Summary
The RNA interference (RNAi) mediated by homology-dependent degradation of the target mRNA with small RNA molecules plays a key role in controlling transcription and translation processes in a number of eukaryotic organisms. The RNAi machinery is also evolutionarily conserved in a wide variety of fungal species, including pathogenic fungi. To elucidate the physiological functions of the RNAi pathway in Cryptococcus neoformans that causes fungal meningitis, here we performed genetic analyses for genes encoding Argonaute (AGO1 and AGO2), RNA-dependent RNA polymerase (RDP1), and Dicers (DCR1 and DCR2) in both serotype A and D C. neoformans. The present study shows that Ago1, Rdp1, and Dcr2 are the major components of the RNAi process occurring in C. neoformans. However, the RNAi machinery is not involved in regulation of production of two virulence factors (capsule and melanin), sexual differentiation, and diverse stress response. Comparative transcriptome analysis of the serotype A and D RNAi mutants revealed that only modest changes occur in the genome-wide transcriptome profiles when the RNAi process was perturbed. Notably, the serotype D rdp1Δ mutants showed an increase in transcript abundance of active retrotransposons and transposons, such as T2 and T3, the latter of which is a novel serotype D-specific transposon of C. neoformans. In a wild type background both T2 and T3 were found to be weakly active mobile elements, although we found no evidence of Cnl1 retrotransposon mobility. In contrast, all three transposable elements exhibited enhanced mobility in the rdp1Δ mutant strain. In conclusion, the RNAi pathway plays an important role in controlling transposon activity and genome integrity of C. neoformans.
Keywords: RNA interference, RNA-dependent RNA polymerase, Dicer, Argonaute, Transposon
1. Introduction
The RNA interference (RNAi) pathway that is mediated by homology-dependent degradation of the target mRNA with small RNA molecules is a central regulatory mechanism controlling transcription and translation in eukaryotic organisms. In this process, a double stranded RNA (dsRNA) produced by RNA-dependent RNA polymerase (RdRP) is first processed into small interfering RNA (siRNA) duplexes of 21–28 nucleotides by the RNase-III-like Dicer and enters the canonical pathway (Bernstein et al., 2001). The siRNA is incorporated into a nuclease complex, the RNA-induced silencing complex (RISC), containing an Argonaute protein with slicer (RNA degrading) activity, and the bound siRNA is then unwound. Next, single stranded siRNA-guided RISC targets homologous mRNAs and confers the silencing activity either by cleaving target mRNAs or by blocking translation.
The RNAi machinery is evolutionarily conserved in a wide variety of fungal species, but not all fungi (Nakayashiki, 2005). In Saccharomyces cerevisiae and Ustilago maydis, none of the RNAi components are present, strongly implicating that the RNAi pathway is dispensable for basic metabolism and growth of at least some fungi. In fungi, the RNA silencing phenomenon has been first reported in Neurospora crassa. N. crassa has a process called quelling , which is a post-transcriptional gene silencing (PTGS) guided by siRNAs (Romano and Macino, 1992). Quelling occurs during vegetative cell growth and can be triggered by introduced transgenes, whose spliced transcripts are specifically degraded by action of the RdRP, Dicer, and Argonaute-containing RISC complex. The physiological roles of quelling appear to be as a defense against viruses and in silencing of active transposons (Cogoni and Macino, 1999a). N. crassa also has another RNA silencing mechanism, called meiotic silencing of unpaired DNA (MSUD) (Shiu et al., 2001). During meiosis of N. crassa, MSUD plays a role in silencing genes that are not paired with its homologous chromosomal partner. MSUD utilizes a distinct set of silencing components from those in the quelling process, and yet the silencing mechanisms are quite similar (Nakayashiki, 2005; Nakayashiki et al., 2006).
RNA silencing has been also discovered in the fission yeast, Schizosaccharomyces pombe, and yet its physiological role is rather different from N. crassa. Argonaute and Dicer are required for heterochromatin assembly at centromeres and the mating type locus of S. pombe (Noma et al., 2004; Sigova et al., 2004; Verdel et al., 2004; Volpe et al., 2002). Defective centromeric heterochromatin formation observed in the ago1Δ, dcr1Δ, and rdp1Δ mutants results in aberrant chromosome segregation in S. pombe. However, the RNA silencing components of the quelling pathway in N. crassa are not involved in maintenance of centromeric heterochromatin assembly (Chicas et al., 2004; Freitag et al., 2004), strongly indicating that the physiological roles of the RNAi components appear to be diverse between fungal species. Notably, some of the S. pombe RNAi components play an siRNA-independent role. In S. pombe, Ago1 and Dcr1, but not RdRP, regulate cell cycle progression and govern cell survival from DNA damage conferred by genotoxic agents through Cdc2-dependent activation of the DNA damage checkpoint (Carmichael et al., 2004b). The S. pombe Ago1 interacts with 14-3-3 proteins to regulate the cyclin-dependent kinase Cdc2 (Stoica et al., 2006). Interestingly, heterologous expression of the human Ago1 homolog can complement the loss of Ago1 function in S. pombe, strongly suggesting that the siRNA-independent roles played by Ago1-like proteins are evolutionarily conserved even in more complex eukaryotes.
Recent analysis reveals that some budding yeasts, which had been presumed to lack the RNAi pathway, indeed have siRNAs and utilize non-canonical Dicer proteins to control Argonaute-like proteins without RdRP. Saccharomyces castellii contains a single Argonaute and a non-canonical Dicer. The S. castellii Dicer, Dcr1, contains an RNaseIII domain, but lacks the N-terminal helicase and DEAD or DEAD-like domains that are generally found in canonical Dicers, such as the human and S. pombe Dicers (Drinnenberg et al., 2009). Therefore, the RNAi pathway appears to be widely conserved in the fungal kingdom.
Human fungal pathogens, including Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus, also retain the RNAi components although their physiological functions are largely unknown. Fungal genome database analyses independently performed by others identified a number of Argonaute, Dicer or Dicer-like proteins, and RdRPs in the pathogens (Drinnenberg et al., 2009; Nakayashiki et al., 2006; Nakayashiki and Nguyen, 2008). Similar to S. castellii, C. albicans contains a single Argonaute and a non-canonical Dicer protein, but not RdRP (Drinnenberg et al., 2009). The filamentous fungus Aspergillus fumigatus contains genes encoding two homologs of Argonaute, Dicer, and RdRP. C. neoformans, which causes fatal meningoencephalitis (Idnurm et al., 2005; Lin and Heitman, 2006), also contains Argonaute, Dicer, and RdRP. However, several interesting features were discovered in the Cryptococcus RNAi machinery. First, serotype A C. neoformans (H99 strain) contains a single Argonaute whereas serotype D C. neoformans (JEC21 strain) contains two Argonaute proteins (Nakayashiki et al., 2006; Nakayashiki and Nguyen, 2008). Second, C. neoformans Dicer proteins do not carry the N-terminal DEAD/DEAH box helicase domain, which is a typical signature of canonical Dicer protein (Nakayashiki and Nguyen, 2008). However, the physiological roles of the RNAi pathway in the human fungal pathogens have been poorly understood. In Mucor circinelloides, which causes invasive maxillofacial zygomycosis (Khan et al., 2009), mutation of dicer-like gene (dcl-1) does not affect siRNA-mediated gene silencing, but causes reduced growth and defective hyphal growth (Nicolas et al., 2007), which could potentially modulate the virulence of the pathogen.
In this study we aimed to characterize the role of the RNAi pathway in growth, differentiation, stress response, and virulence factor regulation of human fungal pathogens by employing C. neoformans as a model system since it has a full suite of the RNAi components. To elucidate the physiological functions of the RNAi pathway in C. neoformans, here we performed genetic analyses for genes encoding Argonaute, RdRP, and Dicers in both serotype A and D C. neoformans, and also employed comparative transcriptome analysis using the RNAi mutants to gain insights into the global regulatory circuit governed by the RNAi pathway. Therefore, this study can improve our understanding of the role of the RNAi genes in human fungal pathogens including C. neoformans.
