Abstract
Tail tendon break time (TTBT), a measure of collagen cross-linking, shown to increase with age differs significantly among inbred strains of mice, indicating underlying genetic influences. This study was aimed to identify quantitative trait loci (QTLs) associated with tail tendon break time at three ages (200, 500, and 800 days of age) for 23 BxD recombinant inbred strains of mice and B6D2F2 mice derived from C57BL/6J and DBA/2J strains. Heritability estimates were calculated, and QTL analyses were conducted using interval-mapping methods. Mean tail tendon break time values were higher in males and increased nonlinearly with age. Eight total QTLs were nominated in the B6D2F2 mice at the three measured ages, with the QTL at 800 days confirmed in the recombinant inbred strains. Allelic effect modeling for the identified QTLs suggests differences in gene action between sexes. Candidate genes in the QTL regions include collagen genes and an advanced glycation end-product receptor. The QTLs identified demonstrate influence at some but not all ages.
Keywords: Mouse, Tail tendon, Quantitative trait loci (QTL), Aging
TAIL tendon break time (TTBT) is a simple and robust assay to measure the strength of tail tendon collagen, the primary component of tendons. TTBT has been widely used as an index of in vivo nonenzymatic cross-linking, which increases with age (1–6). During aging, the extracellular matrix (ECM) undergoes changes believed to result in part from cross-linking of ECM proteins. Elastin, collagen, and other components of the ECM, located in every organ and tissue, are integral in the formation of connective tissues and other cellular functions (7–9). Within the ECM, increased nonenzymatic cross-linking results in shrinking of collagen fibers, stiffened tendons, and decreased overall flexibility. These properties are reflected in reduced solubility and digestibility of tissues (1–4) and may contribute to the physiological declines associated with aging.
TTBT varies among mouse strains, for example, in a comparison of the DBA/2 and C57BL/6 mouse strains, the shorter lived DBA/2 mice have longer TTBT than the C57BL/6 mice at all ages tested (3,5,6). Previous studies have relied extensively on quantitative genetic analyses to characterize polygenic influences on TTBT. Quantitative trait loci (QTLs) are anonymous loci with detectable effects on quantitative traits that are determined by associations between marker loci (known genes or polymorphic regions of the chromosome) and variability in continuously distributed phenotypes. The procedure for their identification was originally developed in agricultural research, particularly in horticulture and livestock (10–12), and has been extended for use in a wide spectrum of quantitative traits including biomedical research on rodents.
Research on QTLs in laboratory animals commonly uses F2 and recombinant inbred (RI) mice. Inbreeding from a genetically heterogeneous F2 population derived from two inbred strains yields RI strains. Animals within each strain are approximately homozygous in like allelic state for all genes (13), but strains differ from each other. The BxD RI strains (derived from a cross between progenitor strains C57BL/6J and DBA/2J) are the most common of many RI series used in QTL studies due to the relatively large number of inbred strains within the BxD series (14,15). The original strains in the BxD RI series were developed by Benjamin Taylor from the Jackson Laboratories (16).
The objective of this study was to identify QTLs influencing TTBT. QTLs related to TTBT were nominated in F2 mice and were tested for verification in BxD RI mouse strains from the same progenitor populations. Age-related effects on TTBT across mice at 200, 500, and 800 days of age were also examined.
METHODS
Animals and Husbandry
All mice (Mus musculus) in this study (C57BL/6J [B6], DBA/2J [D2], 23 BxD RI strains, and B6D2F2 mice) were born and housed in a barrier facility maintained by the Center for Developmental and Health Genetics at the Pennsylvania State University. The cross-sectional analyses included 379 mice at 200 days, 372 mice at 500 days, and 265 mice at 800 days of age for the B6D2F2 (F2 generation of C57BL/6J and DBA/2J cross) and 475 mice at 200 days, 385 mice at 500 days, and 230 mice at 800 days of age for the means for the 23 BxD RI strains. For each BxD RI strain, the numbers of mice per each RI strain and each sex tested at each age were as follows: 200 days, 8–12 animals per sex; 500 days, 5–12 animals per sex; and 800 days, 4–10 animals per sex. These animals were age matched within 10–14 days of one another.
Mice of the same litter and sex were housed four per microisolator cage upon weaning. All mice were maintained on a schedule of 12/12 hour light/dark cycle at a temperature of 21°C ± 2°C with approximately 50% humidity. Mice were fed autoclaved Purina Laboratory Rodent Chow Diet #5010 and water ad libitum. Anaerobic bacteria via the Schaedler microflora (17) was administered to the axenic founding population of the barrier colony. The microbiological status of the colony was monitored regularly by the University of Missouri Research Animal Diagnostic and Investigative Laboratory using their Comprehensive Profile for necropsy and serology. Negative results were consistent for all the viral and bacterial pathogens included in this diagnostic profile. Occasional indicators of “opportunistic” bacteria, Pasteurella pneumotropica and Pseudomonas aeruginosa, were identified, with no or minimal health consequences in immunocompetent mice.
