Abstract
Cyclin-dependent kinase (Cdk) 5 is a unique member of the Cdk family, because Cdk5 kinase activity is detected only in the nervous tissue. Two neuron-specific activating subunits of Cdk5, p35 and p39, have been identified. Overlapping expression pattern of these isoforms in the embryonic mouse brain and the significant residual Cdk5 kinase activity in brain homogenate of the p35−/− mice indicate the redundant functions of the Cdk5 activators in vivo. Severe neuronal migration defects in p35−/−Cdk5 +/− mice further support the idea that the redundant expression of the Cdk5 activators may cause a milder phenotype in p35−/− mice compared with Cdk5−/− mice. Mutant mice lacking either Cdk5 or p35 exhibit certain similarities with Reelin/Dab1-mutant mice in the disorganization of cortical laminar structure in the brain. To elucidate the relationship between Cdk5/p35 and Reelin/Dab1 signaling, we generated mouse lines that have combined defects of these genes. The addition of heterozygosity of either Dab1 or Reelin mutation to p35−/− causes the extensive migration defects of cortical neurons in the cerebellum. In the double-null mice of p35 and either Dab1 or Reelin, additional migration defects occur in the Purkinje cells in the cerebellum and in the pyramidal neurons in the hippocampus. These additional defects in neuronal migration in mice lacking both Cdk5/p35 and Reelin/Dab1 indicate that Cdk5/p35 may contribute synergistically to the positioning of the cortical neurons in the developing mouse brain.
During brain development, cortical neurons are generated in a proliferative zone called the ventricular zone. After final cell division, neurons migrate variable distances before they settle and form unique laminar structures. Several molecules are critical for proper neuronal migration in the developing brain. Gene-targeting experiments in mice identified certain genes including cyclin-dependant kinase (Cdk) 5 and p35 that are involved in the migration of cortical neurons (1, 2). Cdk5 is a serine/threonine kinase with close homology to other Cdks (3–5). Cdk5 is a unique Cdk, because its kinase activity can be detected mainly in postmitotic neurons (6). Association of Cdk5 with a neuron-specific regulatory subunit, either p35 or its isoform p39, is critical for kinase activity (7–9). Cdk5−/− mice exhibit embryonic lethality associated with disruption of the cortical laminar structures in the cerebral cortex, olfactory bulb, hippocampus, and cerebellar cortex (1). Neuronal birthdate labeling by BrdU revealed an inverted pattern of cell layers in the cerebral cortex in Cdk5−/− mice (10). An inverted pattern of layer structure in the cerebral cortex is a well known characteristic of the spontaneous mouse mutants reeler and scrambler/yotari. They exhibit nearly identical phenotypes suggesting that the gene products mutated in these mutants, Reelin and Dab1 respectively, act in a common signaling pathway during cortical development (11–17). Reelin is a secreted extracellular matrix glycoprotein, and Dab1 is an intracellular adapter protein. Recently, ApoER2, VLDLR, and CNR have been shown to be the components of the Reelin receptor (18–21). The critical role of Reelin-induced tyrosine phosphorylation of Dab1 has been demonstrated by using transgenic mice in which the Dab1 protein had all of the potential tyrosine phosphorylation sites mutated (22). Despite these recent discoveries, downstream effectors of Reelin/Dab1 signaling remain to be identified.
Although Cdk5−/− and p35−/− mice demonstrate some similarities with reeler and scrambler/yotari mice, the development of the embryonic cerebral cortex in Cdk5−/− and p35−/− mice also shows significant differences from reeler and scrambler/yotari such as splitting of the preplate (10, 23). In wild-type mice, successive waves of migrating neurons form the cortical plate in an inside-out fashion, splitting the preplate into the marginal zone and subplate. In reeler and scrambler/yotari mutants, the migrating cortical neurons appear incapable of splitting the preplate, and cortical plate neurons stack up in an inverted order beneath the preplate. In Cdk5−/− and p35−/− mice, earlier-born neurons successfully split the preplate; however, late-born neurons stack up in an inverted layer under the subplate (10, 23). Because of the phenotypic similarities and differences between Cdk5/p35 and Reelin/Dab1 mutants, several models have been proposed regarding the relation between Reelin/Dab1 signaling and Cdk5/p35 (24–26). However, there is no evidence that Cdk5/p35 is a downstream effector of Reelin/Dab1 signaling. In the current study, we attempt to clarify the relationship between Cdk5/p35 and Reelin/Dab1 by genetic approaches using mice in which both of these genes have been mutated.
Materials and Methods
Mice.
p35−/− mice were generated by targeted deletion of amino acid residues 148 to the carboxyl terminus by insertion of a neomycin-resistant gene cassette in the p35 gene locus. To construct a targeting vector for the p35 gene, 0.5 kb of NotI–SpeI fragment and 5 kb of SpeI–BamHI fragment from 129/Sv-derived p35 genomic clone (27) were subcloned into XhoI and XbaI/BamHI sites in the pPNT vector, respectively (28). Genotyping of the embryonic stem clones and F1 mice was performed by Southern blot analysis using a 5′ flanking probe (27) and/or by PCR using primers, p35F1 (5′-GTCTCCTCTTCTGTCAAGAAG-3′) and p35R1 (5′-CTCTGCTAGACACATACTGTAC-3′) for the wild-type allele, and p35F1 and PGK-1 (5′-CCATCTGCACGAGACTAGT-3′) for the mutated allele. Mouse lines of p35 and yotari mutants were maintained in C57BL/6 × 129/Sv hybrid background. Cdk5+/− mice were maintained in C57BL/6 background after backcrossing of four generations from C57BL/6 × 129/Sv hybrid. Homozygous reeler mice were bred from heterozygous B6C3Fe-a/a-rl (The Jackson Laboratory). Double-mutant mice were obtained after mating each mouse line, and genotyping for Reelin and Dab1 alleles were performed by PCR (29, 30). For the genotyping of the Cdk5 allele, Cdk5F1 (5′-ATTGTGGCTCTGAAGCGTGTC-3′) and Cdk5R1 (5-CTTGTCACTATGCAGGACATC-3′) primers were used for wild-type allele and Cdk5F1 and PGK-1 for the mutated allele (1).
