Abstract
The type I interferons (IFNs), IFN-α and -β, are key effector molecules of the immune response to viruses. The anti-viral action of IFNs on virus-infected cells and surrounding tissues is mediated by expression of hundreds of IFN-stimulated genes. Viperin (virus inhibitory protein, endoplasmic reticulum-associated, IFN-inducible) is an Interferon stimulated gene (ISG), which is induced by type I, II, and III IFNs or after infection with a broad range of DNA and RNA viruses. Recent evidence indicates that Viperin disrupts lipid rafts to block influenza virus budding and release and interferes with replication of hepatitis C virus by binding to lipid droplets, small organelles involved in lipid homeostasis that are essential for hepatitis C virus replication. Viperin is also induced by nonviral microbial products such as lipopolysaccharide (LPS) and by a wide range of bacteria, suggesting a broader role in innate antimicrobial defenses.
To survive infection the immune system deploys an arsenal of defensive measures to combat invading microbes (Biron 2001; Samuel 2001; Barnes and others 2002). The type I interferons (IFNs) are among the earliest of these defenses. The importance of type I IFNs is highlighted by the enhanced susceptibility of IFN-α/βR-deficient mice to virus infection (Kamijo and others 1994; Hwang and others 1995) and the myriad of strategies employed by viruses to interfere with their production and/or action and the antiviral activity of the IFN-stimulated genes (ISGs) themselves [reviewed in (Katze and others 2002)]. The workhorses of the type I IFN system are these ISGs, several of which have been characterized in detail. The mechanisms by which ISGs restrict viral replication are ill-defined and are covered in this issue. Viperin is one such example. The results of gene-profiling microarray studies showed that the Viperin gene is one of those that is often most highly induced by a range of different viruses (Boudinot and others 2000; Sun and Nie 2004; Helbig and others 2005; Olofsson and others 2005; Severa and others 2006; Chan and others 2008), by the double-stranded RNA analog poly(I-C) (Severa and others 2006), by the double-stranded B-form DNA (poly-dAdT) (Kato and others 2006) and by microbial products, such as lipopolysaccharide (Olofsson and others 2005; Severa and others 2006), in various cell types. While considerable progress has been made in our understanding of how Viperin is turned on in cells, key mechanistic insights into its mode of action are only beginning to be elucidated.
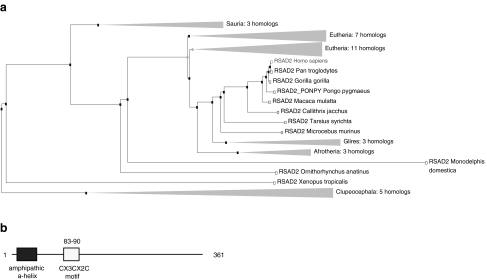
Viperin (virus inhibitory protein, endoplasmic reticulum [ER]-associated, IFN-inducible) was first identified in fish, as virus induced gene (vig1), where it was shown to be induced to high levels during infection of rainbow trout leukocytes with viral hemorrhagic septicemia virus, a fish rhabdovirus (Boudinot and others 2000). Its human homolog, cytomegalovirus-induced gene (cig5) [also known as radical S-adenosyl methionine (SAM) domain-containing 2 in both human and mouse], was similarly identified from primary skin cultures incubated with inactivated human cytomegalovirus (HCMV). While examining the IFN-γ response of human macrophages, Chin and Cresswell (2001) identified a unique IFN-γ inducible gene, fragments of which were identical to cig5, which they subsequently named Viperin. The amino acid sequence of Viperin was then found to be homologous to a rat gene, interferon-inducible gene expressed during bone formation (BEST5), expressed during rat osteoblast differentiation. Viperin has since been found in a wide range of organisms ranging from bony fish to mammals (Fig. 1). Although IFN-γ was capable of inducing expression of Viperin, the type I IFNs were far more potent activators. Additional studies revealed the ability of human cytomegalovirus as well as the HCMV glycoprotein B (gB) protein to potently upregulate Viperin gene expression (Chin and Cresswell 2001).
FIG. 1.
Phylogenetic tree showing the evolutionary conservation of RSAD2 (Viperin) (a) and the domain organization of the human Viperin gene (b). RSAD2, radical S-adenosyl methionine domain-containing 2.