Collagen Denaturation Assay
The experimental schedule for harvesting tissues was randomized with males and females and various strains sacrificed each day. Approximately 2,000 animals were sacrificed over the course of the entire study. Age cohorts were sacrificed together, all 200-day-old animals before 500-day-old animals. The 800-day-old animals were terminated last. Tissues of the B6D2F2 animal group were harvested prior to the BxD RI group for each age. Between 12 and 16 mice were sacrificed daily via cervical dislocation. Tails were collected and placed on ice until the tendons were removed prior to further dissection and harvesting of tissues for other experimental assays reported elsewhere.
Originally devised by Verzar (18) in rats, the TTBT assay was modified by Elden and Boucek (19) to use urea as a denaturation agent. Further modifications to the assay were completed by Olsen and Everitt (20) and Harrison and Archer (2). The procedure used for collagen fiber removal in this study was a modified version of that reported by Harrison and Archer (2). Dorsal tendons were removed from the tail and placed in distilled water to prevent the tendons and tibers from drying out. Individual collagen fibers were separated by forceps in the visual field of a dissecting microscope. Three fibers of intermediate thickness were chosen for each mouse. Each fiber (approximately 1 cm in length) was attached to a 2.0-g weight using 5-0 surgical silk and suspended in a 125-mL Erlenmeyer flask filled with a 7-M urea solution to denature the collagen fibers at 45°C ± 0.1°C. Fiber break times were measured by recording time of initial suspension until the fiber broke, with the weight dropping to the bottom of the flask.
Genotyping
All B6D2F2 mice were genotyped for 96 microsatellite markers spaced at ∼20-cM intervals throughout the mouse genome as previously reported (21). The DNA for genotyping was extracted from tail-tips collected at weaning. Genotypes for the 23 BxD RIs used included 623 microsatellite markers, a subset of markers already determined in the RI lines from the Williams and colleagues database (22) that only included informative markers with duplicate marker genotypes removed.
QTL Analyses
Interval mapping.—
Interval mapping, proposed by Lander and Botstein (23), is used to estimate the position of a QTL between two markers. Interval mapping was originally based on the maximum likelihood approach, but now may simple regression as an alternate method. In the present data, both interval-mapping methods were used. The statistical program R/qtl (24) was utilized for interval mapping for the B6D2F2 data using multiple regression with sex as a covariate to investigate potential sex differences in TTBT. QTL Cartographer (25) was used for interval mapping in the BxD RIs. This program applied the maximum likelihood approach as the interval-mapping method.
QTLs at particular locations on the chromosome can be displayed on a likelihood map using a log of odds ratio (LOD) score plotted against chromosomal position (12). A LOD score is an estimate of the likelihood that two loci lie near each other on the chromosome and with a nonzero probability are transmitted together. Suggestive and significant thresholds of statistical significance (values corresponding to the 37th and 95th percentiles) were determined from 10,000 permutations of the data (26). Permutations resulted in the estimates of the thresholds of 3.1 LOD (suggestive) and 4.9 LOD (significant) for the B6D2F2s. In the BxD RI data, the threshold estimates for suggestive QTLs were LOD scores of 2.6 for all three ages; the significance threshold at 200 days was a LOD score of 3.9 and at 500 and 800 days of age, the threshold was a LOD score of 3.7. A 1.5-LOD drop-off interval is a 95% confidence interval for interpretation of QTL positioning, calculated by identifying the centimorgan position on either side of the QTL peak with a LOD score 1.5 less than the peak score (27).
Interval mapping in the B6D2F2s was conducted on raw data, allowing sex to be used as a covariate. The BxD RI data was adjusted for sex differences by subtracting the difference of mean values between the male and female mean in each RI strain prior to interval mapping. The male mean value was changed to accommodate this by either adding or subtracting the proper value to correct for the sex differences between the males and females. Animals with scores exceeding 4 SDs from the overall group mean were removed as outliers for both the B6D2F2s (n < 15) and the BxD RIs (n < 15). In addition, the BxD 22 strain was excluded at all ages from the analyses as an extreme outlier, with all individual values much greater than 4 SDs from the mean of the other strains. Early mortality loss resulted in the inability to include the BxD 13 strain in the analyses of the 500- and 800-day-old mice. Furthermore, the analyses of the 800-day BxD RI mice did not include the BxD 8, 14, 16, and 33 strains because fewer than four animals of these strains survived to 800 days of age, totaling 19 strains for analysis.
Heritability of TTBT in the BxD RIs (h2RI) was estimated using the sum of squares (SS) from a one-way analysis of variance by strain as the SSbetween strains/SStotal (12).
Candidate gene identification.—
Candidate genes were sought within the QTL regions through first determining the genetic map location of the peak marker locus. Using the 1.5-LOD drop interval (95% confidence interval), candidate genes were identified using Mouse Genome Informatics of The Jackson Laboratories. The physical map location of the peak marker locus was determined to identify whether the suggested genes were applicable. The Perlegen mouse SNP database was used to determine the extent of haplotype differences between B6 and D2 strains. The Perlegen database constructed through resequencing the nuclear DNA genomes of 15 inbred laboratory mouse strains used the publically available sequence of strain C57BL/6J as a standard. With this approach, candidate genes residing within regions of haplotype similarity may be excluded, and single nucleotide polymorphisms of potential functional importance in candidate genes in the regions of haplotype diversity may be identified. The search for candidate genes contributing to QTL effects (Mouse Genome Informatics, http://www.informatics.jax.org; NCBI Entrez Gene, http://www.ncbi.nlm.nih.gov/gene) was also restricted to those whose function was related to tail tendons and potential factors influencing tail tendon aging.
