Abstract
Background: Dietary magnesium intake has been favorably associated with reduced risk of metabolic outcomes in observational studies; however, few randomized trials have introduced a systems-biology approach to explore molecular mechanisms of pleiotropic metabolic actions of magnesium supplementation.
Objective: We examined the effects of oral magnesium supplementation on metabolic biomarkers and global genomic and proteomic profiling in overweight individuals.
Design: We undertook this randomized, crossover, pilot trial in 14 healthy, overweight volunteers [body mass index (in kg/m2) ≥25] who were randomly assigned to receive magnesium citrate (500 mg elemental Mg/d) or a placebo for 4 wk with a 1-mo washout period. Fasting blood and urine specimens were collected according to standardized protocols. Biochemical assays were conducted on blood specimens. RNA was extracted and subsequently hybridized with the Human Gene ST 1.0 array (Affymetrix, Santa Clara, CA). Urine proteomic profiling was analyzed with the CM10 ProteinChip array (Bio-Rad Laboratories, Hercules, CA).
Results: We observed that magnesium treatment significantly decreased fasting C-peptide concentrations (change: −0.4 ng/mL after magnesium treatment compared with +0.05 ng/mL after placebo treatment; P = 0.004) and appeared to decrease fasting insulin concentrations (change: −2.2 μU/mL after magnesium treatment compared with 0.0 μU/mL after placebo treatment; P = 0.25). No consistent patterns were observed across inflammatory biomarkers. Gene expression profiling revealed up-regulation of 24 genes and down-regulation of 36 genes including genes related to metabolic and inflammatory pathways such as C1q and tumor necrosis factor–related protein 9 (C1QTNF9) and pro-platelet basic protein (PPBP). Urine proteomic profiling showed significant differences in the expression amounts of several peptides and proteins after treatment.
Conclusion: Magnesium supplementation for 4 wk in overweight individuals led to distinct changes in gene expression and proteomic profiling consistent with favorable effects on several metabolic pathways. This trial was registered at clinicaltrials.gov as NCT00737815.
INTRODUCTION
Magnesium is an essential mineral in whole grains, leafy green vegetables, legumes, and nuts that acts as a cofactor in hundreds of enzymatic reactions in the human body. A considerable body of evidence indicates that a higher intake of dietary magnesium may favorably affect a cluster of metabolic and inflammatory disorders including insulin resistance (1), hypertension (2), dyslipidemia (3), type 2 diabetes (4), metabolic syndrome (5), and cardiovascular disease (6). Inverse cross-sectional and prospective associations with intermediate metabolic biomarkers for these disorders, including triglycerides (3), low HDL cholesterol (3), fasting insulin (4), and markers of inflammation and endothelial dysfunction (7, 8), have also been reported in observational settings. Experimentally, a diet low in magnesium led to impaired insulin secretion and glucose uptake (9, 10) as well as acute inflammation (11) in animal models, and in vitro studies suggested that the balance of extra- and intracellular Mg2+ in pancreatic β cells may be important in the regulation of insulin secretion (12). However, the underlying molecular mechanisms for the observed metabolic effects of magnesium remain unclear.
Because of limited and conflicting data from randomized clinical trials (13–17), whether magnesium supplementation can be effective in improving metabolic and inflammatory profiles in apparently healthy individuals at risk of metabolic abnormalities remains uncertain. Moreover, none of the few randomized studies have applied a systems-biology approach to examine the mechanistic effects of magnesium supplementation on gene expression and protein profiling. Therefore, to comprehensively investigate potential biological effects of oral magnesium supplementation in relation to metabolic biomarkers and global genomic and proteomic profiles, we conducted a randomized, controlled, crossover trial in apparently healthy, overweight individuals.