2. Materials and methods
2.1. Strains and media
The strains used in this study are listed in Table S1. Each strain was cultured in YPD (yeast extract-peptone-dextrose) medium unless indicated. Agar-based DME (Dulbecco modified Eagle) medium for capsule production, V8 mating medium containing 5% V8 juice (Campbell s Soup Co.), Niger seed or L-DOPA medium for melanin assay were prepared as previously described (Alspaugh et al., 1997; Bahn et al., 2004; Granger et al., 1985; Hicks et al., 2004).
2.2. Disruption of the AGO1, RDP1, DCR1 and DCR2 genes
The genes described in this report were deleted in both serotype D and A C. neoformans strains. For disruption of the AGO1, AGO2, RDP1, DCR1, and DCR2 genes in the serotype D strain background, the JEC43 strain (Wickes and Edman, 1995) was transformed with a disruption allele constructed by overlap PCR as previously described (Moyrand et al., 2004). The primer sequences used are provided in Table S2. The tagged plasmids, pNATSTM and pHYGSTM, used to amplify the selectable marker were kindly provided by Dr. Jennifer Lodge (Saint Louis University School of Medicine). The AGO1, RDP1, DCR1, and DCR2 genes in the serotype A strain background were disrupted in the congenic C. neoformans serotype A MATα (H99) and MATa (KN99) strains by overlap PCR with primers listed in Table S2 and biolistic transformation as described previously (Bahn et al., 2005; Davidson et al., 2002). To screen mutant strains, diagnostic PCR was performed by analyzing the 5′- or 3′-junction of disrupted alleles with screening primers listed in Table S2. Screened transformants were further confirmed by Southern blot analysis with genomic DNA digested with appropriate restriction enzymes and each gene-specific probe amplified by PCR with primers listed in Table S2.
2.3. Mating assays
All strains for mating assays were grown on YPD medium for 16 hr at 30°C. Four microliters of cell culture was mixed in an equal volume and spotted onto V8 mating medium. Strains of opposite mating type were co-cultured on V8 mating medium and incubated at 25°C for up to 8 weeks at room temperature in the dark. Mating mixtures were monitored weekly by observing colony and filamentation morphology using a differential interference contrast (DIC) microscope at 100× magnification.
2.4. Capsule and melanin assay
All strains assayed for capsule and melanin production were initially incubated on YPD medium at 30°C for 16 hr. Qualitative and quantitative measurement of capsule and melanin production were performed as described previously (Bahn et al., 2004; Kim et al., 2010).
2.5. Stress and antifungal drug sensitivity test
To assess stress and antifungal drug sensitivity, cells grown overnight at 30°C in YPD medium were 10-fold serially diluted (1 to 104 dilutions) in distilled, sterilized H2O. Then cell suspensions (4 μl) of each strain were spotted onto solid YPD agar medium containing indicated concentrations of NaCl and KCl (high salt and osmotic stress), hydrogen peroxide (H2O2) and diamide (oxidative stress), hydroxyurea (HU) and methylmethane sulfonate (MMS) (genotoxic stress), thiabendazole (TBZ), methylglyoxal (toxic metabolite stress), cadmium sultate (CdSO4) (heavy metal stress), and amphotericin B, fluconazole, ketoconazole, itraconazole, and fludioxonil (antifungal drug sensitivity). Plates were observed after 2-4 days of incubation at 30°C and photographed.
2.6. DNA microarray and data analysis
To obtain total RNA for DNA microarray analysis of the serotype D wild-type [NE519 (MATα) and NE520 (MATa)] and rdp1Δ mutant [NE517 (MATα) and NE518 (MATa)] strains, cells were grown in YPD liquid medium to a density of 5 x 107 cells/ml. RNA was extracted with TRIZOL Reagent (Invitrogen) following the manufacturer s instructions. Total RNA of the wild-type (WT) and the rdp1Δ mutant strains of each mating type was used for Cy3 and Cy5 labeling, respectively. To obtain total RNA for DNA microarray analysis of the serotype A WT H99 strain and ago1Δ (YSB299) and rdp1Δ (YSB467) mutant strains, cells were grown in 50 ml YPD medium at 30°C for 16 h. Then 5 ml of the overnight culture was inoculated into 100 ml of fresh YPD medium and further incubated for 4 h at 30°C to an optical density at 600 nm (OD600 =1.0). The 100 ml culture was split to two 50 ml culture, washed two times with sterilized H2O, resuspended in either 50 ml of YPD (glucose-rich condition) or YP (glucose-starved condition) medium, and further incubated for 2 h at 30°C. Then the culture was frozen in liquid nitrogen and lyophilized overnight. Three independent cultures for each strain were prepared for total RNA isolation as biological replicates. The total RNA were isolated using the TRIZOL reagent (Molecular Research Center) as described before (Ko et al., 2009). The concentration and purity of total RNA samples were measured by OD260 and gel electrophoresis. For control total RNA, all total RNA prepared from WT and ago1Δ and rdp1Δ mutant cells grown under the conditions described above were pooled as reference RNA and used for Cy3 labeling. Each RNA sample was used for Cy5 labeling. C. neoformans serotype D 70-mer microarray slide containing 7,936 probes (Duke University) was used. The cDNA synthesis, Cy5/Cy3 labeling, hybridization, washing, scanning and data analysis were performed as described previously (Kim et al., 2010; Ko et al., 2009; Maeng et al., 2010). Three independent DNA microarrays with three independent biological replicates were performed, including one dye-swap experiment. Expression profiles for some of genes identified by microarray data were confirmed by Northern blot analysis as described previously (Ausubel et al., 1994). All microarray data generated by this study (Table S3) was submitted to the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and can be accessed under number GSE21178.
2.7. Southern analysis for monitoring transposon activity
Each strain was streaked on YPD medium and incubated for 3 days at 30°C. Three colonies from each was then used to inoculate three flasks of 10 ml of YPD liquid medium and grown for one day at 30°C with agitation. In the next day, 1 ml of this culture was used to inoculate 10 ml of fresh YPD medium. This procedure was repeated ten-times for a period of 10 days. Then, each culture was streaked on YPD medium and incubated for 3 days at 30°C. Ten colonies per culture (total 30 colonies from three cultures) were used for genomic DNA extraction and Southern blot experiment analysis with probes of T2 and T3 transposons and Cnl1 retrotransposon generated by PCR with primers listed in Table S2. For quantitative analysis of transposition events, transposition frequency per each culture was calculated as the following: [(a total number of new transposition events, including hop-in and pop-out events of transposons, from each independent culture)/10-independent colonies] × 100. Then average transposition frequency was calculated from three independent cultures with standard deviation. Transposition events occurring at the identical position of multiple colonies from a single culture were counted as a single event since they may represent the same transposition event between siblings during subcultures. Statistical analysis of transposition frequency was performed by student t-test.
3. Results and Discussion
3.1. Identification of RNAi components in Cryptococcus neoformans
Analysis of Cryptococcal genome databases revealed that C. neoformans contains a full array of candidate genes potentially encoding the RNAi machinery. The serotype D C. neoformans genome contains two paralogous genes for Argonaute (named AGO1 and AGO2) and Dicer (named DCR1 and DCR2), and a single gene for RdRP (named RDP1). In contrast, the serotype A C. neoformans genome contains single genes for Argonaute (AGO1) and RdRP (RDP1), and two paralogous genes for Dicer (DCR1 and DCR2). The serotype A Ago1p protein sequence is more similar to the serotype D Ago1 than Ago2 and was named accordingly.