The initial characterization of Viperin revealed its ability to prevent the replication of HCMV. Over the last several years, Viperin has been shown to have activity against a range of additional viruses, including influenza virus, hepatitis C virus (HCV), dengue virus, alphaviruses, and retroviruses such as human immunodeficiency virus. The mechanism by which Viperin restricts replication of all of these viruses is not fully clear. In the case of HCMV, it was shown that while ectopic expression of Viperin in fibroblasts had no effect on HCMV immediate early viral gene expression (IE1 and IE2), the synthesis of early late, (pp65), late (gB), or true late (pp28) genes was significantly reduced in cells expressing Viperin compared to control cells. Since expression of these proteins is indispensible for productive HCMV infection, these studies indicated that Viperin likely exerted its antiviral effects at a late stage in of the viral life cycle (Chin and Cresswell 2001).
The human Viperin gene encodes a protein of 361 amino acids with a predicted molecular mass of 42.2 kDa. Viperin contains a 42-amino acid residue N-terminal amphipathic a-helix that, in other proteins, is known to bind membranes and induce membrane curvature (Drin and others 2007; Drin and Antonny 2010). This is followed by a CX3CX2C motif, which is found in the superfamily of SAM-dependent radical enzymes. The radical SAM superfamily comprises more than 600 members (Sofia and others 2001), and evidence suggests that the 3 conserved cysteine residues are part of an unusual iron-sulfur cluster that uses SAM as a cofactor to form a radical that is involved in catalysis. Thus, Viperin is also called radical SAM domain-containing 2. Recent studies have provided clear proof that Viperin is indeed a radical SAM enzyme (Duschene and Broderick 2010; Shaveta and others 2010), although how this relates to its antiviral activities is still unclear. A schematic representation of the domain organization of Viperin is shown in Fig. 2. The 42-amino acid N-terminal amphipathic a-helix is critical for the antiviral action of Viperin. This region localizes Viperin to the ER, where it has been shown to dimerize. Expression of Viperin can interfere with the secretion of soluble proteins from cells, although again, the relevance of these findings to its anti-viral effects is not clear.
FIG. 2.
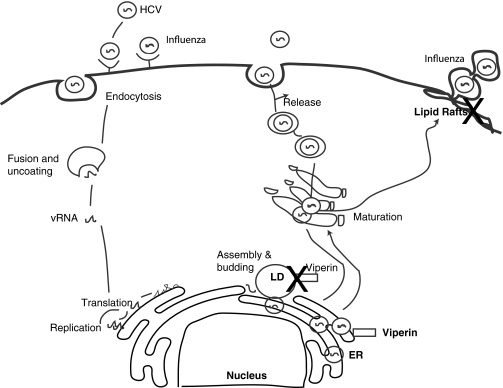
Schematic representation of the antiviral action of Viperin against influenza and HCV. Viperin prevents HCV assembly and/or budding by binding to lipid droplets that are essential for HCV replication. In the case of influenza virus, Viperin binds to farnesyl diphosphate synthase, and thereby disrupts lipid rafts to prevent virus release. The X indicates the site of action of Viperin. HCV, hepatitis C virus; ER, endoplasmic reticulum; LD, lipid droplets; vRNA, viral RNA.
What has emerged is the ability of Viperin to inhibit influenza virus replication and insights into the mechanisms underlying this effect have been uncovered (Wang and others 2007). To infect cells, influenza binds the cell surface receptor, sialic acid, and is then internalized via endocytosis. The low endosomal pH enables membrane fusion, thereby releasing the viral genome into the cytosol and ultimately into the nucleus, where viral mRNA synthesis and genome replication occur. The viral mRNAs enter the cytosol and initiate viral protein synthesis, whereas the transmembrane glycoproteins, hemagglutinin and neuraminidase, traffic to lipid rafts at the plasma membrane. Lipid rafts are essential sites from where viral budding takes place (Zhang and others 2000; Takeda and others 2003; Leser and Lamb 2005). The matrix protein (M1), nuclear export protein, and the nucleoprotein move to the nucleus and form the nucleocapsid. The nucleocapsid translocates the viral genome to the cytosol and finally to the plasma membrane, where the new virus particles are assembled and released. Viperin exerts its effects at the later stages of this life cycle by preventing the release of viral particles and appears to do this by disrupting lipid rafts. Viperin achieves this effect by binding to and inhibiting farnesyl diphosphate synthase (FPPS), an enzyme involved in cholesterol and isoprenoid biosynthesis. It is unclear exactly how this effect on FPPS leads to disruption of lipid rafts, and future studies will no doubt uncover the mechanisms involved. In addition to influenza virus retroviruses (eg, human immunodeficiency virus-1), paramyxoviruses (eg, Sendai and measles) and filoviruses (eg, Ebola) have all been linked to lipid rafts either for their entry into, or exit from cells (Ono and Freed 2005). It is likely therefore that Viperin may target lipid rafts to also interfere with these viruses.