RESULTS
Analyses of TTBT in B6D2F2 Intercross and BxD RI Strains
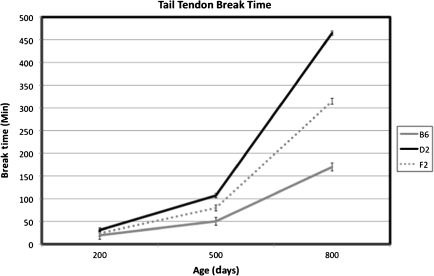
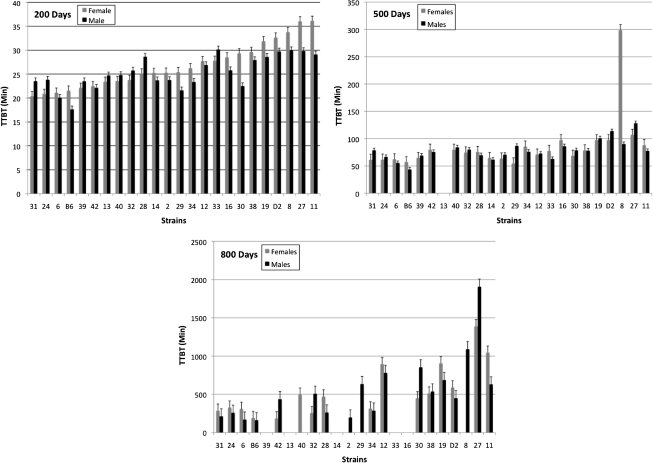
Among the B6D2F2 intercross mice, mean TTBT increased nonlinearly with age from 200 to 800 days of age (Figure 1) when examining the 200-, 500-, and 800-day-old mice from the cross-sectional design of the study. The continuous nature of the distribution of the BxD RI means indicates that TTBT is likely influenced by a polygenic effect rather than a single major gene (Figure 2). The distribution also demonstrates that genetic segregation can yield RIs beyond the range of the progenitors. Further examination of these BxD RI strains through analyses of variance illustrates several significant and marginally significant effects of strain, sex, and age. At 200 days of age, BxD RI strains 11, 27, 29, and 30 show a significant sex difference (p = .006, .032, .058, and .009, respectively). Fewer significant and marginally significant sex differences were present at 500 and 800 days, with BxD 8 significantly different at 500 days (p = .001) and BxD 6 (p = .059) significant at 800 days. The estimates of heritability for TTBT derived for the BxDk RI population for males and females, respectively, were 0.35 and 0.55 at 200 days of age; 0.38 and 0.69 at 500 days of age; and 0.47 and 0.35 at 800 days of age.
Figure 1.
Mean TTBT change with age of the C57BL/6J, DBA2/J, and B6D2F2 animals at 200, 500, and 800 days of age. B6, C57BL/6J; D2, DBA/2J; and F2, B6D2F2. The mean values for 200, 500, and 800 days, respectively, are B6s: 19.6, 50.14, and 169.83; D2s: 30.99, 107.08, and 464.55; and F2: 23.97, 79.63, and 314.69. The values at each age represent different cohorts because the mice were not measured longitudinally.
Figure 2.
Distributions of strain means with standard errors of tail tendon break time (TTBT) at 200, 500, and 800 days of age in the BxD recombinant inbred (RI) strains and progenitor strains (B6 and D2) sorted in order of increasing female TTBT at 200 days of age. Order of the RI strains in graphs at 500 and 800 days are based on the order at 200 days of age for easy comparison. For each RI strain and sex, n = 8–12 animals at 200 days, 5–12 animals at 500 days, and 4–10 animals at 800 days of age. Strains with no values had less than four animals at that age or did not survive to age of testing.
QTL Analysis of TTBT in B6D2F2 Intercross
Linkage analysis was used to associate microsatellite marker genotypes with TTBT. Suggestive or significant QTLs influencing TTBT at 200, 500, and 800 days of age were identified on chromosomes 2, 3, 4, 5, 6, 10, 15, and 17 (Table 1).
Table 1.