SUBJECTS AND METHODS
Study population
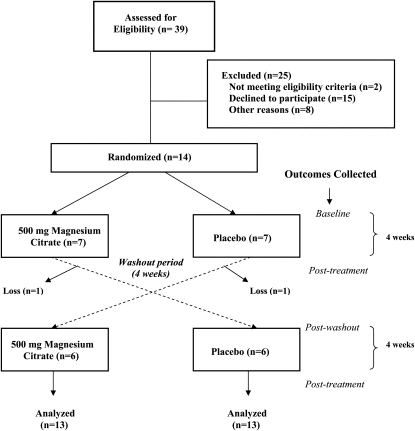
A total of 39 overweight volunteers were screened for eligibility. Inclusion criteria included being aged 30–70 y and having a body mass index (BMI; in kg/m2) ≥25, general good health, mobility, and no dietary restrictions or allergies. After an initial screening and run-in period, 14 participants were randomly assigned to the study treatments (Figure 1). Two participants did not complete the crossover portion of the study because of scheduling conflicts. All participants were asked to maintain their usual diet and not make any significant changes in diet or physical activity over the course of the study. All participants provided written informed consent to participate in the study. The Office for the Protection of Research Subjects at the University of California, Los Angeles (UCLA), Institutional Review Board approved the study protocol and all study procedures.
FIGURE 1.
Flow chart of magnesium-trial enrollment and design.
Study procedures
We used a randomized crossover design to compare 4 wk oral magnesium supplementation (500 mg elemental Mg in the form of magnesium citrate) with 4 wk placebo intake (inactive pills that were identical in appearance to the magnesium pills). The crossover component was added 2 mo into the study to enhance the study efficiency. A 4-wk washout period was included between treatments. Randomization was carried out by using a computer-generated table of random numbers, and participants were randomly assigned to one of 2 treatment sequences. Treatment capsules were prepared and dispensed by independent pharmacists at the UCLA pharmacy according to the computer-generated randomization list. Participants were instructed to take one capsule twice daily that contained either magnesium (total daily dose: 500 mg elemental Mg) or the placebo for 4 wk. Treatment capsules were identical in appearance and were prepackaged in bottles by the pharmacists. Study participants, investigators, and nursing staff were blinded; only the independent pharmacists and study coordinator were aware of the treatment assignment. Study enrollment and follow-up were conducted between June 2007 and March 2009.
At all 4 visits (ie, baseline, postwashout, and 2 posttreatments), participants underwent a clinical examination including a fasting blood collection. A fasting urine collection for proteomics analysis was collected at each posttreatment visit. At the initial screening visit, standing height was measured with a wall-mounted stadiometer (Holtain Ltd, Pembrokeshire, Wales, United Kingdom). Weights (in kg) of participants were measured in hospital gowns with an electronic scale (Scale-Tronix Inc, White Plains, NY). BMI was calculated as the weight (in kg) divided by height (in m2). Participants were instructed to fast for 12 h before visits, refrain from smoking for 1 h before the visit, and perform no vigorous physical activity for ≥12 h before the visit. All study procedures were carried out by trained nursing personnel at the UCLA General Clinical Research Center.
Outcome measures
Primary outcomes for this trial were plasma concentrations of metabolic and inflammatory biomarkers. Secondary outcomes were exploratory in nature and included gene and protein expression profiles. Two changes in study outcomes were made after the trial commencement: 1) the oral-glucose-tolerance test was discontinued because of the high participant time commitment and 2) urine protein measurements were added to enhance the comprehensive biological nature of the study.