When compared with Argonaute and RdRP in other fungi, C. neoformans (Cn) Ago1/2 and CnRdp1 contain evolutionarily conserved domain structures (Fig. S1). Based on Pfam protein domain analysis (http://pfam.sanger.ac.uk/), both CnAgo1 and CnAgo2 proteins contain PAZ and PIWI domains similar to other fungal Argonaute proteins, which belong to the PAZ-Piwi Domain (PPD) family of proteins that is evolutionarily conserved in a wide variety of multicellular eukaryotes and fungi, but not in S. cerevisiae or bacteria. The PAZ (~100 amino acid) and Piwi (~300 amino acid) domains are found at the N- and C-termini of the PPD proteins, respectively. The Piwi domain is also involved in Dicer binding (Hammond et al., 2000). CnRdp1 has an RdRP domain, which is common to all other fungal RdRPs (Fig. S1). In contrast, the domain structure of CnDcr1/2 is rather different from those of other fungal Dicer proteins. Canonical Dicers, such as S. pombe (Sp) Dcr1 and N. crassa (Nc) Dcl2, contains ResIII, Helicase C, dsRNA bind, and Ribonuclease 3 domains. N. crassa Dcl1 has the DEAD box domain instead of the ResIII domain at the N-terminus. Non-canonical Dicer proteins, such as S. castellii (Sc) Dcr1, contain only Ribonuclease 3 and double-strand RNA binding motif domains and are much smaller in size than canonical Dicer proteins. Interestingly, however, CnDcr1/2 proteins have structural characteristics different from those of known canonical and non-canonical Dicers (Fig. S1). First, CnDcr1/2 carries dsRNA binding and Ribonuclease domains, but not Helicase C and DEAD or DEAD-like (ResIII) domains. Second, CnDcr1 and CnDcr2 are smaller in size than SpDcr1 and NcDcl1/2, but bigger than ScDcr1 (Fig. S1).
3.2. The role of Ago1/2, Rdp1, and Dcr1/2 in RNAi
To characterize physiological functions of the RNAi-related genes in C. neoformans, we constructed gene deletion mutants for each gene by using overlap PCR and biolistic transformation. In the serotype D MATα JEC21 strain background, we deleted the AGO1, AGO2, RDP1, DCR1, and DCR2 genes using different dominant selectable marker containing cassettes as described in the Material and Methods. Each MATα mutant strain was then backcrossed with the MATa JEC20 strain and four progeny strains containing the WT or the mutant gene in both mating type background were isolated for each gene. We also constructed an ago1Δ ago2Δ double mutant strain to test if two argonaute proteins have any redundant roles. No alteration of the growth rate was observed in the RNAi mutants at 30 or 37°C on complete or minimal medium.
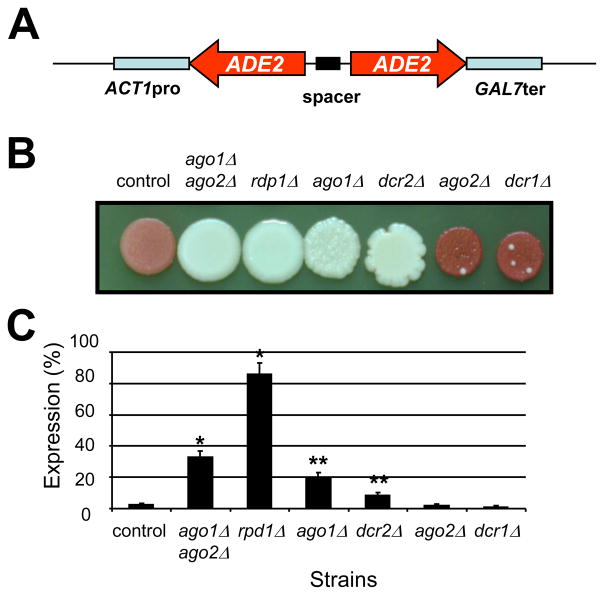
First, we examined whether the potential RNAi components, Ago1/2, Rdp1, and Dcr1/2, are required for the RNAi process by using the RNAi mutants. RNA interference was already shown to be functional in C. neoformans. In their pioneer study, Liu et al. used an episomal plasmid expressing a chimeric hairpin RNA to knock-down the capsule gene CAP59 and the ADE2 gene simultaneously (Liu et al., 2002). Here a hairpin RNA to knock-down the ADE2 gene was integrated into the genome of a serotype D strain because episomal plasmids are unstable in C. neoformans. After transformation, red colonies were chosen and streaked on YPD medium. One transformant displaying the reddest colony-color was chosen for further study (strain NE535). We verified by PCR amplification that the native ADE2 gene was not mutated (data not shown) and demonstrated by back crossing with a ura5 mutant strain that the red phenotype was genetically linked with the URA5 marker. Quantitative RT-PCR confirmed that the ADE2 gene was knocked-down to ~5% of the WT ADE2 expression level (Fig. 1C). We note that the red phenotype was not completely stable, as small sectors of the colonies reverted back to the white WT phenotype (possibly due to recombination between the inverted repeats of the integrated plasmid). Thus, the level of expression of the ADE2 gene measured here probably underestimates the extent of repression by RNAi.
Fig. 1.
The role of the argonaute-, RdRP-, and Dicer-like genes in the RNA interference process of C. neoformans. (A) Map of the pRNAi plasmid used to transform C. neoformans. (B) Coloration of the different mutant strains after 4 days on YPD agar medium at 30°C. (C) ADE2-specific mRNA levels as measured by quantitative RT-PCR. An ade2 knockdown strain was constructed by integration of the pRNAi plasmid (GeneBank HM352736, kindly given by T. Doering) in its genome. This strain (control, strain name) produced red colonies on complete medium and the levels of ADE2-specific mRNA were fiftyfold lower in the control strain than in the WT strain. To test the importance of each putative RNAi-component on the silencing of ADE2, the control ADE2-knockdown strain was crossed with different single and double RNAi mutant strains. As shown here, RDP1 was the only gene completely necessary for ADE2-silencing. AGO1 and DCR2 also play a major role whereas the deletion of either DCR1 or AGO2 appeared to have a very limited impact on silencing. * Student t-test P value <0.001.** Student t-test P value<0.05
Ade2-suppressed NE535 mutant strain was then crossed with the different serotype D RNAi mutant strains. The rdp1Δ mutation completely reversed the red phenotype induced by the presence of the ADE2 specific hairpin RNA (Fig. 1B). Consistently, the expression levels of the ADE2 gene were restored to the WT level (Fig. 1C), indicating that the RNAi pathway induced by a hairpin RNA is nearly completely blocked in the rdp1Δ mutant strain. Ago1 and Dcr2 also appeared to be involved in the RNAi pathway, albeit to a lesser extent than Rdp1, in the RNAi pathway. As shown in Figure 2, ago1Δ and dcr2Δ mutations completely suppressed the color phenotype associated with the ADE2-knockdown. However, the expression levels of ADE2 in the ago1Δ and dcr2Δ genetic backgrounds were only partially restored (Fig. 1C). Finally, deletion of AGO2 or DCR1 had no influence on the red color phenotype of the ADE2-knockdown strain (Fig. 1B). Quantitative RT-PCR experiments confirmed that these genes have little influence in the RNAi pathway induced by a hairpin RNA (Fig. 1C). However, the ADE2-expression levels in the ago1Δ ago2Δ double mutant strain were higher than those in the ago1Δ or ago2Δ single mutants, suggesting that AGO2 might play a minor role in the RNAi pathway.
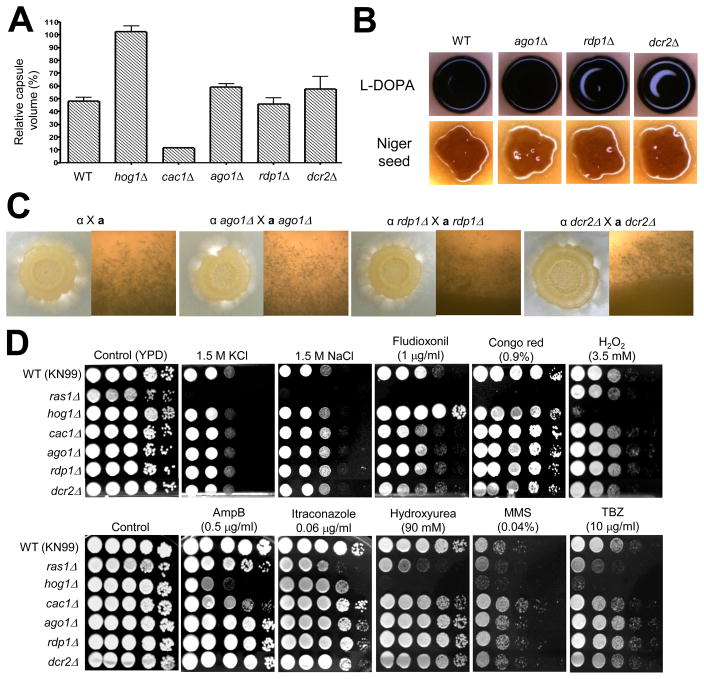
Fig. 2.