Multiple Regression Model for TTBT at Different Ages of B6D2F2 Mice
| Age (days) | Chromosome | Peak Marker | Source of Variance | 1.5-LOD (confidence interval) cM | % Variance | p Value (loci) | Increasing Allele | Allelic Model* |
| 200 | Sex | 8.5 | .00 | |||||
| 2 | D2Mit300 | Chromosome 2 at 46 cM | 7.0–109.0 | 2.1 | .01 | D2 | ♀ H ♂ A | |
| 3 | D3Mit42 | Chromosome 3 at 55 cM | 34.4–64.4 | 3.4 | .00 | D2 | H | |
| 5 | D5Mit227 | Chromosome 5 at 15 cM | 9.0–39.0 | 4.2 | .00 | ♀ B6 ♂ D2 | ♀A ♂ R | |
| Chromosome 5 at 15 cM: sex | 2.1 | .01 | ||||||
| 10 | D10Mit36 | Chromosome 10 at 29 cM | 22.0–38.0 | 5.3 | .00 | ♂ B6 ♀ D2 | ♀H ♂ D | |
| Chromosome 10 at 29 cM: sex | 4.6 | .00 | ||||||
| 17 | D17Mit93 | Chromosome 17 at 46 cM | 26.2–56.7 | 2.4 | .01 | D2 | A | |
| Model total | 19.3 | |||||||
| 500 | Sex | 6.8 | .01 | |||||
| 6 | D6Mit111 | Chromosome 6 at 64 cM | 48.5–70.5 | 5.2 | .00 | B6 | A | |
| 15 | D15Mit105 | Chromosome 15 at 36 cM | 17.8–52.8 | 3.2 | .00 | D2 | D | |
| Model total | 10.3 | |||||||
| 800 | ||||||||
| 4 | D4Mit190 | Chromosome 4 at 79 cM | 3.9–79.0 | 7.1 | .00 | D2 | R | |
| Model total | 7.1 |
Notes: 1.5-LOD, cM, support interval of 1.5-LOD drop in cM determined from interval mapping; A, additive; D, dominant (dominant effect of the allele associated with an increase in TTBT); H, heterosis; R, recessive (recessive effect of the allele associated with an increase in TTBT); See Figure 3 for graphs of the allelic models for each identified QTL. The increasing allele is noted followed by the allelic model.
Listed as combined sexes unless different for sex as noted.
At 200 days of age, a QTL on chromosome 10 at marker D10Mit36 was considered significant, with remaining QTLs on chromosomes 2 (D2Mit30), 3 (D3Mit42), 5 (D5Mit227), and 17 (D17Mit93) identified as suggestive. At 500 days of age, two suggestive QTLs were identified on chromosomes 6 (D6Mit111) and 15 (D15Mit105), and data from 800 days of age identified one suggestive QTL located on chromosome 4 with the D2 allele on the QTL associated with increased TTBT. Table 1 shows the increasing allele associated with either increased or decreased TTBT for each nominated QTL.
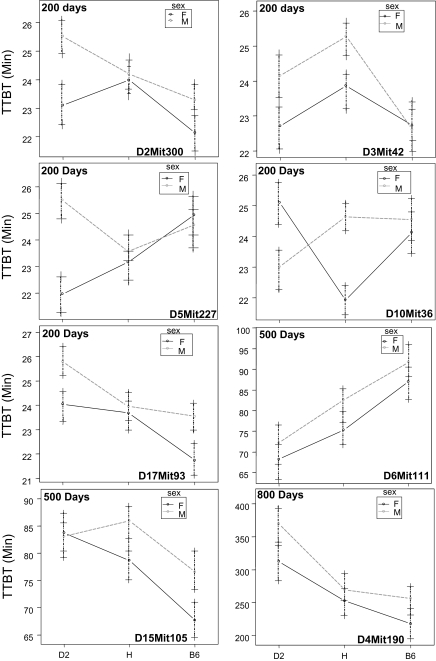
Figure 3 shows the modes of gene action for males and females of the D2 and B6 alleles at the peak marker for each QTL identified. At 200 days of age, the allelic effects of three of five QTLs were different between the sexes. The remaining QTLs at 200 days of age and those at 500 and 800 days of age show the same effects between sexes. The characteristics of the QTLs including 1.5-LOD confidence intervals, variance attributed to each QTL, p value, and allelic model for the three age groups are summarized in Table 1.
Figure 3.
Allelic effects of the markers closest to the quantitative trait loci (QTL) peak in the B6D2F2 intercross at 200, 500, and 800 days of age. Values are means ± standard error for males and females separately. B6, homozygous for C57BL/6J; H, heterozygous; and D2, homozygous for DBA/2J.
The multiple regression models used in the R/qtl program included sex as a covariate for each age group analysis. The effects of the five QTLs at 200 days of age were retained in the regression model at different levels of significance: chromosome 2 (p < .05), chromosomes 5 and 17 (p < .01), and chromosomes 3 and 10 (p < .001). In addition, two statistically significant Sex × QTL interactions were identified at 200 days of age; one on chromosome 5 (p < .01) and the other on chromosome 10 (p < .001). At 500 days of age, the QTLs on chromosomes 6 and 10 were statistically significant in the regression model (p < .01), whereas the QTL on chromosome 4 at 800 days of age was retained in the model at p < .001 level of statistical significance.
It is notable that the identified QTLs are different in each age group. Figure 4 displays an overlapping view of the three ages illustrating changes of the QTLs over time from young adult mice to elderly mice.
Figure 4.