Biochemical assays
Fasting blood samples were collected by venipuncture at all visits according to a standardized protocol. Insulin and C-peptide concentrations were measured on an immunoassay analyzer (Immulite 2000; Siemens Healthcare Diagnostics, Deerfield, IL) by using chemiluminescent immunoassay technology. Parathyroid hormone concentrations were measured by using electrochemiluminescence technology (Elecsys 2010; Roche Diagnostics, Indianapolis, IN). Glycated hemoglobin concentrations were measured by ion exchange HPLC (Variant II; Bio-Rad Laboratories, Hercules, CA). Glucose, magnesium, calcium, and triglyceride concentrations were measured with an Olympus AU5400 automated chemistry analyzer (Beckman Coulter Inc, Miami, FL). Glucose concentrations were measured by using the hexokinase glucose-6-phosphate dehydrogenase method; magnesium concentrations were measured by using the direct xylidyl blue complex method; calcium concentrations were measured by using the Arsenazo III endpoint method; and triglyceride concentrations were measured with a series of coupled enzymatic reactions by using lipase, glycerol kinase, and glycerol oxidase. High-sensitivity C-reactive protein (hs-CRP), interleukin-6 (IL-6), tumor necrosis factor-α receptor 2 (TNF-α), soluble intercellular adhesion molecule-1 (sICAM-1), soluble vascular cell adhesion molecule-1 (sVCAM-1), E-selectin, and leptin concentrations were measured with enzyme-linked immunoabsorbent assays (hs-CRP: ALPCO, Salem, NH; IL-6, sICAM-1, sVCAM-1, E-selectin, and leptin: R&D Systems, Minneapolis, MN; TNF-α: Invitrogen Corp, Carlsbad, CA).
Interassay CVs for each analyte on the basis of quality-control materials that covered a range of concentrations were 3.4–5.3% for insulin, 5.0% for C-peptide, 0.9–2.8% for glycated hemoglobin, 2.1–5.4% for parathyroid hormone, 1.2–1.8% for glucose, 1.2–2.1% for triglycerides, 2.2–4.2% for magnesium, 1.0–2.4% for calcium, 3.5–4.5% for leptin, 11.6–13.8% for hs-CRP, 6.5–9.6% for IL-6, 8.2–9.7% for TNF-α, 4.4–6.8% for sICAM-1, 5.5–7.8% for sVCAM-1, and 7.3–8.7% for E-selectin.
RNA extraction and microarray hybridization
The Paxgene blood RNA system (PreAnalytix; Qiagen/BD, Franklin Lakes, NJ) was used to collect 2.5 mL whole blood for RNA extraction and stabilization and was stored at −20°C for future RNA extraction. RNA isolation and purification were performed according to the manufacturer's instructions. RNA concentrations and the ratio of A260 to A280 were measured with the use of a NanoDrop-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE); the acceptable ratio of A260 to A280 was 1.9–2.1. All hybridizations were performed with the Human Gene ST 1.0 array chip (Affymetrix, Santa Clara, CA). Briefly, 0.2 μg total RNA/μL was used to synthesize double-stranded complementary DNA with the Superscript Choice System (Life Technologies, Carlsbad, CA). Real-time quantitative polymerase chain reaction was used to validate the results obtained from oligonucleotide microarrays for 2 genes, ion channel transient receptor potential membrane melastatin 6 (TRPM6) and 7 (TRPM7), which may play an essential role in intestinal and renal magnesium absorption. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference gene with the Human GAPD (GAPDH) Endogenous Control assay (Applied Biosystems, Carlsbad, CA).
Urine surface-enhanced laser desorption/ionization time-of-flight mass spectrometry
Participants were asked to collect 60 mL of their first urine void of the day in a sterile plastic container the morning of each posttreatment visit. Samples were kept cool until stored at −80°C for future proteomic analyses. Urine samples were thawed on ice and filtered through a 1-μm membrane before dilution into binding buffer for magnetic-bead solid-phase extraction [either C18 reversed phase (10% acetonitrile + 0.1% trifluoroacetic acid) or weak cation exchange (0.2 mol ammonium acetate (pH 4) + 0.01% Triton X-100]. Beads were washed to remove nonspecifically bound proteins and extracted with solvent. Extracts were mixed with matrix-assisted laser desorption/ionization matrix and spotted on 96-well plate targets for high-resolution time-of-flight mass measurement (prOTOF2000; Perkin-Elmer, Waltham MA). Another aliquot of filtered patient urine was diluted and applied to surface-enhanced laser desorption CM10 ProteinChip arrays (Bio-Rad Laboratories; weak cation exchange surface). The arrays were read in a PBS-IIc linear mass spectrometer (Ciphergen Biosystems, Fremont CA).