The role of the RNAi components in virulence factor production, mating, and stress response of C. neoformans. (A) Capsule synthesis levels of the following strains were quantitatively measured by using hematocrit as previously described (Kim et al., 2010): the serotype A MATa WT strain (KN99) and cac1Δ (YSB79), hog1Δ (YSB81), ago1Δ (YSB300), rdp1Δ (YSB306), and dcr2Δ (YSB305) mutant strains. The Y axis indicates the relative capsule volume, which is percent ratio of length of packed cell volume phase versus length of total loading volume phase. (B) Melanin production of the following strains was visualized on L-DOPA or Niger seed medium: the serotype A MATa WT strain (KN99) and cac1Δ (YSB79), hog1Δ (YSB81), ago1Δ (YSB300), rdp1Δ (YSB306), and dcr2Δ (YSB305) mutant strains. The hog1Δ and cac1Δ mutants were used as positive and negative controls, respectively, in both capsule and melanin production (Bahn et al., 2004; Bahn et al., 2005). (C) Each serotype A α mating type strain [WT H99, ago1Δ (YSB299), rdp1Δ (YSB624), and dcr2Δ (YSB304)] was cocultured with a mating type strains [WT KN99, ago1Δ (YSB300), rdp1Δ (YSB307), and dcr2Δ (YSB305)] on V8 medium (pH 5.0) at room temperature in the dark. Representative edges of the mating patches were photographed at 100x magnification after 21 d incubation. (D) Each C. neoformans strain (the WT KN99 strain and ras1Δ [YSB73], cac1Δ [YSB79], hog1Δ [YSB81], ago1Δ (YSB300), rdp1Δ (YSB306), and dcr2Δ (YSB305)) was grown overnight at 30°C in liquid YPD medium, 10-fold serially diluted (1 10−4 dilutions), and spotted (3 μl of dilution) on YPD agar medium containing the indicated concentrations of KCl, NaCl, fludioxonil, Congo red, hydrogen peroxide (H2O2), amphotericin B, itraconazole, hydroxyurea, methylmethane sulfonate (MMS), or TBZ. Cells were incubated at 30°C for 72 h and photographed.
These results are in good agreement with other independent findings that Ago1, Rdp1, and Dcr2 play a major role in transgene-mediated co-suppression (termed sex-induced silencing, SIS) in serotype A C. neoformans (personal communication, Yen-Ping Hsueh and Joseph Heitman). Heitman and colleagues demonstrated that Dcr2 is the major dicer protein for controlling SIS triggered by a tandem insertion of multiple copies of a transgene (SXI2a-URA5), and yet Dcr1 plays only a minor role. Taken together, Ago1, Rdp1, Dcr2 are the key RNAi components in C. neoformans.
3.3. The role of the RNAi components in virulence factor production and mating of C. neoformans
Because we confirmed the role of the Ago1, Rdp1, and Dcr2 in the RNAi process, we next examined other possible physiological and cellular functions of RNAi in C. neoformans. To investigate the roles of the RNAi components in the serotype A C. neoformans strain, we also isolated the ago1Δ, rdp1Δ, dcr1Δ, and dcr2Δ mutants in the H99 strain background as described in Materials and Methods.
First, we investigated whether the RNAi machinery plays a role in controlling biosynthesis of the antiphagocytic polysaccharide capsule and the antioxidant melanin, which are two major virulence factors of C. neoformans (Lin and Heitman, 2006). We analyzed the capsule size of the serotype D RNAi mutants as previously described (Janbon et al., 2001) but no difference was observed to be associated with any of the mutations generated, including rdp1Δ or ago1Δ ago2Δ double mutation. Similarly, dot blot analysis of the capsule structure using anti GXM antibodies (Moyrand et al., 2007) did not reveal any difference between the RNAi mutants and WT strains (data not shown). Similarly the serotype A ago1Δ, rdp1Δ, and dcr2Δ mutants appeared to produce WT levels of capsule (Fig. 2A). Next we examined the role of the RNAi mutants in melanin production by visual inspection of melanin accumulation in cells on L-DOPA or Niger seed agar medium. The serotype D and A C. neoformans RNAi mutants produced WT levels of melanin (data not shown and Fig. 2B). Taken together, we conclude that the RNAi machinery is not involved in capsule and melanin production of both serotype A and D C. neoformans strains.
Second, we examined the role of the RNAi components in morphological differentiation of C. neoformans. In the bilateral mating cross between the serotype A MATα and MATa ago1Δ, rdp1Δ, or dcr2Δ mutants, no obvious mating deficiency was observed and hyphae with clamp connection, basidia, and spores were observed (Fig. 2C). Similarly, the serotype D RNAi mutants, including ago1Δ ago2Δ or rdp1Δ mutants, also exhibited WT levels of mating efficiency (data not shown). Furthermore, the dcr1Δ mutant was as normal in virulence factor production and mating as the WT strain (data not shown). All of these data suggest that the RNAi machinery is not involved in sexual differentiation of C. neoformans, but additional studies to examine mating in further detail are clearly warranted.
3.4. The role of the RNAi components in diverse stress responses of C. neoformans
Next we tested whether the RNAi machinery play any roles in survival of C. neoformans under diverse environmental conditions. In response to osmotic and salt shock, high temperature (37~40°C), cell wall/membrane destabilizing agents, oxidative stress, heavy metal, and antifungal drug treatment, the serotype A C. neoformans ago1Δ, rdp1Δ, and dcr2Δ mutants exhibited WT levels of resistance or sensitivity as opposed to ras1Δ, hog1Δ, and cac1Δ mutants which were used as controls since these strains were reported to display differential sensitivity to diverse environmental stresses and antifungal drugs (Ko et al., 2009; Maeng et al., 2010) (Fig. 2D and Fig. S2). We also observed that the serotype D RNAi mutants, including rdp1Δ or ago1Δ ago2Δ mutants, were as resistant as the WT strain to most of stresses we tested here (data not shown).
In the fission yeast S. pombe, ago1+ and dcr1+ are essential for initiation and/or maintenance of the DNA replication and damage checkpoints. Therefore, the S. pombe ago1Δ, dcr1Δ, and rdp1Δ mutants exhibit hypersensitivity to the microtubule-destabilizing drug TBZ (thiabendazole) and the DNA damaging agent HU (hydroxyurea) (Carmichael et al., 2004a). In the filamentous fungus N. crassa, RNA interference pathways are also involved in DNA damage repair (Cogoni and Macino, 1999b; Lee et al., 2009; Nolan et al., 2008). In contrast to the case in S. pombe and N. crassa, the serotype A C. neoformans RNAi mutants did not exhibit increased sensitivity to TBZ, HU, or MMS (Fig. 2D). Similarly, the serotype D C. neoformans RNAi mutants did not show any increased sensitivity to TBZ or HU (data not shown). Deletion of DCR1 did not affect cellular stress response to any of environmental stresses we tested (data not shown). Taken together, we conclude that the RNAi machinery is not involved in survival or proliferation of C. neoformans in response to a variety of environmental stresses.