Biochemical Analyses.
For Western blot analysis, whole brains were homogenized in RIPA buffer (150 mM NaCl/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/10 μg/ml leupeptin/10 μg/ml aprotinin/1 mM phenylmethylsulfonyl fluoride/50 mM Tris⋅HCl, pH 8.0). The homogenates were centrifuged at 12,000 × g for 20 min at 4°C. Protein from the supernatant (20 μg) was subjected to Western blot analysis (31). To detect p35 protein, two anti-p35 rabbit polyclonal antibodies, which recognize a peptide corresponding to either the N terminus (amino acids 13–33) or the carboxyl terminus (amino acids 280–307) of human p35 (ref. 32; K.I., unpublished data) were used. Western blots were developed by using enhanced chemiluminescence (BM Chemiluminescence, Roche Molecular Biochemicals). Cdk5 immunoprecipitation was performed by using anti-Cdk5 antibody (C-8, Santa Cruz Biotechnology; ref. 31). Kinase activity of the Cdk5 immunoprecipitate was measured by using KSPXK peptide, which represents two KSP repeat sequences corresponding to the C terminus of human high-molecular-weight neurofilament protein (31, 33).
Immunohistochemical Study and in Situ Hybridization.
Mice were perfused intracardially with 4% (vol/vol) paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). Then 10-μm cryostat sections were stained with 0.9% toluidine blue solution for Nissl staining. For immunohistochemistry, antibodies were diluted in PBS/0.01% Triton X-100 and 5% BSA. Anti-IP3R mAb (clone 4C11, ref. 34) was used at 1:10 dilution. Secondary antibody was visualized by using diaminobenzidine reaction product as specified by Vectastain Elite protocol (Vector Laboratories). Monoclonal anti-calbindin D-28K antibody (Sigma) was used at 1:1000. For fluorescent staining, FITC-conjugated anti-mouse IgG (Jackson ImmunoResearch) was used. In situ hybridization was performed by using either 35S-labeled or digoxigenin-labeled probe as described (35, 36). p39 cDNA clones were obtained by screening the adult mouse-brain cDNA library (Stratagene) by using a reverse transcription–PCR-generated human p39 cDNA fragment (nucleotides 664-1038, GenBank accession no. U34051) as a probe and verified by sequencing (37).
Results
p35−/− Mice Exhibit a Mild Phenotype Because of the Residual Cdk5 Kinase Activity in the Developing Brain.
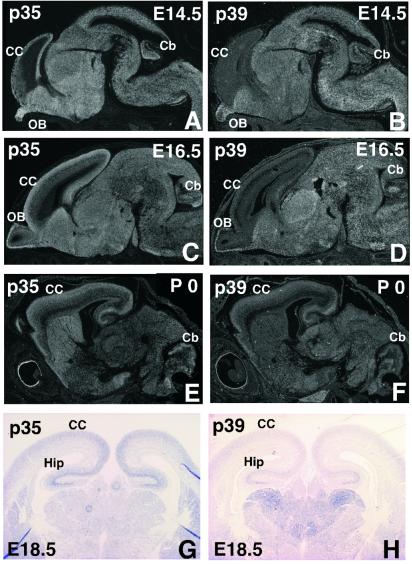
To study the expression pattern of two Cdk5 activators in the brain, we performed a comparative study of p35 and p39 mRNA expression in embryonic (embryonic days (E) 13.5, 14.5, and 16.5) and newborn mouse brains by in situ hybridization with 35S-labeled anti-sense probe on parasagittal sections (Fig. 1 A–F). At E13.5, only p35 but not p39 is detected in the brain including cerebral cortex (data not shown). In general, an overlapping expression of these two activators is observed in most of the brain areas except the cerebral cortex. Apparently, very low expression of p39 and a dominant expression of p35 is detected in the cerebral cortex at E14.5 and E16.5. At birth, both isoforms are expressed to a similar degree in the cerebral cortex. Coronal sections from E18.5 embryonic brain hybridized with digoxigenin-labeled anti-sense p35 and p39 probes indicated expression of p35 and p39 in the cerebral cortex (Fig. 1 G and H). These results are comparable to those obtained in a rat study (38).
Figure 1.
Overlapping expression of Cdk5 activating subunits. Comparison of p35 and p39 gene expression in sagittal brain sections from E14.5 and E16.5 embryos and newborns with 35S-labeled riboprobes (A–F) and in coronal sections from E18.5 embryos with digoxigenin-labeled probes (G and H) by in situ hybridization. Expressions of p35 and p39 overlap in the brain except in the cerebral cortex at E14.5 and E16.5. CC, cerebral cortex; Cb, cerebellum; OB, olfactory bulb; Hip, hippocampus.