Statistical analyses
Baseline characteristics of participants were reviewed. We log-transformed all skewed variables and calculated geometric means. For all other continuous variables, arithmetic means were presented. The Mann-Whitney-Wilcoxon's exact test and Pearson's chi-square test were used to calculate P values for baseline differences between the magnesium and placebo treatments. We compared differences in changes in biochemical markers between magnesium and placebo treatments by using generalized linear models (with Proc Mixed in SAS software, version 9.2; SAS Institute, Cary, NC) and the Wilcoxon's signed rank sum test. All statistical analyses were conducted with SAS software (version 9.2; SAS Institute).
Microarray data analyses were performed with the Partek Genomics Suite version 6.5 software (Partek Genomics, St Louis, MO). Post-treatment measurements of RNA were compared for differential expression. Affymetrix CEL files (Affymetrix) were imported into the Partek Genomics Suite software (Partek Genomics) by using the default Partek normalization variables. Probe-level data were preprocessed, including background correction, normalization, and summarization, by using robust multiarray average analysis adjusted for probe sequence and guanine cytosine (GC) content (GC robust multiarray average). Subsequent data normalization was performed across all arrays by using quantile normalization. The background-adjusted, normalized perfect match (PM) values were compiled and summarized (within each probe set by using the median polish technique) to generate a single measure of expression. A list of differentially expressed genes across the magnesium and placebo treatments was generated by using analysis of variance (ANOVA). For comparison analyses, thresholds for selecting significant genes were set at a relative fold change >1.2-fold signal intensity, and a statistically statistical difference was set at P < 0.05. Cluster analysis was performed on the selected genes, and principal-component analysis was conducted to identify major effects that influenced the expression values in each treatment. ProteinChip Data Manager software (version 2.0; Bio-Rad Laboratories) was used for proteomics analysis to compile all spectra and automatically detect quantified mass peaks. ANOVA tests were conducted to test for significant differences in protein profiles between magnesium and placebo treatments. Receiver operating characteristic curves and the corresponding area under the curve were generated as a measure of discrimination to determine whether the set of protein peaks identified by proteomic profiling distinguished between magnesium and placebo treatments.
RESULTS
Baseline characteristics of study participants are summarized in Table 1. The mean age of study participants (71% men and 29% women) was 44 y. All participants were overweight or obese with a BMI ranging from 26 to 32. Forty-three percent of participants were classified as hypomagnesemic (defined as serum Mg concentration <1.6 mEq/L) at baseline. After the washout period, the same proportion of participants exhibited hypomagnesaemia, and mean serum magnesium concentrations returned to baseline values (data not shown), which suggested that the washout period was successful. Although we observed small differences in several of the biochemical measurements across the magnesium and placebo treatments at baseline, none of these differences were significant at the 0.05 level.