A lack of any obvious phenotypes in the RNAi mutants has been also reported in the budding yeast S. castellii (Drinnenberg et al., 2009). Deletion of AGO1 and DCR1 genes does not result in any defects in viability on rich or minimal media at a range of temperatures, mating, sporulation, or chromosome stability of S. castellii (Drinnenberg et al., 2009). Furthermore, the S. castellii ago1Δ and dcr1Δ mutants do not show any altered sensitivity to HU and TBZ, similar to the C. neoformans RNAi mutants, strongly indicating that the role of Argonaute and Dicer proteins in DNA replication and cell cycle progression is limited to certain fungi such as S. pombe and N. crassa.
3.5. Transcriptome analysis of the serotype D RNAi mutant strains
A lack of any obvious phenotype in both serotype A and D RNAi mutants prompted us to look at the genome-wide transcript profiles of the mutants by using DNA microarray analysis. We performed transcriptome analysis on four progeny generated from back-crossing of the original rdp1Δ mutant serotype D strain with the WT strain. Comparison was made in MATα and MATa backgrounds. As a first analysis, we considered genes that were regulated similarly in both mating types.
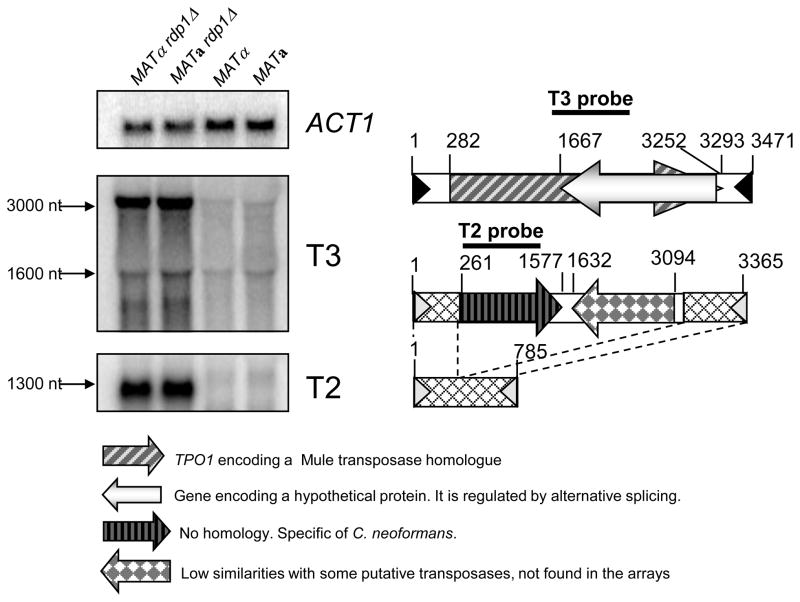
As shown on the Table 1 and Table S4, very little modification was observed and most of the RDP1-dependent regulation appeared to be negative ones. Actually, no change was observed with the marked exception of transposable elements. As shown in Table 1, nine independent tags present in the serotype D arrays and specific for different paralogous hypothetical genes encoding putative transposases that yielded a much stronger signal when probed with the rdp1Δ cDNAs than with the WT ones. These genes encode hypothetical proteins possessing a Mule transposase domain (pfam10551). Analysis of their genomic environments revealed a transposon-like structure and the presence of a 144 bp long terminal inverted repeat (Fig. 3). As the T1 and T2 DNA transposons have been already identified in C. neoformans (unpublished data, Cruz and Heitman) and annotated in Genebank (accession numbers AY145838 and AY145839 respectively), we named this new transposon T3. Interestingly, another gene in this transposon family exhibited higher transcript levels in the rdp1Δ mutant strains. This gene encodes a putative protein showing no homology to any known proteins and it is transcribed from the opposite strand of the transposase gene. Northern analysis confirmed higher transcript levels for both T3 genes (TPO1 and XP_568503/4) in the rdp1Δ mutant compared to the WT strain (Fig. 3). Five completely identical copies of the T3 transposon and 2 copies containing one gap were identified in the JEC21 genome (Table S5). Blastn analysis revealed the presence of 6 additional partial versions of T3. In the serotype A C. neoformans genome, the T3-like transposon appeared to be missing. Furthermore, none of the serotype A C. neoformans genes exhibited significant homology to the two serotype T3 genes. However, a serotype A gene, CNAG_02253.2 (80 amino acids), showed partial homology to the Mule transposable domain of the TPO1 (XP_572854, 932 amino acids). CNAG_02253.2 was the only serotype A gene that shows partial homology to the Mule transposable domain. Based on the serotype A microarray data, however, transcript levels of CNAG_02253.2 were not significantly higher in the serotype A rdp1Δ mutants than the WT strain (Table S3).
Table 1.
Genes regulated by Rdp1 in the serotype D C. neoformans
| Gene Locus (Tag ID on DNA chip) | Sero A ID CNAG | Sero A H99 | Sero D JEC21 | Comments on the encoded protein | |
|---|---|---|---|---|---|
| rdp1/ WT | rdp1/ WT a | rdp1/ WT α | |||
| 181.m08309 | 1.163 | 4.613 | 4.84 | Hypothetical proteins with MULE transposase domain part of the transposable element T3 | |
| 184.m04347 | none | 4.539 | 4.433 | ||
| 179.m00071 | 0.086 | 5.44 | 4.249 | ||
| 180.m00376 | 1.278 | 4.845 | 3.642 | ||
| 1621.seq.158 | 0.524 | 10.4 | 4.329 | Hypothetical proteins with MULE transposase domain part of the transposable element T3 | |
| 184.m05148 | 0.978 | 5.267 | 4.669 | Hypothetical transposase domain-containing proteins (Transposase family tnp2) | |
| 1631.seq.008 | 0.194 | 5.628 | 3.219 | Hypothetical proteins with MULE transposase domain part of the transposable element T3 | |
| 1702.seq.061 | −0.565 | 4.192 | 3.746 | Hypothetical proteins sharing no homologies, and specific to C. neoformans. This tag recognizes also the Transposable element T2 (strain C20F2) GenBank: AY145845.1 | |
| 1642.seq.047 | −0.037 | 4.01 | 4.035 | Hypothetical proteins with MULE transposase domain part of the transposable element T3 | |
| 163.m06565 | 03896 | 0.252 | 3.898 | 3.878 | Hypothetical transposase domain-containing proteins (Transposase family tnp2) |
| 177.m03398 | 0.685 | 3.563 | 3.56 | Hypothetical proteins sharing no homologies, and specific to C. neoformans. This tag recognizes also the Transposable element | |
| 177.m03397 | 0.361 | 3.36 | 3.439 | T2 (strain C20F2) GenBank: AY145845.1 | |
| 1642.seq.046 | 0.104 | 3.422 | 2.493 | Hypothetical proteins with MULE transposase part of the transposable element T3 | |
| 1682.seq.124 | −0.951 | 3.454 | 3.209 | Hypothetical protein with MULE transposase domain part of the transposable element T3 | |
| 1704.seq.045 | 07523 | −0.58 | 3.279 | 3.208 | Not annotated in Genebank. Blastx analysis revealed a hypothetical protein sharing similarities with the Coprinopsis cinerea hypothetical protein XP_001832197.1 |
| 164.m02817 | 0.271 | 3.221 | 3.166 | Hypothetical proteins sharing no homologies, and specific to C. neoformans. This tag recognizes also the Transposable element T2 (strain C20F2) GenBank: AY145845.1 | |
| 163.m06563 | −5.45 | 2.945 | 3.045 | Hypothetical proteins with MULE transposase domain part of the transposable element T3 | |
| 1712.seq.016 | −1.578 | 2.931 | 2.426 | Hypothetical proteins with MULE transposase domain part of the transposable element T3 | |
| 181.m08012 | 0.167 | 2.808 | 2.742 | Hypothetical proteins sharing no homologies, and specific to C. neoformans. This tag recognizes also the Transposable element | |
| 184.m04750 | 2.382 | 2.657 | 3.113 | T2 (strain C20F2) GenBank: AY145845.1 | |
| 164.m02163 | 1.848 | 2.938 | 2.595 | Hypothetical proteins antisense of the mule transposase part of the transposable element T3 | |
| 1681.seq.118 | 0.173 | 2.297 | 2.356 | Transposase ; Crypton transposon (chromosome 6). Not annotated although present in the strain JEC21. Annotated only in the strain B-3501 | |
| 184.m04637 | 0.927 | 1.971 | 1.915 | RNA-dependent DNA polymerases (slacs 132 kDa protein orf 2) present in retrotransposable element | |
| 167.m03164 | 0.333 | 1.773 | 1.416 | RNA-dependent DNA polymerase (slacs 132 kDa protein orf 2) present in retrotransposable element | |
| 1742.seq.060 | 0.174 | 2.535 | 1.58 | RNA-dependent DNA polymerases (slacs 132 kDa protein orf 2) present in retrotransposable element | |
| 1751.seq.002 | 0.103 | 1.609 | 2 | RNA-dependent DNA polymerase (slacs 132 kDa protein orf 2) present in retrotransposable element | |
| 1671.seq.034 | −0.108 | 1.646 | 1.831 | Not annotated in Genebank. Blastx analysis revealed an hypothetical protein sharing similarities with the Coprinopsis cinerea hypothetical protein XP_001829802.1. | |
| 184.m05114 | 05640 | −0.227 | 1.685 | 2.286 | Protein sharing similarities with the Saccharomyces cerevisiae protein Smf1p (Divalent metal ion transporter) |
| 180.m00245 | 06877 | 0.498 | 1.557 | 1.608 | RNA-dependent DNA polymerase (slacs 132 kDa protein orf 2) present in retrotransposable element |
| 1661.seq.033 | 9.758 | 1.417 | 1.196 | Not annotated in GeneBank. Blastx analysis revealed an hypothetical protein sharing similarities with the C. neoformans strain JEC21 protein XP_572518.1 belonging to the DDE superfamily endonuclease | |
| 1701.seq.190 | 0.491 | 1.815 | 1.906 | Blastx analysis revealed an hypothetical protein sharing similarities with the ATP-dependent DNA helicase Pif1 from Microsporum canis | |
| 1741.seq.124 | 0.09 | 1.312 | 1.272 | Hypothetical protein sharing no sequence homology. Not annotated in JEC21 although present. Annotated only in the strain B- 3501 | |
| 162.m02616 | 04538 | 0.096 | 1.108 | 1.021 | Ubiquitin-like conjugating enzyme |
| 176.m02142 | −0.599 | 0.981 | 1.259 | Hypothetical protein | |
Fold increase (rdp1Δ/WT) is log2 scale.