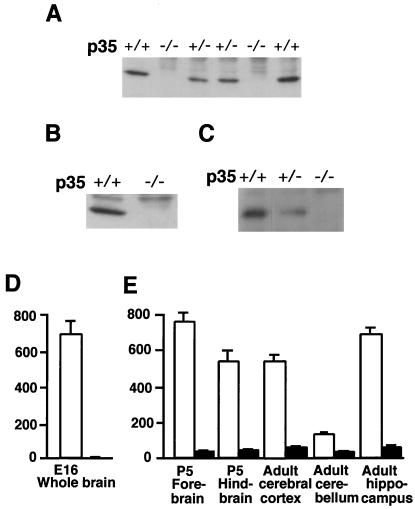
We generated p35−/− mice by targeted disruption of the p35 gene, and complete inactivation of p35 was confirmed by Northern blot analysis (data not shown) and also by Western blot analysis using brain homogenates from p35+/+ and p35−/− mice (Fig. 2 A–C). Unlike Cdk5−/− mice that die during perinatal period (1), p35−/− mice were found to be viable and fertile. Some of the mice experience sudden death in adult age possibly because of spontaneous seizures. Histological abnormalities of laminar structure in p35−/− mice are evident in the cerebral cortex (data not shown), whereas the other Cdk5 activator p39 is not expressed overtly until E18.5 (Fig. 1). p35−/− mice also exhibit subtle abnormalities in laminar structures of the olfactory bulb (data not shown), hippocampus, and cerebellum (Fig. 3 C and D and Fig. 5G), in which mRNA expressions of both p35 and p39 are detected (Fig. 1 A–D). This phenotype is almost identical to another p35−/− mouse line (2). To determine the biochemical basis for the phenotypic differences observed between p35−/− and Cdk5−/− mice, we performed Cdk5-specific kinase assays by using Cdk5 immunoprecipitates of brain homogenate from Cdk5−/− and p35−/− mice with littermate wild-type controls (Fig. 2 D and E). As we reported in the earlier study, no detectable Cdk5 kinase activity was observed in Cdk5−/− brain homogenate (1). On the other hand, p35−/− brain homogenates exhibit substantial residual Cdk5 kinase activity, corresponding to 10 and 20% in the cerebral cortex and the cerebellum, respectively, compared with the wild-type controls. The cerebellum of p35−/− mice has much higher residual activity, indicating more contribution of p39 (or yet another unidentified Cdk5 activator) to Cdk5 kinase activity in the cerebellum that develops postnatally.
Figure 2.
p35−/− mice retain residual Cdk5 kinase activity. (A–C) Western blot analysis of p35 protein of the brain homogenate (A and B) and immunoprecipitate with anti-Cdk5 antibody (C) from the indicated genotype at postnatal day (P) 2 by using p35 antibodies that recognize the carboxyl terminus of p35 protein (A and C) and the N terminus of p35 protein (B). No p35 protein was detected in p35−/− brain, and a reduced amount of p35 protein was detected in the p35+/− brain (A and C). (D and E) Cdk5 kinase activity in brain homogenates from Cdk5+/+ (white bar) and Cdk5−/− (D) and p35+/+ (white bar) and p35−/− (black bar) homogenates (E). The diagram represents mean ± standard deviation (n = 3) of Cdk5 activity. Cdk5 kinase activity is expressed as pmol of phosphate incorporated per hr per mg protein. Approximately 10 and 20% residual Cdk5 kinase activity is detected in the cerebral cortex and cerebellum of p35−/− brain, respectively, but none in the Cdk5−/− brain.
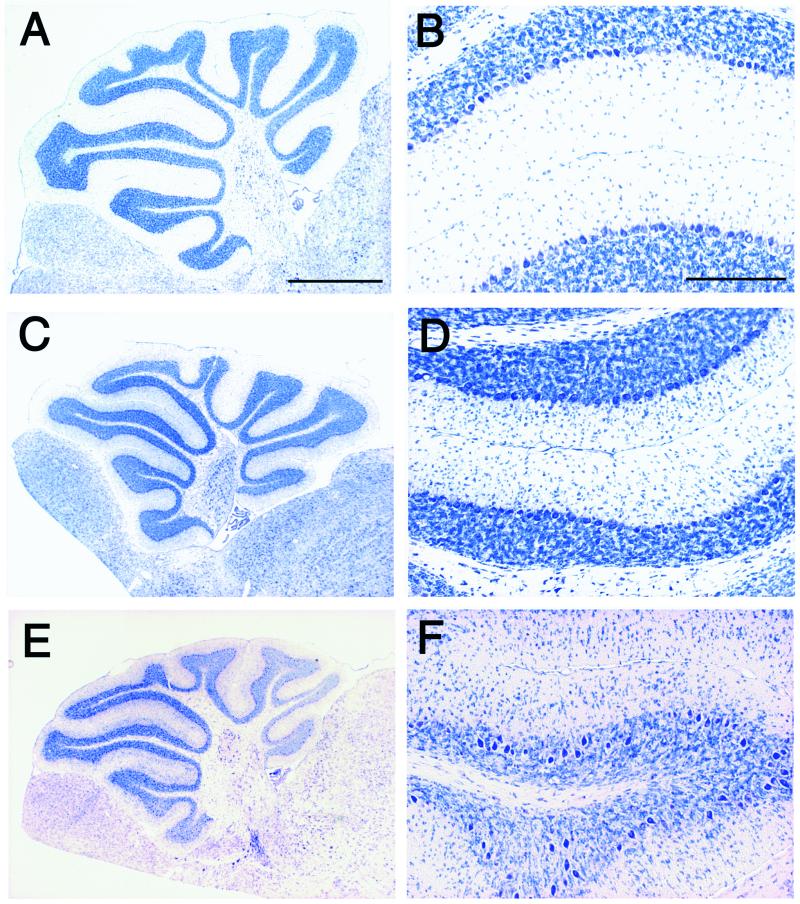
Figure 3.