TABLE 1.
Characteristics of overweight participants enrolled in a randomized crossover trial of magnesium supplementation compared with placebo at baseline (n = 14)1
| All (n = 14) | Magnesium (n = 7) | Placebo (n = 7) | P | |
| Demographic characteristics | ||||
| Age (y) | 44.4 ± 13.02 | 47.0 ± 13.8 | 41.9 ± 12.7 | 0.38 |
| BMI (kg/m2) | 28.2 ± 1.8 | 28.3 ± 1.6 | 28.1 ± 2.2 | 0.98 |
| Sex (% male) | 71 | 57 | 86 | 0.11 |
| Metabolic biomarkers | ||||
| Insulin (μU/mL) | 7.5 ± 5.7 | 7.6 ± 6.6 | 7.3 ± 5.0 | 0.99 |
| C-peptide (ng/mL) | 1.9 ± 1.2 | 2.0 ± 1.4 | 1.8 ± 1.0 | 0.92 |
| Hb A1c (%) | 5.5 ± 0.5 | 5.3 ± 0.4 | 5.7 ± 0.5 | 0.07 |
| Parathyroid hormone (pg/mL) | 45.6 ± 11.8 | 43.6 ± 12.8 | 47.6 ± 11.3 | 0.51 |
| Fasting glucose (mg/dL) | 85.0 ± 7.2 | 87.4 ± 7.7 | 82.2 ± 6.0 | 0.24 |
| Serum magnesium (mEq/L) | 1.58 ± 0.12 | 1.60 ± 0.12 | 1.57 ± 0.12 | 0.57 |
| Calcium (mg/dL) | 8.2 (6.7, 9.9)3 | 9.2 (9.0, 9.3) | 7.3 (4.8, 11.1) | 0.32 |
| Leptin (pg/mL) | 9222 (6710, 12,675) | 11,844 (6795, 20,642) | 7442 (476, 11,358) | 0.31 |
| Triglycerides (mg/dL) | 108.6 (87.2, 135.2) | 104.1 (64.5, 168.0) | 113.3 (94.0, 136.7) | 0.42 |
| Inflammatory markers | ||||
| hs-CRP (mg/L) | 1.30 (0.84, 2.00) | 0.97 (0.49, 1.91) | 1.67 (0.87, 3.22) | 0.31 |
| IL-6 (pg/mL) | 1.63 (1.19, 2.24) | 1.23 (0.67, 2.28) | 2.07 (1.47, 2.92) | 0.18 |
| TNF-α (pg/mL) | 11.2 (9.4, 13.5) | 11.0 (7.1, 17.2) | 11.4 (10.0, 13.0) | 0.51 |
| sICAM-1 (ng/mL) | 136.6 (116.4, 160.3) | 130.8 (99.8, 171.3) | 141.8 (110.3, 182.2) | 0.73 |
| sVCAM-1 (ng/mL) | 434.4 (362.8, 520.0) | 485.5 (379.9, 620.5) | 394.8 (292.2, 533.5) | 0.45 |
| E-selectin (ng/mL) | 34.1 (29.1, 40.1) | 30.7 (24.0, 39.4) | 37.3 (29.1, 47.8) | 0.45 |
Hb A1c, glycated hemoglobin; hs-CRP, high-sensitivity C-reactive protein; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α; sICAM-1, soluble intercellular adhesion molecule-1; sVCAM-1, soluble vascular cell adhesion molecule-1. P values for differences between magnesium and placebo treatments were calculated by using Mann-Whitney-Wilcoxon's exact test (continuous) and Pearson's chi-square test (categorical).
Mean ± SE (all such values).
Geometric mean; 95% CI in parentheses (all such values).
Metabolic and inflammatory biomarkers
The effects of magnesium and placebo treatments on metabolic and inflammatory biomarkers are shown in Table 2. We observed improvements in several metabolic biomarkers after magnesium treatment including a decrease in C-peptide concentrations (change: −0.4 ng/mL after magnesium treatment compared with 0.05 ng/mL after placebo treatment; P = 0.004) and a nonsignificant decrease in fasting insulin concentrations (change: −2.2 μU/mL after magnesium treatment compared with 0.0 μU/mL after placebo treatment; P = 0.25). There were no consistent patterns across markers of inflammation or endothelial dysfunction in response to magnesium supplementation, although we did observe a significant increase in IL-6 concentrations after magnesium treatment (0.23 pg/mL after magnesium treatment compared with −0.37 pg/mL after placebo treatment; P = 0.03).
TABLE 2.