The ability of Viperin to disrupt lipid rafts does not explain its antiviral activity against other viruses, however. Viperin limits replication of HCV, a virus that does not bud from lipid rafts (Helbig and others 2005; Hinson and Cresswell 2009a, 2009b). HCV is a positive-sense, single-stranded RNA virus that is packaged into an enveloped virus particle (Dustin and Rice 2007; Randall and others 2007). Its genome encodes one open reading frame translated into a single polyprotein with ∼3,000 amino acids, which is both co- and post-translationally cleaved at the membrane of the ER by host and viral proteases into 3 structural (core, E1, and E2) and 7 nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). To initiate its life cycle, HCV particles are internalized by clathrin-dependent endocytosis. Viral and cellular membranes are then thought to fuse at the acidic compartment and the genome is then released into the cytoplasm. The viral proteins are processed from a polyprotein translated from the genomic RNA. For viral RNA replication, a membrane-associated replication complex, composed of viral nonstructural proteins, replicating RNA, and cellular membranes, is formed. Enveloped HCV virions carrying a newly synthesized viral genome appear to form by budding into the ER lumen and then they leave the cells through the secretory pathway. The HCV core protein initially localizes to the cytosolic face of the ER. After maturation, the core relocalizes to lipid droplets. Lipid droplets are cellular lipid storage organelles not only involved in lipid storage and trafficking but also now implicated in a variety of metabolic diseases.
Chronic HCV infection affects host lipid metabolism, and induces lipid droplet accumulation in the liver [reviewed in (Fukasawa 2010)]. The core protein of HCV recruits the HCV nonstructural proteins and the replication complex to lipid droplets. Several of the NS proteins contain amphipathic α-helices that are thought to facilitate interactions with the ER (Elazar and others 2003, 2004; Suzuki and others 2005). The N-terminal α-helices of Viperin also localize Viperin to the lipid droplets (Hinson and Cresswell 2009a, 2009b). It is likely that Viperin associates with lipid droplets as they form from the outer leaflet of ER membranes. Exactly how Viperin interferes with HCV replication within these organelles is unclear. The number, size, or location of lipid droplets does not appear to change when Viperin is present; however, similar studies have not been done in HCV-infected cells. It is possible that in the context of infection, Viperin alters lipid droplet formation or the ability of the HCV proteins to localize to this organelle. This is an attractive possibility given the ability of Viperin to alter ER membranes and lead to curvature of ER membranes (Drin and others 2007; Drin and Antonny 2010). Perhaps Viperin alters lipid droplet formation or morphology during infection. Another possibility is that Viperin changes the lipid content of these organelles through its actions on FPPS. Given that lipid droplets are derived from the ER where cholesterol synthesis occurs, it is possible that Viperin affects the type or quantity of lipids that accumulate in the ER membrane leaflet and thus affect the lipid content of lipid droplets.
Regardless of the exact mechanism of action of Viperin, its high inducibility in various cell types indicates that this gene plays a central role in antiviral defenses. The regulation of Viperin gene expression is somewhat complicated. Its expression has been shown to be dependent on IFN production and autocrine/paracrine IFN signaling in a number of settings. A variety of pathways have now been described leading to transcriptional regulation of IFN-β [reviewed in (Bowie and Unterholzner 2008; Hornung and Latz 2010)]. Viperin has been shown to be induced downstream of toll like receptor (TLR)3, TLR4, retinoic acid induced gene (RIG)-I, and melanoma differentiation associated (MDA5), as well as downstream of cytosolic DNA sensing pathways, which use some of these same components to turn on IFNB gene expression. In the case of RIG-I signaling triggered by Sendai virus, mitochondrial associated antiviral signaling (MAVS), TANK (TRAF family member-associated NFKB activator) binding kinase (TBK1), and a combination of interferon regulatory factor (IRF)3 and 7 drive IFNB production, which then signals via the IFNa/bR to induce Viperin. Viperin is not induced in bone marrow macrophages derived from alpha/beta IFN (IFN-α/β) receptor-deficient mice (Severa and others 2006). The key factor regulating Viperin promoter activity in this system is the interferon-stimulated gene factor 3 (ISGF3) complex and not IRF3 itself. Lipopolysaccharide and poly(I-C) also induce Viperin expression through an ISGF3-mediated pathway (Severa and others 2006).