(A) Log of odds ration (LOD) plots of interval mapping in the 200-, 500-, and 800-day old B6D2F2 intercross; black line is 200 days, gray solid line is 500 days, and dashed black line is 800-day age group. Thresholds of significance marked by horizontal dotted lines were determined by 10,000 permutations. The suggestive and significant thresholds for TTBT were 3.1 and 4.8 for 200 days of age, 3.1 and 4.9 for 500 days of age, and 3.1 and 4.9 for 800 days of age, respectively. (B) Separate LOD plots of interval mapping of the 200, 500, and 800 days of age in the BxD RI strains. Thresholds of significance marked by the horizontal dotted lines were determined by 10,000 permutations. The suggestive thresholds for TTBT at all three ages were 2.6. The significant thresholds for TTBT were 3.9 at 200 days of age and 3.7 for 500 and 800 days of age.
QTL Verification Testing and Linkage Analysis in BxD RI Strains
Consistent with other QTL studies in the literature (28), statistical methods to verify QTLs nominated in the B6D2F2 mice and confirm in the BxD RI mice were performed. For each QTL nominated in the B6D2F2 analyses, the 1.5-LOD drop-off interval was calculated (Table 1), followed by identification of the markers in the BxD RIs that fell within these support intervals. Analysis of variance was performed on single markers using TTBT strain means. Furthermore, the statistically significant relationships within each interval were corrected for multiple comparisons (ie, the number of markers within the interval in the BxD RI database) by the Bonferroni method (28). A one-tailed test was used as a conservative method for multiple comparisons because comparisons are not independent due to linkage of microsatellite markers. Following the adjustment for multiple comparisons, the effect on chromosome 4 remained statistically significant (p < .05), providing confirmation of the QTL at 800 days of age from the B6D2F2 intercross results in the BxD RI mice. The effects of the QTLs at 200 and 500 days of age were not significant in the RIs after adjusting for multiple comparisons.
Interval mapping was also used to search for QTLs (not nominated in the F2 analyses) influencing TTBT in the BXD RI strains. One QTL at 800 days of age on chromosome 1 was identified in a combined analysis of males and females. This QTL, positioned at 59.7 cM, had a peak LOD score of 3.40, with the 1.5-LOD drop-off interval from 54.2 to 62.2 cM. Like most QTLs identified in this study, this QTL is genome-wide suggestive. The D2 allele contributed to increased TTBT for this QTL.
Candidate Genes
Candidate genes were identified in two of the eight support intervals for the QTLs nominated. The Col11a1 gene located on chromosome 3 was situated within the identified 1.5-LOD support interval, coding for an alpha subunit of Type XI collagen. Type XI collagen is expressed in the tail during mouse development and controls matrix assembly in the ECM (29–31). The region of the support interval of the QTL on chromosome 4 contains three candidate genes: Col8a2, Col9a2, and Ddost. Col8a2 and Col9a2 are both alpha subunits of Type VIII and Type IX collagens, respectively. The function of the Type VIII alpha 2 subunit entails interactions with proteins in the ECM (32), whereas the Type IX alpha 2 subunit is involved at the attachment site of bone and tendon (33). The tail contains coccygeal (caudal) vertebrae (34) and the collagen attaches to the bone and ECM. Therefore, Type IX collagen from the attachment site may be intertwined with the other tail tendon components. The dolichyl-di-phosphooligosaccharide-protein glycotransferase gene, Ddost located on chromosome 4, is located near the peak of the chromosome 4 QTL identified at 800 days. It is a receptor for advanced glycation end products (35), which accumulate with age in collagen from B6 and D2 mice along with an increase in TTBT (6,36). The Ddost gene appears to be a strong candidate gene for a reproducible age-related QTL.
DISCUSSION
Mouse tail tendon fiber denaturation in urea measured as TTBT has long been used as a biomarker of aging, although the underlying genetic basis for interstrain variation has not yet been determined. The results of this study show an age-related decline in heritability and changes in identifiable QTLs. The QTLs identified at 200 days exhibited a greater influence on TTBT than the QTLs at 500 days of age, with the least influence at 800 days of age. These results are consistent with environmental or other nongenetic factors, for example, disease, affecting the TTBT at the oldest age group.
The total phenotypic variance accounted for by the QTLs with the covariates was 19.3%, 10.3%, and 7.1% for 200, 500, and 800 days, respectively, with most of the phenotypic variance attributable to sex (8.5% at 200 days and 6.8% at 500 days of age). Furthermore, the effects of the QTLs contribute a small proportion of the estimated heritabilities for each age group. These results indicate that many genes may contribute to TTBT, presumably many genes with small effect size contributing to the total variation.
The results presented here are in agreement with previous studies documenting an increase with age in mean TTBT for each strain, with the shorter lived DBA/2 mice exhibiting higher break times than the C57BL/6 animals (3). Sell and Monnier (6) reported TTBT values and tissue pentosidine (an advanced glycated end product) concentrations increasing with age in DBA/2 and C57BL/6 mice, with the rate of increase in TTBT occurring faster in DBA/2 mice (6). They also found that dietary restriction significantly inhibited the age-related increase of both TTBT and pentosidine formation in DBA/2 mice, but only affected TTBT in C57BL/6 mice. Furthermore, dietary restriction also had differential effects on lysyl oxidase–mediated cross-linking and nonenzymatic glycation of collagen (36). Though both increased with age, nonenzymatic glycation of collagen was attenuated by dietary restriction, whereas lysyl oxidase–mediated cross-linking was not. These data suggest that nonenzymatic glycation may be an important variable influencing TTBT, though possibly not the primary factor influencing change in TTBT with age. Genetic variation in the generation and metabolism of advanced glycation end products may therefore influence the TTBT. The finding that the Ddost gene is associated with TTBT is consistent with this hypothesis.