Fig. 3.
Increased transcript levels of transposable elements in the serotype D rdp1Δ mutants. Northern blot analysis showed basal expression levels of T2 and T3 transposons in the serotype D WT [NE519 (MATα) and NE520 (MATa)] and increased levels in rdp1Δ mutant [NE517 (MATα) and NE518 (MATa)]. Diagrams of T3 and T2 (long and short versions) transposons are depicted at the right panel.
Another family of genes appeared to be abundant in the serotype D rdp1Δ mutant strains. These genes encode proteins showing no homology with any proteins of known function. The tags specific to these genes were included in a transposon named T2 in strain C20F2 (Cruz et al, unpublished data). In the genome of JEC21 this transposon exists as a long and a short version (Fig. 3). Both versions have the same 17 bp TIR sequences. The short version does not contain any putative gene. The 5′ ends of both versions are identical over 270 bp. At the other end the sequence is degenerated between the two versions although 73% identity over 441 bp does exist. Four almost identical copies of the longer version are present in the JEC21 genome and Blastn analysis revealed the presence of six additional truncated or degenerated copies (Table S6). Four identical copies of the short version are present in the JEC21 genome and Blastn analysis revealed the presence of three additional truncated or degenerated copies (Table S6). The serotype A C. neoformans appears not to contain any T2-like transposons in its genome.
A third class of transposons affected by the rdp1Δ mutation included a gene encoding protein sharing similarity to an RNA-dependent DNA polymerase (SLACS 132 kDa protein ORF2). Its transcript levels increased about fourfold in the rdp1Δ mutant strains compared to the WT. This gene was annotated as part of the non-LTR retrotransposon Cnl1 (C. neoformans LINE-1) (Goodwin and Poulter, 2001). Blastn analysis revealed the presence of eight very similar copies of this gene and 17 partial ones. Most, but not all, of these are located at subtelomeric regions (Table S7). The tBlastn analysis revealed a number of additional related retrotransposons, mostly located close to telomeres and to the mating locus (data not shown) (Loftus et al., 2005). The serotype A C. neoformans contains eleven genes sharing minor homology to the SLACS, but only one gene, CNAG_06863.2 that shares the highest homology to the serotype D SLACS, was found to be slightly overexpressed in the serotype A ago1Δ (~twofold) and rdp1Δ (~1.5-fold) mutants (Table S3).
One last family of transposons affected by the rdp1Δ mutant was represented by the XM_769017 gene (1681.seq.118), of which expression levels were more than fourfold increased in the rdp1Δ mutant compared to the WT strain. This gene encodes a putative tyrosine recombinase, which is part of the previously identified Crypton transposable element Cn1 (Goodwin et al., 2003), and is present in two copies in the JEC21 genome. Two serotype A C. neoformans genes (CNAG_02719.2 and 04497.2) exhibit only limited homology to the XM_769017.
Other genes overexpressed in the rdp1Δ mutants encode proteins sharing homology with proteins implicated directly in transposable element metabolism. A protein encoded by the gene XM_567974 (184.m05148 and 163.m06565), which is more than sixteenfold overexpressed in the rdp1Δ mutant strains compared to the WT strains, contains a transposase family Tnp2 domain. Although it is similar to the transposase gene from Schistosoma mansoni TRC1 transposon (DeMarco et al., 2006), we found no evidence of conserved inverted repeats flanking this gene. A serotype A C. neoformans gene, CNAG_00878.2, exhibits only limited homology to XM_567974. Finally, a gene encoding a protein homologous to proteins belonging to the DDE superfamily of integrases/endonucleases was shown to be nearly threefold overexpressed in the rdp1Δ mutant strain. These proteins have been shown to be involved in the integration/transposition of most LTR retrotransposons and many DNA transposons (Kulkosky et al., 1992).
Transcripts of two other genes appear to be abundant in the rdpΔ1 mutants. One encodes a putative divalent metal ion transporter and the other a ubiquitin-like conjugating enzyme. Finally, a gene encoding a protein homologous to a Pif1 helicase is about threefold more expressed in the rdp1Δ mutant strain. Although no direct link between this activity and transposon metabolism has been ever demonstrated, this activity plays an essential role in the replication of both nuclear and mitochondrial DNA of S. pombe (Pinter et al., 2008).
The increase in transposon- or retrotransposon-transcript levels in RNAi mutants has also been reported in other organisms. In Trypanosoma brucei, deletion of the gene encoding Argonaute protein Ago1 leads to an increase in transcript abundance of retrotransposons, such as Ingi and SLACS, at both the transcriptional and post-transcriptional levels (Shi et al., 2004). Furthermore, a recent transcriptome analysis of the WT, ago1Δ, and dcr1Δ strains of S. castellii by using high-throughput sequencing of polyadenylated RNA revealed that transcripts for two ORFs, the Y subtelomeric repeats and Ty transposable elements, were significantly enriched in the ago1Δ and dcr1Δ mutants whereas any changes in transcript abundance for other S. castellii ORFs were minimal (Drinnenberg et al., 2009).