Metabolic and inflammatory biomarkers before and after 4 wk of 500 mg Mg and placebo treatments in overweight and obese participants enrolled in a randomized crossover trial of magnesium supplementation1
| Magnesium |
Placebo |
||||||
| Baseline (n = 13) | Post (n = 13) | Change | Baseline (n = 13) | Post (n = 13) | Change | P | |
| Metabolic biomarkers | |||||||
| Insulin (μU/mL) | 6.9 ± 5.62 | 4.8 ± 3.7 | −2.2 | 7.4 ± 4.3 | 7.4 ± 3.7 | 0 | 0.25 |
| C-peptide (ng/mL) | 1.9 ± 1.2 | 1.5 ± 0.9 | −0.4 | 1.9 ± 0.9 | 2.0 ± 0.9 | 0.1 | 0.004 |
| Hb A1c (%) | 5.5 ± 0.5 | 5.5 ± 0.5 | 0.05 | 5.6 ± 0.5 | 5.4 ± 0.5 | −0.1 | 0.08 |
| Parathyroid hormone (pg/mL) | 47.5 ± 11.7 | 55.9 ± 16.1 | 8.5 | 47.1 ± 8.8 | 41.3 ± 10.5 | −5.8 | 0.04 |
| Fasting glucose (mg/dL) | 87.9 ± 11.7 | 86.7 ± 12.3 | −1.2 | 84.3 ± 8.7 | 88.4 ± 12.7 | 4.1 | 0.44 |
| Serum Mg (mEq/L) | 1.57 ± 0.12 | 1.67 ± 0.11 | 0.10 | 1.56 ± 0.10 | 1.59 ± 0.06 | 0.03 | 0.23 |
| Calcium (mg/dL) | 9.2 (9.1, 9.3)3 | 9.2 (9.1, 9.3) | 0.0 | 8.1 (6.6, 10.0) | 9.2 (9.1, 9.3) | 1.1 | 0.26 |
| Leptin (pg/mL) | 9226 (6702, 12,700) | 8212 (5076, 13,282) | −1014 | 8465 (5783, 12,390) | 8331 (5648, 12,289) | −134 | 0.58 |
| Triglycerides (mg/dL) | 120.3 (87.6, 165.3) | 124.3 (86.7, 178.3) | 4.0 | 102.8 (85.6, 123.5) | 112.9 (84.4, 151.0) | 10.1 | 0.49 |
| Inflammatory markers | |||||||
| hs-CRP (mg/L) | 0.98 (0.67, 1.43) | 1.33 (0.80, 2.20) | 0.35 | 1.37 (0.90, 2.10) | 1.04 (0.68, 1.58) | −0.33 | 0.50 |
| IL-6 (pg/mL) | 1.55 (1.05, 2.28) | 1.78 (1.19, 2.65) | 0.23 | 1.67 (1.23, 2.27) | 1.30 (0.94, 1.81) | −0.37 | 0.03 |
| TNF-α (pg/mL) | 10.8 (8.9, 13.2) | 10.6 (9.0, 12.6) | −0.2 | 12.4 (11.1, 13.9) | 10.0 (8.6, 11.7) | −2.4 | 0.21 |
| sICAM-1 (ng/mL) | 138 (115, 166) | 135 (115, 158) | −3.6 | 142 (123, 164) | 144 (123, 168) | 1.7 | 0.32 |
| sVCAM-1 (ng/mL) | 456 (388, 534) | 429 (369, 498) | −26.8 | 417 (354, 491) | 406 (350, 471) | −10.8 | 0.64 |
| E-selectin (ng/mL) | 34.1 (28.7, 40.4) | 34.1 (27.3, 41.0) | 0.0 | 38.2 (32.0, 45.7) | 36.0 (30.1, 42.1) | −2.2 | 0.15 |
Hb A1c, glycated hemoglobin; hs-CRP, high-sensitivity C-reactive protein; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α; sICAM-1, soluble intercellular adhesion molecule-1; sVCAM-1, soluble vascular cell adhesion molecule-1. P values for differences in changes between magnesium and placebo treatments were calculated by using Wilcoxon's signed rank sum test.
Mean ± SE (all such values).
Geometric mean; 95% CI in parentheses (all such values).
Differential gene expression
We detected a list of 58 differentially regulated genes after 4 wk of magnesium treatment compared with after 4 wk of placebo treatment by using a threshold set at a relative difference of 1.2-fold and statistical difference at P < 0.05 (Table 3). Of these genes, 36 genes were down-regulated, and 22 genes were up-regulated, after 4 wk of treatment. Several genes closely linked to metabolic and inflammatory pathways were down-regulated, including C1q and tumor necrosis factor related protein 9 (C1QTNF9), which is a gene that encodes a glycoprotein secreted by the adipose tissue that plays a role in insulin and glucose metabolism, and pro-platelet basic protein [chemokine (C-X-C) motif] ligand (PPBP), which is a platelet-derived growth factor that belongs to the CXC chemokine family involved in the activation of neutrophils.