Defects in neuronal migration in p35−/− mice are accentuated by Cdk5 heterozygosity. Parasagittal sections of cerebella of wild-type littermates (A and B) and p35−/− (C and D) at P21, and p35−/−Cdk5+/− at P35 (E and F) are stained with toluidine blue. B, D, and F are higher magnification of A, C, and E, respectively. In the p35−/− cerebellum, the alignment of Purkinje cells is disturbed slightly in some areas, and significant numbers of granule cells are found in the molecular layer (D). In the cerebellum of p35−/−Cdk5+/− mice, defects in the alignment of Purkinje cells are accentuated in the most areas (E and F). Purkinje cells are found occasionally in the granule cell layer, and increased numbers of granule cells are trapped in the molecular layer (F). (Bar in A, 1 mm; bar in B, 200 μm.)
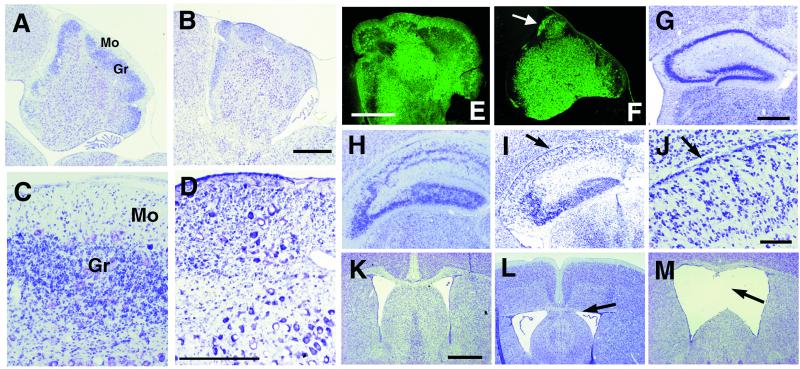
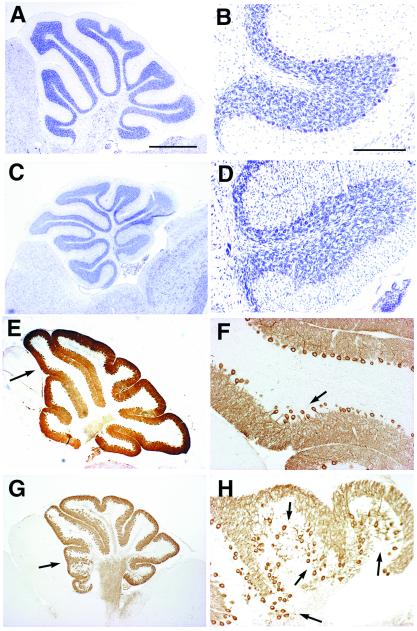
Figure 5.
Double-null mutant mice for p35 and Reelin/Dab1 display additional defects. (A–F) Comparison of the histological appearance of cerebella between Dab1yot/yot (A, C, and E) and p35−/−Dab1yot/yot at P22 (B, D, and F) in parasagittal sections stained with toluidine blue (A–D) and with anti-calbindin antibody (E and F). The molecular layer is defective in the p35−/−Dab1yot/yot cerebellum (B, D, and F). The arrow in F indicates the molecular layer with dendrites of Purkinje cells. (Bar in B and E, 500 μm; bar in D, 200 μm.) Mo, molecular layer; Gr, granule cell layer. (G–M) Coronal sections of Nissl staining at the level of hippocampus (G–J) and of septum (K–M) from wild type (K), p35−/− (G and L), and Dab1yot/yot (H) at P21 and p35−/−Dab1yot/yot (M) and p35−/−rl/rl (I and J) at P15. Accentuated disorganization of the pyramidal cell layer is observed in p35−/−rl/rl (arrows in I and J). The upper part of the septum is hypoplastic in p35−/− (arrow in L). Separation of the upper part of the septum is observed in p35−/−Dab1yot/yot mice (arrow in M). (Bar in G, 400 μm; bar in J, 80 μm; bar in K, 200 μm.)
To understand whether this residual Cdk5 kinase activity contributes to the mild phenotype of p35−/− mice, we generated a p35−/−Cdk5+/− mouse line and compared its histological phenotype with that of p35−/− mice. The p35−/−Cdk5+/− mice showed additional abnormalities in the cerebellum. In the cerebellum of p35−/− mice, the alignment of Purkinje cells in the Purkinje cell layer was disturbed in some areas at which Purkinje cells form 2–3 cell-width layers (Fig. 4 E and F). Additionally, significant numbers of granule cells are found in the molecular cell layer (Fig. 3D). In the cerebellum of p35−/−Cdk5+/− mice, extensive migration defects of Purkinje and granule cells are present (Fig. 3 E and F). Ectopic Purkinje cells are found frequently in the granule cell layer, and greater numbers of granule cells are observed in the molecular layer in the cerebellum of p35−/−Cdk5+/− mice (Fig. 3F).
