry C, Leprince O, Ly Vu B, Rogniaux H, Buitink J. The regulatory gamma subunit SNF4b of the sucrose non-fermenting-related kinase complex is involved in longevity and stachyose accumulation during maturation of Medicago truncatula seeds. Plant J. 2007;51:47–59. doi: 10.1111/j.1365-313X.2007.03116.x. [DOI] [PubMed] [Google Scholar]
- Saglio PH, Pradet A. Soluble sugars, respiration, and energy charge during aging of excised maize root tips. Plant Physiol. 1980;66:516–519. doi: 10.1104/pp.66.3.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove D. The expansin superfamily. Genome Biol. 2005;6:242. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarda X, Tousch D, Ferrare K, Cellier F, Alcon C, Dupuis JM, Casse F, Lamaze T. Characterization of closely related delta-TIP genes encoding aquaporins which are differentially expressed in sunflower roots upon water deprivation through exposure to air. Plant Mol Biol. 1999;40:179–191. doi: 10.1023/A:1026488605778. [DOI] [PubMed] [Google Scholar]
- SAS (2000) SAS Institute Inc. Version 8.1
- Sato S, Isobe S, Asamizu E, Ohmido N, Kataoka R, Nakamura Y, Kaneko T, Sakurai N, Okumura K, Klimenko I, Sasamoto S, Wada T, Watanabe A, Kohara M, Fujishiro T, Tabata S. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.) DNA Res. 2005;12:301–364. doi: 10.1093/dnares/dsi018. [DOI] [PubMed] [Google Scholar]
- Schneider A. Variability of maize seed imbibition rates as influenced by seed size distribution and coating application. Agronomie. 1998;18:247–260. doi: 10.1051/agro:19980401. [DOI] [Google Scholar]
- Singh KB, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5:430–436. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Sinha AK, Ghildyal BP (1979) Emergence force of crop seedlings. Plant Soil 51:153–156
- Sung DY, Kaplan F, Lee KJ, Guy CL. Acquired tolerance to temperature extremes. Trends Plant Sci. 2003;8:179–187. doi: 10.1016/S1360-1385(03)00047-5. [DOI] [PubMed] [Google Scholar]
- Tamura N, Yoshida T, Tanaka A, Sasaki R, Bando A, Toh S, Lepiniec L, Kawakami N. Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol. 2006;47:1081–1094. doi: 10.1093/pcp/pcj078. [DOI] [PubMed] [Google Scholar]
- Tardieu F, Granier C. Quantitative analysis of cell division in leaves: methods, developmental patterns and effects of environmental conditions. Plant Mol Biol. 2000;43:555–567. doi: 10.1023/A:1006438321386. [DOI] [PubMed] [Google Scholar]
- Tekrony DM, Bustamam T, Egli DB, Pfeiffer TW. Effects of soybean seedsize, vigor and maturity on crop performance in row and hill plots. Crop Sci. 1987;27:1040–1045. doi: 10.2135/cropsci1987.0011183X002700050043x. [DOI] [Google Scholar]
- Teulat B, Zoumarou-Wallis N, Rotter B, Ben Salem M, Bahri H, This D. QTL for relative water content in field grown barley and their stability across Mediterranean environments. Theor Appl Genet. 2003;108:181–188. doi: 10.1007/s00122-003-1417-7. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. So what’s new in the field of plant cold acclimation? Lots! Plant Physiol. 2001;125:89–93. doi: 10.1104/pp.125.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoquet P, Gherardi M, Journet EP, Kereszt A, Ane JM, Prosperi JM, Huguet T. The molecular genetic linkage map of the model legume Medicago truncatula: an essential tool for comparative legume genomics and the isolation of agronomically important genes. BMC Plant Biol. 2002;2:1. doi: 10.1186/1471-2229-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Imamura A, Watanabe A, Nakabayashi K, Ohamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, Iuchi S, Kobayashi M, Yamaguchi S, Kamiya Y, Nambara E, Kawakami N. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin in Arabidopsis seeds. Plant Physiol. 2008;146:1368–1385. doi: 10.1104/pp.107.113738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondelli A, Francia E, Barabaschi D, Aprile A, Skinner JS, Stockinger EJ, Stanca AM, Pecchioni N. Mapping regulatory genes as candidates for cold and drought stress tolerance in barley. Theor Appl Genet. 2006;112:445–454. doi: 10.1007/s00122-005-0144-7. [DOI] [PubMed] [Google Scholar]
- Tunnacliffe A, Wise MJ. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94:791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- Utz HF, Melchinger MF (1996) PLABQTL: a program for composite interval mapping of QTL. J Quant Loci 2
- Valdes VM, Bradford KJ, Mayberry KS. Alleviation of thermodormancy in coated lettuce seeds by seed priming. HortScience. 1985;20:1112–1114. [Google Scholar]
- Van Buskirk HA, Thomashow MF. Arabidopsis transcription factors regulating cold acclimation. Physiol Plantarum. 2006;126:72–80. doi: 10.1111/j.1399-3054.2006.00625.x. [DOI] [Google Scholar]
- Vocansson A, Jeuffroy MH. Agronomic performance of different pea cultivars under various sowing periods and contrasting soil structures. Agron J. 2008;100:748–759. doi: 10.2134/agronj2005.0301. [DOI] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Vriezen WH, Achard P, Harberd NP, Van der Straeten D. Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J. 2004;37:505–516. doi: 10.1046/j.1365-313X.2003.01975.x. [DOI] [PubMed] [Google Scholar]
- Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: an overview. Environ Exp Bot. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- Walker-Simmons M. Enhancement of ABA responsiveness in wheat embryos by high temperature. Plant Cell Environ. 1988;11:769–775. doi: 10.1111/j.1365-3040.1988.tb01161.x. [DOI] [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Wang R, Bai Y, Tanino K. Effect of seed size and sub-zero imbibition-temperature on the thermal time model of winterfat (Eurotia lanata (Pursh) Moq.) Environ Exp Bot. 2004;51:183–197. doi: 10.1016/j.envexpbot.2003.10.001. [DOI] [Google Scholar]
- Weiser RL, Wallner SJ, Waddell JW. Cell wall and extensin mRNA changes during cold acclimation of pea seedlings. Plant Physiol. 1990;93:1021–1026. doi: 10.1104/pp.93.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Ann Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Zhang B, Foley RC, Singh KB. Isolation and characterization of two related Arabidopsis ocs-element bZIP binding proteins. Plant J. 1993;4:711–716. doi: 10.1046/j.1365-313X.1993.04040711.x. [DOI] [PubMed] [Google Scholar]
- Zhang ZH, Qu XS, Wan S, Chen LH, Zhu YG. Comparison of QTL controlling seedling vigour under different temperature conditions using recombinant inbred lines in rice (Oryza sativa) Ann Bot. 2005;95:423–429. doi: 10.1093/aob/mci039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Choi H-K, Cook DR, Shoemaker RC. Bridging model and crop legumes through comparative genomics. Plant Physiol. 2005;137:1189–1196. doi: 10.1104/pp.104.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]