3.6. The role of the RNAi machinery in mobility of transposable elements
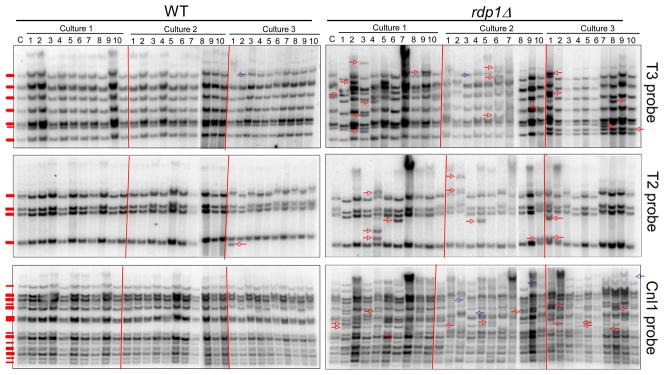
In the serotype D background, most of the transcriptome modifications are related to transposon metabolism. Thus, the high transcript levels of different transposases suggest that some transposons might be more active in the RNAi mutant strains. We analyzed the transposition events of Cnl1 retrotranposon and T2/T3 transposons after prolonged cultures of WT and rdp1Δ strains by Southern blot (see Material and Methods). As shown in Figure 4, the transposons T2 and T3 were found to be weakly mobile in the wild type strain as only one transposition event per transposon was identified out of total 30 colonies tested from three independent cultures. In contrast, mobility of T2 and T3 transposons (particularly T3) appeared to be much enhanced in the rdp1Δ mutant (Figure 4). The Cnl1 retrotransposon we tested did not move at all in the wild-type strain as no transposition event was observed in any of the colonies tested. In contrast, in the rdp1Δ mutant strain changes in the retrotransposon hybridization patterns after prolonged growth were numerous in all three independent cultures (Figure 4), suggesting that mobility of the Cnl1 retrotransposon is much enhanced in the rdp1Δ mutant background. Quantitative measurement of transposon mobility events showed that transposition frequency of T3 transposon and Cnl1 retrotransposon was significantly higher in the rdp1Δ mutant than the wild-type strain (Table 3). All these data suggest that the RNAi pathway control the mobility of transposons and retrotransposons in serotype D C. neoformans.
Fig. 4.
Mobility of T2 and T3 transposons and Cnl1 retrotransposon in the WT (MATa) and rdp1Δ mutant strain backgrounds. Analysis of the transposition events by Southern blot analysis was conducted with genomic DNA isolated after prolonged cultures (10 subcultures) of the serotype D WT strain (NE520) and rdp1Δ mutant (NE518) and compared to the reference strain (C, before subculture). Ten colonies per each culture were analyzed. From each independent culture (10 colonies), only novel transposition events were indicated as arrows. “Hop-in” and “pop-out” events of transposons were indicated as red and blue arrows, respectively. Transposition events occurring at the identical position of multiple colonies from a single culture were counted as a single event since they may represent the same transposition event between siblings during subcultures. Genomic DNA was cleaved with PstI for T2, XhoI for T3, and PstI for Cnl1, separated on 0.8% agarose gel, transferred to Nylon membranes, and hybridized with each specific probe generated by PCR with primers listed in Table S2.
Table 3.
Transposition frequency in the rd1pΔ mutant compared to the wild-type strain.
| Probe | Strain | Transposition frequency (%)a | P valueb |
|---|---|---|---|
| T2 | WT | 3.33±5.77 | 0.0955 |
| rdp1Δ | 33.33±11.55 | ||
| T3 (TPO1) | WT | 3.33±5.78 | 0.0026 |
| rdp1Δ | 56.67±11.55 | ||
| Cnl1 retrotransposon | WT | 0 | 0.0034 |
| rdp1Δ | 56.67±15.28 |
Transposition frequency (%) was calculated as the following: [average of a total number of new transposition events (sum of total number of red and blue arrows in Figure 4 in a single culture) per 10-independent colonies from each independent culture]×100 ± standard deviation.
P value was obtained by student t-test.
3.7. Transcriptome analysis of the serotype A RNAi mutant strains
To gain further insight into the role of the RNAi machinery in C. neoformans, we also performed comparative transcriptome analysis of the ago1Δ and rdp1Δ mutants of the serotype A strains by using DNA microarray analysis. From a total of 7,936 spots we monitored on the serotype D DNA microarray chips, 966 genes displayed differential expression patterns compared to the WT strain H99 at statistically significant levels (ANOVA test, P <0.05). Both the ago1Δ and rdp1Δ mutants exhibited similar transcriptome patterns to those of the WT strains in both normal and no-glucose conditions (Fig. S3), demonstrating that the RNAi machinery does not greatly affect transcriptome patterns that are regulated by glucose-starvation stress.
Under unstressed, glucose-rich conditions, basal expression levels of a total of 72 genes were changed statistically significantly in the ago1Δ and rdp1Δ mutants compared to the WT under normal growth conditions, whereas expression levels of 54 genes were changed significantly in the ago1Δ and rdp1Δ mutants compared to the WT under no glucose conditions (Fig. S3). Similar to the results from the transcriptome analysis of the serotype D RNAi mutants, only a small number of genes (18 genes) exhibited more than a twofold induction or reduction in the ago1Δ or rdp1Δ mutants (Table 2). A majority of them appear to encode proteins of unknown functions in C. neoformans, which did not have any orthologs in other fungi. Among these, expression of two genes, CNAG_01256.2 and CNAG_07505.2, was found to be reduced more than fourfold (six- and eightfold, respectively) in both the ago1Δ and the rdp1Δ mutant. Both genes appear to encode proteins of unknown functions. Furthermore, transcript levels of these two genes were not abundant in the serotype D rdp1Δ mutant (Table 2). We performed Northern blot analysis to verify expression patterns of CNAG_01256.2 and 07505.2. Transcript levels of CNAG_07505.2 were almost undetectable even after long exposure (data not shown). Similarly, CNAG_01256.2 appeared to be expressed at a very low level and yet its expression levels were found to be slightly lower in the ago1Δ and rdp1Δ mutants than the WT strain (Fig. S4), which is in agreement with the microarray data.
Table 2.
Genes regulated by Ago1 and Rdp1 in the serotype A C. neoformans
| SeroD CnID | SeroA CnID | ScID | Serotype A |
Function (KOG predicted function) | |
|---|---|---|---|---|---|
| ago1Δ/ WTα | rdp1Δ /WTα | ||||
|
Under glucose-rich condition | |||||
| 163.m06442 | 01256 | None | −2.265 | −2.128 | Unknown (Zn-finger protein) |
| 163.m02872 | 01024 | None | −2.20 | −0.392 | Unknown |
| 181.m07823 | 00049 | None | −1.28 | 0.664 | Unknown (Sentrin-specific cystein protease) |
| 162.m02614 | 07779 | YPL069C | 1.159 | 1.17 | D-glycerate 3-kinase |
| 167.m03679 | 02488 | None | 1.31 | 2.546 | Unknown (Methionyl-tRNA formyltransferase) |
| 181.m08143 | 00397 | None | 1.329 | 0.931 | Pyruvate dehydrogenase |
|
Under glucose-starved condition | |||||
| 179.m00054 | 07505 | None | −3.765 | −3.704 | Unknown (Nuclear protein containing WD40 repeat) |
| 181.m08459 | 00738 | None | −1.026 | −1.688 | Unknown (Flavonol reductase/cinnamoyl-CoA reductase) |
| 179.m00600 | 01474 | None | −1.591 | −3.14 | Unknown (Cysteine proteinase Cathepsin L) |
| 180.m00181 | 06629 | None | −1.874 | −0.94 | Unknown (Serine/threonine protein phosphatase 2A) |
| 163.m06474 | 01286 | None | −0.931 | −1.667 | Unknown (Leucine rich repeat proteins) |
| 163.m02895 | 01001 | None | −0.211 | −1.563 | Unknown (Polypeptide release factor 3) |
| 186.m03966 | 04036 | None | −0.054 | −1.514 | Unknown (Heat shock transcription factor) |
| 184.m05097 | 05546 | None | −0.12 | −1.064 | Unknown (Predicted membrane protein) |
| 167.m05909 | 02319 | None | −0.973 | −1.047 | Unknown (DNA-dependent protein kinase) |
| 177.m02995 | 03440 | None | −0.785 | −1.034 | Unknown (Telomerase elongation inhibitor/RNA maturation protein PINX1) |
| 181.m08838 | 00052 | None | 1.039 | 1.186 | Unknown |
| 163.m06568 | 00885 | None | 1.667 | 1.389 | Unknown (Histones H3 and H4) |
Fold increase (ago1Δ/WT or rdp1Δ/WT) is log2 scale.