Figure 4.
Migration defects of cerebellar cortical neurons in p35−/− mice are accentuated by Dab1 heterozygosity. Comparative histology of cerebella of Dab1yot/+ (A and B), p35−/− (E and F), and p35−/−Dab1yot/+ (C, D, G, and H) at P21 in the parasagittal sections stained with toluidine blue (A–D) and with anti-IP3R antibody (E–H). B, D, F, and H are higher magnifications of A, C, E, and G, respectively. Arrows in E and G indicate magnified areas in F and H, respectively. Subtle disturbances of the alignment of Purkinje cells are observed in p35−/− (arrows in F). Extensive migration defects of Purkinje cells and granule cells are observed in p35−/−Dab1yot/+ compared with p35−/− cerebellum (D, G, and H). (Bar in A, 1 mm. A, C, E, and G are identical magnification. Bar in B, 200 μm.)
p35 and Reelin/Dab1 Double Mutants Exhibit Additional Migration Defects of Cortical Neurons.
To elucidate the relationship between Cdk5/p35 and Reelin/Dab1 signaling, we generated double-mutant mice for p35 and Reelin/Dab1 genes. The viability of p35−/− mice provides an opportunity to analyze double-mutant mice lacking p35 and Reelin or Dab1 in the mature brain. This feature is important particularly for the analysis of brain structures such as the cerebellum, in which most development occurs postnatally. We first analyzed p35−/−Dab1yot/+ mice. Because mouse lines of both p35−/− and Dab1yot/+ are maintained in a129/Sv × C57BL/6 hybrid background, a bias of genetic background will not be a factor in the phenotypic analysis. Clear effects of the addition of heterozygosity of Dab1 mutation to the p35−/− genotype are detected in the cerebellar cortex. As compared with the subtle migration defects of the granule and Purkinje cells in the p35−/− cerebellum (Figs. 3D and 4 E and F), additional migration defects of cerebellar cortical neurons, granule cells, and Purkinje cells are observed in p35−/− Dab1yot/+ mice (Fig. 4 C and D). To examine the location of Purkinje cells and the appearance of their dendrites, we stained sections with anti-IP3R antibodies, specific markers for Purkinje cells. In the cerebellum of p35−/− mice, Purkinje cells that are localized abnormally within the granule cell layer extend their dendrites into the molecular layer (Fig. 4F). The alignment of Purkinje cells is disturbed in the most areas in p35−/−Dab1yot/+ cerebellum. Dendrites of the abnormally localized Purkinje cells in the granule cell layer occasionally do not extend to the molecular layer. Instead, their dendrites are tangled and irregular in shape (Fig. 4H). Identical defects in the migration of cerebellar cortical neurons are detected in the p35−/−rl/+ cerebellum (data not shown). In addition to the accentuated migration defects of cerebellar cortical neurons, pyramidal cell layers of hippocampus are less compact and split into two layers in p35−/− Dab1yot/+ and p35−/−rl/+ mice (data not shown). This separation of pyramidal neurons in the hippocampus is characteristic of the reeler-phenotype (Fig. 5H).
We next analyzed the double-null mice for p35 and Dab1. In the cerebellum of yotari mouse (Dab1yot/yot), cerebella are hypoplastic and lack typical foliation. The majority of Purkinje cells are clumped in central clusters; however, a small but significant number of mutant Purkinje cells successfully complete migration to the Purkinje cell layer as reported in the reeler mouse (39). Therefore, thin but significant molecular layer and granule cell layers are detected with scattered Purkinje cells between these layers (Fig. 5 A, C, and E). In the cerebellum of p35−/−Dab1yot/yot mice, the extensive disturbance of Purkinje-cell migration results in narrowing of the molecular layer that consists of dendrites of Purkinje cells (Fig. 5 B, D, and F). A decrease in the number of granule cells also is observed (Fig. 5 B and D). Identical defects in the cerebellum are also observed in p35−/−rl/rl mice (data not shown). In addition to the cerebellar phenotypes, apparent disorganization in the hippocampus, dilated lateral ventricles, and hypogenesis of the septum are observed in p35−/−Dab1yot/yot and p35−/−rl/rl mice (Fig. 5 I, J, and M). Pyramidal neurons are scattered in the hippocampus in both double-null mutants (Fig. 5 I and J).