Most QTLs identified in this study were nominated in the F2 population and not confirmed in the RI mouse strains, with the only confirmation at 800 days of age through statistical tests correcting for multiple comparisons. Although it was confirmed through statistical tests, it is more surprising that the one QTL present at 800 days was not confirmed in the RIs. The RI strains that died prior to 800 days of age differed in genotype at the location where the QTL was identified, indicating that not all the important variance resided in this allele. The surviving RI strains also varied in their genotype between the B6 and D2 alleles at this location. Furthermore, the smaller sample size of the BxD RIs (n = 23 strain means used) contributes to low power, presumably limiting detection of QTLs in the analyses (26). As a result, QTLs with small effects may be undetectable in the BxD RI mouse population. However, lack of verification is not conclusive evidence that the QTLs do not exist: they suggest hypotheses for further research such as genotypic selective breeding and further investigations of candidate genes. Therefore, all QTLs nominated were examined.
Gender also influenced the action of QTLs. The regression model indicated a Sex × QTL interaction at 200 days for QTLs on chromosomes 5 and 10. Examination of allelic effects (Figure 3) for the QTL on chromosome 5 revealed differences for males and females with females exhibiting an additive effect and males expressing heterosis (hybrid vigor) in the direction of shorter TTBT. The QTL on chromosome 10 presents significantly different effects for the males and females, with males showing dominance, whereas the females display heterosis in a decreasing direction (shorter TTBT). These are the only QTLs noted in the regression and allelic effects models for the three age groups that confirm significant sex differences. The mechanisms underlying these sex-specific effects are unknown and could be caused by interactions of mitochondrial or sex-linked genes or hormonal influence of the polymorphic genes.
Candidate Genes
The search for candidate genes contributing to QTL effects (Mouse Genome Informatics, http://www.informatics.jax.org; NCBI Entrez Gene, http://www.ncbi.nlm.nih.gov/gene) was restricted to those whose function was already established as related to TTBT physiology. These identified candidate genes were within the 1.5-LOD support interval of the QTLs identified and were in a region of haplotype diversity between C57BL/6 and DBA/2 strains.
TTBT largely reflects the ability of the tendon structure to withstand tensile force, which is primarily due to the presence and arrangement of collagen fibrils in the ECM of the tail tendon. Tendon fibrils are predominately, though not exclusively, comprised Type I collagen (37). The gene for Type I collagen would thus be an obvious candidate gene. However, the results did not nominate or confirm a QTL in the regions of Type I collagen genes (alpha 1: chromosome 11, 56.0 cM and alpha 2: chromosome 6, 0.68 cM; Mouse Genome Informatics, http://www.informatics.jax.org), suggesting that C57BL/6J and DBA/2J strains may not be polymorphic for this gene or that it does not affect TTBT. No polymorphisms exist in the Col1a1 gene between the C57BL/6J and DBA/2J strains, although 15 single nucleotide polymorphisms were identified in the Col1a2 gene: 1 in a synonymous region that does not change the polypeptide sequence and 14 in intronic regions of the gene (NCBI Entrez SNP, http://www.ncbi.nlm.nih.gov/snp). Type VI collagen is present during mouse tail development as well as among thick collagen fibrils in the adult tendon. (38) Type XI collagen is also expressed in the tail during mouse development. The process whereby the long parallel collagen fibrils are deposited into the ECM has not been completely described, but it is believed that a variety of molecules may affect tail tendon structure (31). Growth differentiation factor 5 deficiency increased the proportion of medium diameter collagen fibrils in tail tendon (39). No QTLs were identified in the regions that encode growth differentiation factor 5. However, the effects from QTLs that reside elsewhere but still influence this gene, for example, through regulatory mechanisms, cannot be excluded.
Developmental Change
The different QTLs identified across the life span suggest that the effects of aging are polygenic. None of the QTLs detected at 200 days influenced TTBT at 500 or 800 days, nor did the QTLs identified at 500 days influence TTBT at 800 days of age. These findings imply that the QTLs and underlying genes influencing TTBT at 200 days of age may be “turned off” or are not detectable at 500 or 800 days, whereas the QTLs and underlying genes at 500 days may not have been “turned on” or do not have a significant effect at 200 days. Between 200 and 500 days of age, the QTLs identified at 500 days exhibit greater influence on the trait, whereas the effect of the QTLs at 200 days of age declines. At 800 days, the influence of the QTLs detected at 500 days has decreased, resulting in nonsignificant or absent QTLs, whereas evidence for a new QTL appeared, which was not detectable at earlier ages.