It is also known that some ISGs, including Viperin, are induced directly by IRF-3 itself (Grandvaux and others 2002). vesicular stomatitis virus (VSV) directly induces Viperin expression because treatment with anti-IFN antibody has no effect on its induction. Similar studies were conducted with the fish vig-1 gene, where neither anti-IFN antibodies nor cycloheximide, which blocked de novo protein synthesis, interfered with its induction (Boudinot and others 2000). Key insights into this direct pathway of Viperin induction were recently revealed. Direct induction of Viperin was shown to be mediated by a pathway involving MAVS, the adapter molecule downstream of RIG-I. MAVS is localized to both mitochondria and peroxisomes and it is from this latter location that direct induction of Viperin occurs (Dixit and others 2010). Peroxisomal MAVS functions early to establish an immediate although transient anti-viral state within the cell, which can halt or delay viral replication until the more robust and sustained antiviral response driven by the mitochondrial MAVS pathway come into play. VSV appears to utilize this peroxisomal MAVS mechanism to turn on Viperin gene expression.
It is not surprising that viral evasion mechanisms exist to counteract Viperin itself. This has recently been revealed in the case of Japanese encephalitis virus (JEV) (Chan and others 2008). Viperin is highly induced by JEV and the Viperin protein is degraded in JEV-infected cells through a proteasome-dependent mechanism. Overexpression of Viperin had no effect on JEV replication, whereas the proteasome inhibitor MG132 sustained the protein level and antiviral effect of Viperin in JEV-infected cells. Consistent with this model, knockdown of Viperin by RNA interference had no effect on JEV replication. These findings suggest that even though Viperin gene expression is highly induced by JEV, it is negatively regulated at the protein level to counteract its antiviral effect.
Future studies will likely unveil additional immune evasion strategies employed by viruses to subvert the action of Viperin. Despite the considerable progress that has been made in our understanding of how Viperin controls virus infection, a key mechanistic understanding of its mode of action awaits further clarification. Viperin-deficient mice have been generated; however, studies to examine their susceptibility to viruses have not yet been revealed. In addition to its induction by viruses and viral nucleic acids, the ability of other pathogens, and nonviral microbial products such as LPS to induce this gene suggests a broader role for Viperin in innate defenses. In this regard, Viperin has recently been shown to be induced in neutrophils and macrophages and is localized to the ER and lipid droplet-like vesicles in neutrophils. In this context, it will be of great interest to further elaborate the molecular basis for Viperin action in these cells and delineate its role in the regulation of the immune response to other classes of pathogens such as bacteria that also induce Viperin to high levels.
Acknowledgments
This work was supported by NIH grants AI083713 and AI067497 (to K.A.F.).
Author Disclosure Statement
No competing financial interests exist.
References
- Barnes B. Lubyova B. Pitha PM. On the role of IRF in host defense. J Interferon Cytokine Res. 2002;22(1):59–71. doi: 10.1089/107999002753452665. [DOI] [PubMed] [Google Scholar]
- Biron CA. Interferons alpha and beta as immune regulators—a new look. Immunity. 2001;14(6):661–664. doi: 10.1016/s1074-7613(01)00154-6. [DOI] [PubMed] [Google Scholar]
- Boudinot P. Riffault S. Salhi S. Carrat C. Sedlik C. Mahmoudi N. Charley B. Benmansour A. Vesicular stomatitis virus and pseudorabies virus induce a vig1/cig5 homologue in mouse dendritic cells via different pathways. J Gen Virol. 2000;81(Pt 11):2675–2682. doi: 10.1099/0022-1317-81-11-2675. [DOI] [PubMed] [Google Scholar]
- Bowie AG. Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8(12):911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL. Chang TH. Liao CL. Lin YL. The cellular antiviral protein Viperin is attenuated by proteasome-mediated protein degradation in Japanese encephalitis virus-infected cells. J Virol. 2008;82(21):10455–10464. doi: 10.1128/JVI.00438-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin KC. Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci USA. 2001;98(26):15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E. Boulant S. Zhang Y. Lee AS. Odendall C. Shum B. Hacohen N. Chen ZJ. Whelan SP. Fransen M. Nibert ML. Superti-Furga G. Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141(4):668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G. Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584(9):1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- Drin G. Casella JF. Gautier R. Boehmer T. Schwartz TU. Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14(2):138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Duschene KS. Broderick JB. The antiviral protein Viperin is a radical SAM enzyme. FEBS Lett. 2010;584(6):1263–1267. doi: 10.1016/j.febslet.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin LB. Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- Elazar M. Cheong KH. Liu P. Greenberg HB. Rice CM. Glenn JS. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J Virol. 2003;77(10):6055–6061. doi: 10.1128/JVI.77.10.6055-6061.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazar M. Liu P. Rice CM. Glenn JS. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol. 2004;78(20):11393–11400. doi: 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa M. Cellular lipid droplets and hepatitis C virus life cycle. Biol Pharm Bull. 2010;33(3):355–359. doi: 10.1248/bpb.33.355. [DOI] [PubMed] [Google Scholar]