Discussion
Our results indicate overlapping expression patterns of p35 and p39 in the developing mouse brain, particularly in the olfactory bulb, hippocampus, and cerebellar cortex, in which p35−/− mice display subtle abnormalities. In the cerebral cortex, however, only p35 appeared to be expressed highly during the embryonic stage of E13.5 to E16.5. The extent of the abnormalities in the laminar structure in the cerebral cortex of p35−/− mice is milder than in Cdk5−/− mice (refs. 1, 2, 10, and 23; T.O. and A.B.K., unpublished data). In the p35−/− mice, layer I of the cerebral cortex appears normal, but in Cdk5−/− mice it is very thin (1, 2, 10). In addition, the final position of the subplate at birth is quite different between Cdk5−/− and p35−/− mice (10, 23). This alteration may be attributed to delayed activation of Cdk5 by p39. This deduction is supported by higher expression of p39 mRNA observed in the cerebral cortex at birth. We also have demonstrated substantial residual Cdk5 kinase activity in p35−/− brains. These findings are in contrast to the reported undetectable Cdk5 kinase activity in the p35−/− mice by other investigators (2). This discrepancy is probably because of different substrates used in these assays. We used the synthetic KSPXK repeat polypeptide derived from human high-molecular-weight neurofilament protein, whereas Chae et al. (2) used histone H1 as a substrate. Histone H1 is a general substrate for other cdc2 kinases, whereas KSPXK polypeptide is a more restricted substrate for Cdk5 (31, 33). The gene-expression pattern and biochemical analysis of p35−/− brain indicate that the redundant functions of the activators p35 and p39 (or yet another unidentified Cdk5 activator) in vivo may be responsible for the phenotypic differences between Cdk5−/− and p35−/− mice. This deduction is supported by the observed accentuation of migration defects of cerebellar cortical neurons in p35−/−Cdk5+/− mice compared with p35−/− mice. We reported earlier a complete migration arrest of Purkinje cells in Cdk5−/− mice and also in Cdk5+/+/Cdk5−/− chimeric mice (40). Moreover, an inward migration of granule cells was found also to be Cdk5-dependent in a cell-autonomous fashion (40). These results indicate that the migration of cerebellar cortical neurons largely depends on Cdk5 kinase activity.
As a second objective of the present studies, we attempted to determine whether Cdk5/p35 and Reelin/Dab1 interact in a common signal-transduction pathway. Our results demonstrate that the addition of heterozygosity of Dab1 mutation to the p35−/− genotype results in severe migration defects of granule and Purkinje cells in the cerebellum. Similar results are seen also in p35−/−rl/+ mice indicating that this phenomenon depends on additive gene mutations. This effect is similar to the addition of Cdk5 heterozygosity to p35−/− genotype. The further deterioration in granule-cell migration by the addition of heterozygosity of Dab1 or Reelin is unexpected; granule cells secrete Reelin and lack Dab1 expression, and therefore they do not seem to be direct target cells of Reelin signaling (11). We next analyzed mice null for both p35 and Reelin/Dab1. The number of Purkinje cells in the Purkinje cell layer in these mutants is deceased significantly resulting in a defective molecular layer in the cerebellum (see Fig. 5 B, D, and F). Decreased numbers of granule cells in the double-null mutants are presumably a consequence of the accentuation of defects in Purkinje-cell migration. It is likely that an inadequate mitogenic support from Purkinje cells (41) may result in a suppression of the numbers of granule cells in double-null mutants. A similar reduction of granule cells was also reported when Purkinje cells were ablated experimentally during development (42). In the present study of double mutants, we observed typical additive effects on the migration defects in the cerebellar cortical neurons. This alteration presumably is related to the relatively high residual Cdk5 kinase activity in the cerebellum (see Fig. 2E).
It has been reported that the phenotype of mutant mice for both Reelin and Dab1 resemble the phenotypes of individual mutants of these genes (17). The lack of additional defects in the double mutants supports the model that Reelin and Dab1 act on the same signaling pathway (17). Because the double-null mice for p35 and Reelin/Dab1 demonstrate an accentuation of migration defects in several types of cortical neurons, it is unlikely that Cdk5/p35 and Reelin/Dab1 act on the same signal pathway. It is obvious also that Cdk5/p35 and Reelin/Dab1 contribute synergistically to the positioning of the cortical neurons in certain areas of the developing mouse brain such as the cerebellar cortex and hippocampus.
The precise roles of Cdk5/p35 kinase and Reelin/Dab1 signaling in the migratory process of cortical neurons still are unknown. Several studies indicate that Cdk5/p35 kinase may regulate actin dynamics and/or microtubules. The localization of Cdk5 and p35 in the growth cone and interactions with rac and Pak1 suggest that Cdk5/p35 kinase may be involved in the modulation of actin cytoskeletal dynamics (43). Cdk5/p35 also may regulate microtubules, because microtubule-associated protein tau and MAP2 are good substrates of Cdk5/p35 kinase (44–46). Interestingly, the causative genes for human lissencephaly, Lis1 and doublecortin, are implicated in microtubule reorganization (47–49). Another possible role of Cdk5/p35 kinase is regulation of cell adhesion mediated by N-cadherin during cortical development (50). Reelin/Dab1 signaling also may influence the microtubule dynamics or cell adhesion. It could influence the positioning of target cells by providing a stop signal through modification of microtubule dynamics. Another model was proposed for Reelin function in which Reelin may trigger homotypic adhesion mechanisms among target neurons. Recent identification of CNR, a family of cadherin-related neural receptors, as a possible component of Reelin receptors supports this theory (21). mRNA levels for Reelin and Dab1 are not altered in the brain of Cdk5−/− mice (41). Although Dab1 protein level is increased in the reeler brain, levels of Reelin and Dab1 proteins are not altered in Cdk5−/− brains (T.O. and A.B.K., unpublished data). Therefore, it is unlikely that Cdk5/p35 regulates the levels of either transcription or translation of Reelin and Dab1 genes. Although in vitro studies indicate that serine/threonine residue(s) of Dab1 can be phosphorylated by Cdk5/p35 kinase (20), there is no evidence that Cdk5/p35 phosphorylates Dab1 in vivo. It has been shown that the Reelin-induced phosphorylation of tyrosine residue(s) of Dab1 is essential for its function (22). However, the significance of the phosphorylation of serine/threonine residue(s) remains to be investigated. Our present studies indicate the possibility of interactions of Cdk5/p35 kinase and Reelin/Dab1 either in signaling pathways or in indirect regulation of common targets. The identification of the downstream molecules of the Reelin-signaling pathway and also of the unknown targets of Cdk5 kinase will provide us with insights into the process of the neuronal migration and their precise positioning in the developing brain.