Low statistical power is likely the reason only one QTL was identified as significant (on chromosome 10) and one confirmed QTL (on chromosome 4). Regardless, all identified regions were targeted for potential candidate genes and are worth examining further. Perhaps, QTLs do not interact independently and need to be identified in groups. This would require a greater number of RI strains (greater than 23) to increase the power to identify QTLs whether independently or in groups. In summary, TTBT-influencing QTLs have been identified whose influence is evident at some but not other ages. The heritability estimates of TTBT for females were calculated to be greater than males in all age groups and all values decrease over time, consistent with the identification of more QTLs at 200 days of age than in the older age groups.
FUNDING
This study was supported by grants AG-14731 and AG-000276 at the Pennsylvania University and University of Texas Health Science Center at San Antonio grant T32 AG021890 from the National Institute on Aging of the National Institutes of Health.
Acknowledgments
We are grateful to Dr. D. A. Blizard, M. Miedel, L. Kareha, and N. Wells for excellent help with tissue harvesting and break time data collection. We also thank Dr. A. Lionikas for advice with QTL analyses, Dr. D. Lang with advice on heritability analyses, and anonymous reviewers for helpful feedback and comments.
References
- 1.Verzar F. Aging of the collagen fibre. Int Rev Connect Tissue Res. 1964;2:243–300. doi: 10.1016/b978-1-4831-6751-0.50012-4. [DOI] [PubMed] [Google Scholar]
- 2.Harrison DE, Archer JR. Measurement of changes in mouse tail collagen with age: temperature dependence and procedural details. Exp Gerontol. 1978;13:75–82. doi: 10.1016/0531-5565(78)90033-5. [DOI] [PubMed] [Google Scholar]
- 3.Higgins KA, Stout JT, Heller DA, Parker RF. Individual variability in tail tendon fiber break time in three age cohorts of different strains of mice. Exp Gerontol. 1991;26:467–477. doi: 10.1016/0531-5565(91)90035-k. [DOI] [PubMed] [Google Scholar]
- 4.O’Brien M. Functional anatomy and physiology of tendons. Clin Sports Med. 1992;11:505–520. [PubMed] [Google Scholar]
- 5.Heller DA, McClearn GE. A longitudinal genetic study of tail tendon fibre break time. Age Ageing. 1992;21:129–134. doi: 10.1093/ageing/21.2.129. [DOI] [PubMed] [Google Scholar]
- 6.Sell DR, Monnier VM. Age-related association of tail tendon break time with tissue pentosidine in DBA/2 vs C57BL/6 mice: the effect of dietary restriction. J Gerontol Biol Sci. 1997;52A:B, 277–B284. doi: 10.1093/gerona/52a.5.b277. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura N, Hart DA, Frank CB, et al. Efficient transfer of intact oligonucleotides into the nucleus of ligament scar fibroblasts by HVJ-cationic liposomes is correlated with effective antisense gene inhibition. J Biochem. 2001;129:755–759. doi: 10.1093/oxfordjournals.jbchem.a002916. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura F, Syama K. An amino acid derived from aldol crosslink of elastin and collagen: structure, distribution, aging, and two models of hyperglycemia. Arch Biochem Biophys. 1996;325(2):167–173. doi: 10.1006/abbi.1996.0021. [DOI] [PubMed] [Google Scholar]
- 9.Magnussun SP, Hansen P, Kjaer M. Tendon properties in relation to muscular activity and physical training. Scand J Med Sci Sports. 2003;13:211–223. doi: 10.1034/j.1600-0838.2003.00308.x. [DOI] [PubMed] [Google Scholar]
- 10.Tanksley SD, Medina-Gilho H, Rick CM. Use of naturally-occurring enzyme variation to detect and map genes controlling quantitative traits in an interspecific backcross of tomato. Heredity. 1982;49:11–25. [Google Scholar]
- 11.Edwards MD, Stuber CW, Wendel JF. Molecular-marker-facilitated investigations of quantitative-trait loci in maize. Numbers, genomic distribution and types of gene action. Genetics. 1987;116:113–125. doi: 10.1093/genetics/116.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falconer DS, MacKay TFC. Introduction to Quantitative Genetics. 4th ed. Harlow, UK: Longmans Green; 1996. Components of variance; pp. 1pp. 22–125. [Google Scholar]
- 13.Bailey DW. Recombinant-inbred strains, an aid to finding identity, linkage, and function of histocompatability and other genes. Transplantation. 1971;11:324–327. doi: 10.1097/00007890-197103000-00013. [DOI] [PubMed] [Google Scholar]
- 14.Gora-Maslak G, McClearn GE, Crabbe JC, Phillips TJ, Belknap JK, Plomin R. Use of recombinant inbred strains to identify quantitative trait loci in psychopharmacology. Psychopharmacology. 1991;104:413–424. doi: 10.1007/BF02245643. [DOI] [PubMed] [Google Scholar]
- 15.McClearn GE, Plomin R, Gora-Maslak G, Crabbe JC. The gene chase in behavioral science. Psychol Sci. 1991;2(4):222–229. [Google Scholar]