- Grandvaux N. Servant MJ. tenOever B. Sen GC. Balachandran S. Barber GN. Lin R. Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76(11):5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig KJ. Lau DT. Semendric L. Harley HA. Beard MR. Analysis of ISG expression in chronic hepatitis C identifies Viperin as a potential antiviral effector. Hepatology. 2005;42(3):702–710. doi: 10.1002/hep.20844. [DOI] [PubMed] [Google Scholar]
- Hinson ER. Cresswell P. The antiviral protein, Viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc Natl Acad Sci USA. 2009a;106(48):20452–20457. doi: 10.1073/pnas.0911679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson ER. Cresswell P. The N-terminal amphipathic alpha-helix of Viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem. 2009b;284(7):4705–4712. doi: 10.1074/jbc.M807261200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V. Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10(2):123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- Hwang SY. Hertzog PJ. Holland KA. Sumarsono SH. Tymms MJ. Hamilton JA. Whitty G. Bertoncello I. Kola I. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92(24):11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo R. Shapiro D. Gerecitano J. Le J. Bosland M. Vilcek J. Biological functions of IFN-gamma and IFN-alpha/beta: lessons from studies in gene knockout mice. Hokkaido Igaku Zasshi. 1994;69(6):1332–1338. [PubMed] [Google Scholar]
- Kato H. Takeuchi O. Sato S. Yoneyama M. Yamamoto M. Matsui K. Uematsu S. Jung A. Kawai T. Ishii KJ. Yamaguchi O. Otsu K. Tsujimura T. Koh CS. Reis e Sousa C. Matsuura Y. Fujita T. Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Katze MG. He Y. Gale M., Jr. Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2(9):675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Leser GP. Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342(2):215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Olofsson PS. Jatta K. Wagsater D. Gredmark S. Hedin U. Paulsson-Berne G. Soderberg-Naucler C. Hansson GK. Sirsjo A. The antiviral cytomegalovirus inducible gene 5/Viperin is expressed in atherosclerosis and regulated by proinflammatory agents. Arterioscler Thromb Vasc Biol. 2005;25(7):e113–e116. doi: 10.1161/01.ATV.0000170130.85334.38. [DOI] [PubMed] [Google Scholar]
- Ono A. Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- Randall G. Panis M. Cooper JD. Tellinghuisen TL. Sukhodolets KE. Pfeffer S. Landthaler M. Landgraf P. Kan S. Lindenbach BD. Chien M. Weir DB. Russo JJ. Ju J. Brownstein MJ. Sheridan R. Sander C. Zavolan M. Tuschl T. Rice CM. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104(31):12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severa M. Coccia EM. Fitzgerald KA. Toll-like receptor-dependent and -independent Viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J Biol Chem. 2006;281(36):26188–26195. doi: 10.1074/jbc.M604516200. [DOI] [PubMed] [Google Scholar]
- Shaveta G. Shi J. Chow VT. Song J. Structural characterization reveals that Viperin is a radical S-adenosyl-L-methionine (SAM) enzyme. Biochem Biophys Res Commun. 2010;391(3):1390–1395. doi: 10.1016/j.bbrc.2009.12.070. [DOI] [PubMed] [Google Scholar]
- Sofia HJ. Chen G. Hetzler BG. Reyes-Spindola JF. Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29(5):1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BJ. Nie P. Molecular cloning of the Viperin gene and its promoter region from the mandarin fish Siniperca chuatsi. Vet Immunol Immunopathol. 2004;101(3–4):161–170. doi: 10.1016/j.vetimm.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Suzuki R. Sakamoto S. Tsutsumi T. Rikimaru A. Tanaka K. Shimoike T. Moriishi K. Iwasaki T. Mizumoto K. Matsuura Y. Miyamura T. Suzuki T. Molecular determinants for subcellular localization of hepatitis C virus core protein. J Virol. 2005;79(2):1271–1281. doi: 10.1128/JVI.79.2.1271-1281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M. Leser GP. Russell CJ. Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci USA. 2003;100(25):14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Hinson ER. Cresswell P. The interferon-inducible protein Viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2(2):96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Zhang J. Pekosz A. Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J Virol. 2000;74(10):4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]