Acknowledgments
We thank A. Nagy for R1 embryonic stem cells, R. Mulligan for the pPNT vector, the Roche Company (U.K.) for the gift of ganciclovir, and T. Kojima and H. Miyashita for technical advice. We thank P. Zelenka for critical reading of this manuscript. This work was supported by Grants-in-Aid from the Ministry of Education, Science, and Culture, Japan (to T.O.) and National Institute of Dental and Craniofacial Research, Division of Intra-mural Research (to A.B.K.).
Abbreviations
- Cdk
cyclin-dependant kinase
- E
embryonic day
- P
postnatal day
References
- 1.Ohshima T, Ward J M, Huh C-G, Longnecker G, Veeranna, Pant H C, Brady R O, Martin L J, Kulkarni A B. Proc Natl Acad Sci USA. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chae T, Kwon Y T, Bronson R, Dikkes P, Li E, Tsai L-H. Neuron. 1997;18:29–42. doi: 10.1016/s0896-6273(01)80044-1. [DOI] [PubMed] [Google Scholar]
- 3.Hellmich M R, Pant H C, Wada E, Betty J F. Proc Natl Acad Sci USA. 1992;89:10867–10871. doi: 10.1073/pnas.89.22.10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lew J, Winkfein R J, Paudel H K, Wang J H. J Biol Chem. 1992;267:25922–25926. [PubMed] [Google Scholar]
- 5.Meyerson M, Enders G H, Wu C L, Su L K, Gorka C, Nelson C, Harlow E, Tsai L-H. EMBO J. 1992;11:2909–2917. doi: 10.1002/j.1460-2075.1992.tb05360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai L-H, Takahashi T, Caviness VS, Jr, Harlow E. Development (Cambridge, UK) 1993;119:1029–1040. doi: 10.1242/dev.119.4.1029. [DOI] [PubMed] [Google Scholar]
- 7.Lew J, Huang Q Q, Qi Z, Winkfein R J, Aebersold R, Hunt T, Wang J H. Nature (London) 1994;371:423–426. doi: 10.1038/371423a0. [DOI] [PubMed] [Google Scholar]
- 8.Tsai L-H, Delalle I, Caviness V S, Jr, Chae T, Harlow E. Nature (London) 1994;371:419–423. doi: 10.1038/371419a0. [DOI] [PubMed] [Google Scholar]
- 9.Tang D, Yeung J, Lee K-Y, Matsushita M, Matsui H, Tomizawa K, Hatase O, Wang J H. J Biol Chem. 1995;270:26897–26903. doi: 10.1074/jbc.270.45.26897. [DOI] [PubMed] [Google Scholar]
- 10.Gilmore E C, Ohshima T, Goffinet A M, Kulkarni A B, Herrup K. J Neurosci. 1998;18:6370–6377. doi: 10.1523/JNEUROSCI.18-16-06370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Arcangelo G, Miao G G, Chen S C, Soares H D, Morgan J I, Curran T. Nature (London) 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa M, Miyata T, Nakajima K, Yagyu K, Seike M, Ikenaka K, Yamamoto H, Mikoshiba K. Neuron. 1995;14:899–912. doi: 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 13.Sweet H O, Bronson R T, Johnson K R, Cook S A, Davisson M T. Mamm Genome. 1996;7:798–802. doi: 10.1007/s003359900240. [DOI] [PubMed] [Google Scholar]
- 14.Yoneshima H, Nagata E, Matsumoto M, Yamada M, Nakajima K, Miyata T, Ogawa M, Mikoshiba K. Neurosci Res. 1997;29:217–223. doi: 10.1016/s0168-0102(97)00088-6. [DOI] [PubMed] [Google Scholar]
- 15.Sheldon M, Rice DS, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell B W, Cooper J A, Goldowitz D, Curran T. Nature (London) 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 16.Rice D S, Sheldon M, D'Arcangelo G, Nakajima K, Goldowitz D, Curran T. Development (Cambridge, UK) 1998;125:3719–3729. doi: 10.1242/dev.125.18.3719. [DOI] [PubMed] [Google Scholar]
- 17.Howell B W, Herrick T M, Cooper J A. Genes Dev. 1999;13:643–648. doi: 10.1101/gad.13.6.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer R E, Richardson J A, Herz J. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 19.Hiesberger T, Trommsdorff M, Howell B, Goffinet A, Mumby M, Cooper J, Herz J. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 20.D'Arcangelo G, Ramin H, Keshvara L, Rice D, Sheldon M, Curran T. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 21.Senzaki K, Ogawa M, Yagi T. Cell. 1999;99:635–647. doi: 10.1016/s0092-8674(00)81552-4. [DOI] [PubMed] [Google Scholar]
- 22.Howell B W, Herrick T M, Hildebrand J D, Zhang Y, Cooper J A. Curr Biol. 2000;10:877–885. doi: 10.1016/s0960-9822(00)00608-4. [DOI] [PubMed] [Google Scholar]