- 16.Taylor BA. Recombinant inbred strains: use in gene mapping. In: Morse HC, editor. Origins of Inbred Mice. New York: Academic Press; 1978. pp. 4pp. 23–438. [Google Scholar]
- 17.Dewhirst FE, Chien CC, Paster BJ, et al. Phylogeny of the defined murine micriobiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3, 287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verzar F. Changes of thermoelastic properties of tendon fibers in aging. Experientia. 1955;11(6):230. [PubMed] [Google Scholar]
- 19.Elden HR, Boucek RJ. Investigations of the aging process by physical-chemical means—summary. In: Shock NW, editor. Biological Aspects of Aging. 1962. pp. 334–342. Columbia University Press. [Google Scholar]
- 20.Olsen GG, Everitt AV. Retardation of the ageing process in collagen fibres from the tail tendon of the old hypophysectomized rat. Nature. 1965;206(981):307–308. doi: 10.1038/206307b0. [DOI] [PubMed] [Google Scholar]
- 21.Vandenbergh DJ, Heron K, Peterson R, et al. Simple tests to detect errors in high-throughput genotype data in the molecular laboratory. J Biomol Tech. 2003;14:9–16. [PMC free article] [PubMed] [Google Scholar]
- 22.Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high resolution consensus maps for complex trait analysis. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-11-research0046. RESEARCH0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lander E, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:1, 85–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Basten CJ, Zeng Z-B. Windows QTL Cartographer 2.5. Raleigh, NC: Department of Statistics, North Carolina State University; 2006. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Accessed June 15 , 2006. [Google Scholar]
- 26.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broman KW. Review of statistical methods for QTL mapping in experimental crosses. Lab Anim. 2001;30:44–52. [PubMed] [Google Scholar]
- 28.Lionikas A, Blizard DA, Gerhard GS, et al. Genetic determinants of weight of fast- and slow-twitch skeletal muscle in 500-day-old mice of the C57BL/6J and DBA/2J lineage. Physiol Genomics. 2005;21:184–192. doi: 10.1152/physiolgenomics.00209.2004. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Lacerda DA, Warman ML, et al. A fibrillar collagen gene, Coll11a1, is essential for skeletal morphogenesis. Cell. 1995;80(3):423–430. doi: 10.1016/0092-8674(95)90492-1. [DOI] [PubMed] [Google Scholar]
- 30.Iyama K, Sumiyoshi H, Khaleduzzaman M, Matsuo N, Ninomiya Y, Yoshioka H. Differential expression of two exons of the alpha1(XI) collagen gene (Col11a1) in the mouse embryo. Matrix Biol. 2001;20(1):53–61. doi: 10.1016/s0945-053x(00)00130-x. [DOI] [PubMed] [Google Scholar]
- 31.Canty EG, Yinhui L, Meadows RS, Shaw MK, Holmes DF, Kadler KE. Coalignment of plasma membrane channels and protrusions (fibripositors) specifies the parallelism of tendon. J Cell Biol. 2004;165(4):553–563. doi: 10.1083/jcb.200312071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutmuller M, Bruijn JA, de Heer E. Collagen types VIII and X, two non-fibrillar short-chain collagens. Structure homologies, functions and involvement in pathology. Histol Histopathol. 1997;12(2):557–566. [PubMed] [Google Scholar]
- 33.Fukuta S, Oyama M, Kavalkovich K, Fu FH, Niyibizi C. Identification of types II, IX and X collagens at the insertion site of the bovine Achilles tendon. Matrix Biol. 1998;17(1):65–73. doi: 10.1016/s0945-053x(98)90125-1. [DOI] [PubMed] [Google Scholar]
- 34.Shinohara H. The musculature of the mouse tail is characterized by metameric arrangements of bicipital muscles. Okajimas Folia Anat Jpn. 1999;76(4):157–169. doi: 10.2535/ofaj1936.76.4_157. [DOI] [PubMed] [Google Scholar]
- 35.Thornalley PJ. Cell activation by glycated proteins. AGE receptors, receptor recognition factors and functional classification of AGEs. Cell Mol Biol (Noisy-le-grand) 1998;44(7):1013–1023. [PubMed] [Google Scholar]
- 36.Reiser KM. Influence of age and long-term dietary restriction on enzymatically mediated crosslinks and nonenzymatic glycation of collagen in mice. J Gerontol. 1994;49(2):B71–B79. doi: 10.1093/geronj/49.2.b71. [DOI] [PubMed] [Google Scholar]
- 37.McBride DJ, Choe V, Shapiro JR, Brodsky B. Altered collagen structure in mouse tail tendon lacking the alpha 2(I) chain. J Mol Biol. 1997;270(2):275–284. doi: 10.1006/jmbi.1997.1106. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe M, Kobayashi M, Fujita Y, et al. Association of type VI collagen with D-periodic collagen fibrils in developing tail tendons of mice. Arch Histol Cytol. 1997;60(5):427–434. doi: 10.1679/aohc.60.427. [DOI] [PubMed] [Google Scholar]
- 39.Clark RT, Johnson TL, Schalet BJ, et al. GDF-5 deficiency in mice leads to disruption of tail tendon form and function. Connect Tissue Res. 2001;42(3):175–186. doi: 10.3109/03008200109005648. [DOI] [PubMed] [Google Scholar]