ScID refers to the gene name in the SGD (Saccharomyces Genome Database)
In conclusion, the present study aimed to identify and functionally characterize the components of the RNAi machinery, including Ago1/2, Rdp1, and Dcr1/2, in C. neoformans by analyses of gene-deletion mutants and their transcriptome profiles. Ago1, Rdp1, and Dcr2 were found to play a major role in the RNAi process in C. neoformans and yet were not involved in cell viability at a range of temperatures, nutritional conditions, and stress conditions, production of virulence factors such as capsule and melanin, and mating. Furthermore, only modest changes were observed in the genome-wide transcriptome patterns of C. neoformans when the RNAi process was perturbed. The only notable difference was an increase in transcript-abundance of several endogenous transposons such as T2 and T3 (newly identified in this study) in the rdp1Δ mutant of serotype D C. neoformans. Although the T2 and T3 transposons were both active in the wild type background, their mobility was further enhanced when Rdp1 is missing. Moreover, whereas the Cnl1 retrotransposons do not appear to be mobile in the wild-type background, the impairment of the RNAi pathway results in a dramatic increase in the number of transposition events. Therefore, it is clear that the RNAi machinery plays very important roles in limiting transposon- and retrotransposon-activity, thus maintaining genome integrity. In future studies, the small number of RNAi-dependent genes uniquely found in C. neoformans through DNA microarray analysis needs to be further characterized to explore the as yet unknown physiological roles of the RNAi machinery.
Supplementary Material
Acknowledgments
We thank Tamara Doering, Jenny Lodge, and Christina Hull for coordinating the community microarray consortium. We thank Tamara Doering for providing us the pRNAi plasmid. This work was supported by the Korea Research Foundation grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2007-331-C00223) and in part by the National Research Foundation of Korea (NRF) grants funded by the Korea government (MEST) (No. 2009-0063344)(to Y.S.B). This work was supported in part by the ANR (Erapathogenomic program) to G.J. This work was also supported in part by RO1 grant AI39115 from the NIH/NIAID (to J.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 1997;11(23):3206–17. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Greene Publishing Associates and John Wiley & Sons; New York, N.Y.: 1994. [Google Scholar]
- Bahn YS, Hicks JK, Giles SS, Cox GM, Heitman J. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot Cell. 2004;3(6):1476–91. doi: 10.1128/EC.3.6.1476-1491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell. 2005;16(5):2285–300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Carmichael JB, Provost P, Ekwall K, Hobman TC. ago1 and dcr1, two core components of the RNA interference pathway, functionally diverge from rdp1 in regulating cell cycle events in Schizosaccharomyces pombe. Mol Biol Cell. 2004a;15(3):1425–35. doi: 10.1091/mbc.E03-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael JB, Provost P, Ekwall K, Hobman TC. Ago1 and Dcr1, two core components of the RNA interference pathway, functionally diverge from Rdp1 in regulating cell cycle events in Schizosaccharomyces pombe. Mol Biol Cell. 2004b;15(3):1425–35. doi: 10.1091/mbc.E03-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Cogoni C, Macino G. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 2004;32(14):4237–43. doi: 10.1093/nar/gkh764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999a;399(6732):166–9. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999b;286(5448):2342–4. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D'Souza C, Wang P, et al. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148(Pt 8):2607–15. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- DeMarco R, Venancio TM, Verjovski-Almeida S. SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily. BMC Evol Biol. 2006;6:89. doi: 10.1186/1471-2148-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326(5952):544–50. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M, Lee DW, Kothe GO, Pratt RJ, Aramayo R, Selker EU. DNA methylation is independent of RNA interference in Neurospora. Science. 2004;304(5679):1939. doi: 10.1126/science.1099709. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Butler MI, Poulter RT. Cryptons: a group of tyrosine-recombinase-encoding DNA transposons from pathogenic fungi. Microbiology. 2003;149(Pt 11):3099–109. doi: 10.1099/mic.0.26529-0. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. The diversity of retrotransposons in the yeast Cryptococcus neoformans. Yeast. 2001;18(9):865–80. doi: 10.1002/yea.733. [DOI] [PubMed] [Google Scholar]
- Granger DL, Perfect JR, Durack DT. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Invest. 1985;76(2):508–16. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hicks JK, D'Souza CA, Cox GM, Heitman J. Cyclic AMP-dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2004;3(1):14–26. doi: 10.1128/EC.3.1.14-26.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, Heitman J. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol. 2005;3(10):753–64. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- Janbon G, Himmelreich U, Moyrand F, Improvisi L, Dromer F. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol Microbiol. 2001;42(2):453–67. doi: 10.1046/j.1365-2958.2001.02651.x. [DOI] [PubMed] [Google Scholar]
- Khan ZU, Ahmad S, Brazda A, Chandy R. Mucor circinelloides as a cause of invasive maxillofacial zygomycosis: an emerging dimorphic pathogen with reduced susceptibility to posaconazole. J Clin Microbiol. 2009;47(4):1244–8. doi: 10.1128/JCM.02030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13(23):3070–80. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Ko YJ, Maeng S, Floyd A, Heitman J, Bahn YS. Comparative transcriptome analysis of the CO2 sensing pathway via differential expression of carbonic anhydrase in Cryptococcus neoformans. Genetics. 2010 doi: 10.1534/genetics.110.118315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko YJ, Yu YM, Kim GB, Lee GW, Maeng PJ, Kim S, Floyd A, et al. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell. 2009;8(8):1197–217. doi: 10.1128/EC.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12(5):2331–8. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459(7244):274–7. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- Liu H, Cottrell TR, Pierini LM, Goldman WE, Doering TL. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics. 2002;160(2):463–70. doi: 10.1093/genetics/160.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307(5713):1321–4. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng S, Ko YJ, Kim GB, Jung KW, Floyd A, Heitman J, Bahn YS. Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans. Eukaryot Cell. 2010;9(3):360–78. doi: 10.1128/EC.00309-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyrand F, Chang YC, Himmelreich U, Kwon-Chung KJ, Janbon G. Cas3p belongs to a seven-member family of capsule structure designer proteins. Eukaryot Cell. 2004;3(6):1513–24. doi: 10.1128/EC.3.6.1513-1524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyrand F, Fontaine T, Janbon G. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol Microbiol. 2007;64(3):771–81. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H. RNA silencing in fungi: mechanisms and applications. FEBS Lett. 2005;579(26):5950–7. doi: 10.1016/j.febslet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H, Kadotani N, Mayama S. Evolution and diversification of RNA silencing proteins in fungi. J Mol Evol. 2006;63(1):127–35. doi: 10.1007/s00239-005-0257-2. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H, Nguyen QB. RNA interference: roles in fungal biology. Curr Opin Microbiol. 2008;11(6):494–502. doi: 10.1016/j.mib.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Nicolas FE, de Haro JP, Torres-Martinez S, Ruiz-Vazquez RM. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet Biol. 2007;44(6):504–16. doi: 10.1016/j.fgb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Nolan T, Cecere G, Mancone C, Alonzi T, Tripodi M, Catalanotto C, Cogoni C. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 2008;36(2):532–8. doi: 10.1093/nar/gkm1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36(11):1174–80. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol. 2008;28(21):6594–608. doi: 10.1128/MCB.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6(22):3343–53. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Tschudi C, Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol Cell Biol. 2004;24(1):420–7. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107(7):905–16. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Sigova A, Rhind N, Zamore PD. A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004;18(19):2359–67. doi: 10.1101/gad.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447(7145):735–8. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- Stoica C, Carmichael JB, Parker H, Pare J, Hobman TC. Interactions between the RNA interference effector protein Ago1 and 14-3-3 proteins: consequences for cell cycle progression. J Biol Chem. 2006;281(49):37646–51. doi: 10.1074/jbc.M604476200. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wickes BL, Edman JC. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol Microbiol. 1995;16(6):1099–109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.