- 23.Kwon Y T, Tsai L-H. J Comp Neurol. 1998;395:510–522. doi: 10.1002/(sici)1096-9861(19980615)395:4<510::aid-cne7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Goffinet A M. Nature (London) 1997;389:668–669. doi: 10.1038/39453. [DOI] [PubMed] [Google Scholar]
- 25.Gilmore E C, Herrup K. Curr Biol. 2000;10:R162–R166. doi: 10.1016/s0960-9822(00)00332-8. [DOI] [PubMed] [Google Scholar]
- 26.Homayouni R, Curran T. Curr Biol. 2000;10:R331–R334. doi: 10.1016/s0960-9822(00)00459-0. [DOI] [PubMed] [Google Scholar]
- 27.Ohshima T, Kozak C A, Nagle J W, Pant H C, Brady R O, Kulkarni A B. Genomics. 1996;35:372–375. doi: 10.1006/geno.1996.0370. [DOI] [PubMed] [Google Scholar]
- 28.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 29.D'Arcangelo G, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Curran T. J Neurosci. 1997;17:23–41. doi: 10.1523/JNEUROSCI.17-01-00023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kojima T, Nakajima K, Mikoshiba K. Mol Brain Res. 2000;75:121–127. doi: 10.1016/s0169-328x(99)00313-7. [DOI] [PubMed] [Google Scholar]
- 31.Veeranna, Shetty K T, Amin N, Grant P, Albers R W, Pant H C. Neurochem Res. 1996;21:629–636. doi: 10.1007/BF02527763. [DOI] [PubMed] [Google Scholar]
- 32.Uchida T, Ishiguro K, Ohnuma J, Takamatsu M, Yonekura S, Imahori K. FEBS Lett. 1994;355:35–40. doi: 10.1016/0014-5793(94)01163-x. [DOI] [PubMed] [Google Scholar]
- 33.Shetty K T, Link W T, Pant H C. Proc Natl Acad Sci USA. 1993;90:6844–6848. doi: 10.1073/pnas.90.14.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda N, Niinobe M, Nakahira K, Mikoshiba K. Dev Biol. 1989;133:67–76. doi: 10.1016/0012-1606(89)90297-2. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida K, Cleaveland E S, Nagle J W, French S, Yaswen L, Ohshima T, Brady R O, Pentchev P G, Kulkarni A B. DNA Cell Biol. 1996;15:873–882. doi: 10.1089/dna.1996.15.873. [DOI] [PubMed] [Google Scholar]
- 36.Braissant O, Wahli W. Endocrinology. 1998;139:2748–2754. doi: 10.1210/endo.139.6.6049. [DOI] [PubMed] [Google Scholar]
- 37.Nilden F, Backstrom A, Bark C. Biochim Biophys Acta. 1998;1398:371–376. doi: 10.1016/s0167-4781(98)00073-6. [DOI] [PubMed] [Google Scholar]
- 38.Zheng M, Leung C L, Liem R K H. J Neurobiol. 1998;35:141–159. doi: 10.1002/(sici)1097-4695(199805)35:2<141::aid-neu2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Mariani J, Crepel F, Mikoshiba K, Changeux J P, Sotelo C. Philos Trans R Soc London B. 1977;281:1–28. doi: 10.1098/rstb.1977.0121. [DOI] [PubMed] [Google Scholar]
- 40.Ohshima T, Gilmore E C, Longenecker G, Jacobowitz D M, Brady R O, Herrup K, Kulkarni A B. J Neurosci. 1999;19:6017–6026. doi: 10.1523/JNEUROSCI.19-14-06017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wechsler-Reya R J, Scott M P. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 42.Smeyne R T, Chu T, Lewin A, Bian F, Crisman S S, Kunsch C, Lira S A, Oberdick J. Mol Cell Neurosci. 1995;6:230–251. doi: 10.1006/mcne.1995.1019. [DOI] [PubMed] [Google Scholar]
- 43.Nikolic M, Chou M M, Lu W, Mayer B J, Tsai L H. Nature (London) 1998;395:194–198. doi: 10.1038/26034. [DOI] [PubMed] [Google Scholar]
- 44.Paudel H K, Lew J, Ali Z, Wang J H. J Biol Chem. 1993;268:23512–23518. [PubMed] [Google Scholar]
- 45.Kobayashi S, Ishiguro K, Omori A, Takamatsu M, Arioka M, Imahori K, Uchida T. FEBS Lett. 1993;335:171–175. doi: 10.1016/0014-5793(93)80723-8. [DOI] [PubMed] [Google Scholar]
- 46.Hosoi T, Uchiyama M, Okumura E, Saito T, Ishiguro K, Uchida T, Okuyama A, Kishimoto T, Hisanaga S. J Biochem (Tokyo) 1995;117:741–749. doi: 10.1093/oxfordjournals.jbchem.a124771. [DOI] [PubMed] [Google Scholar]
- 47.Sapir T, Elbaum M, Reiner O. EMBO J. 1997;16:6977–6984. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gleeson J G, Lin P T, Flanagan L A, Walsh C A. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 49.Francis F, Koularkoff A, Boucher D, Chafey P, Schaar B, Vinet M C, Friocourt G, McDonnell N, Reiner O, Kahn A, et al. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 50.Kwon Y T, Gupta A, Zhou Y, Nikolic M, Tsai L-H. Curr Biol. 2000;10:363–372. doi: 10.1016/s0960-9822(00)00411-5. [DOI] [PubMed] [Google Scholar]