TABLE 3.
Differentially expressed genes after 4 wk of magnesium treatment compared with placebo treatment (n = 14)1
| Probe-set identifier | Gene name | Gene symbol | Fold change | P | Function |
| Down-regulated genes | |||||
| 8027348 | Zinc finger protein 730 | ZNF730 | −1.23 | 0.005 | DNA binding; metal ion binding; zinc ion binding |
| 7995258 | Zinc finger protein 267 | ZNF267 | −1.24 | 0.041 | DNA binding; metal ion binding; zinc ion binding |
| 8165496 | Tubulin, beta 2C | TUBB2C | −1.21 | 0.042 | GTP binding; GTPase activity; MHC class I protein binding; nucleotide binding; unfolded protein binding |
| 8135625 | Capping protein (actin filament) muscle Z-line, α-2 | CAPZA2 | −1.22 | 0.022 | Protein binding; actin binding; protein tyrosine kinase activity; kinase binding; syntaxin binding; nitric oxide synthase binding |
| 8105111 | F-box protein 4 | FBXO4 | −1.21 | 0.001 | Protein binding; ubiquitin-protein ligase activity |
| 7962013 | ERGIC and golgi 2 | ERGIC2 | −1.20 | 0.046 | Protein binding |
| 7988605 | COP9 constitutive photomorphogenic homolog subunit 2 | COPS2 | −1.24 | 0.049 | Protein binding; signal transducer activity |
| 7954344 | Organic anion transporter LST-3b | LST-3TM12 | −1.26 | 0.018 | Transporter activity |
| 8100971 | Pro-platelet basic protein [chemokine (C-X-C motif) ligand 7] | PPBP | −1.27 | 0.033 | Glucose transmembrane transporter activity; chemokine activity; growth factor activity |
| 7948148 | Olfactory receptor, family 5, subfamily M, member 10 | OR5M10 | −1.23 | 0.011 | Receptor activity; olfactory receptor activity |
| 7908376 | Regulator of G-protein signaling 18 | RGS18 | −1.31 | 0.007 | Signal transducer activity; GTPase activator activity; regulator of G-protein signaling activity |
| 8127391 | Eyes shut homolog (Drosophila) | EYS | −1.28 | 7.73E-05 | Calcium ion binding |
| 7968052 | C1q and tumor necrosis factor–related protein 9 | C1QTNF9 | −1.22 | 0.021 | Insulin and glucose metabolism; hormone activity |
| 8159977 | Relaxin 2 | RLN2 | −1.23 | 0.008 | Hormone activity |
| 7947332 | IMP1 inner mitochondrial membrane peptidase-like | IMMP1L | −1.23 | 0.011 | Peptidase activity; serine-type peptidase activity |
| 8140864 | Cytochrome P450, family 51, subfamily A, polypeptide 1 | CYP51A1 | −1.22 | 0.021 | Electron carrier activity; heme binding; metal ion binding; monooxgenase activity; sterol 14-demethylase activity |
| 8160020 | Chromosome 9 open reading frame 38 | C9orf38 | −1.21 | 0.003 | — |
| 8151766 | — | — | −1.20 | 0.001 | — |
| 7898732 | — | — | −1.46 | 0.003 | — |
| 7951131 | — | — | −1.37 | 0.003 | — |
| 8174373 | — | — | −1.23 | 0.004 | — |
| 8147744 | — | — | −1.22 | 0.006 | — |
| 8119894 | — | — | −1.24 | 0.009 | — |
| 7969701 | — | — | −1.21 | 0.010 | — |
| 7944361 | — | — | −1.26 | 0.015 | — |
| 7955987 | — | — | −1.29 | 0.015 | — |
| 8172195 | — | — | −1.58 | 0.016 | — |
| 8067944 | — | — | −1.27 | 0.021 | — |
| 8079613 | — | — | −1.25 | 0.022 | — |
| 7963588 | — | — | −1.22 | 0.028 | — |
| 8102606 | — | — | −1.26 | 0.029 | — |
| 8042976 | — | — | −1.22 | 0.031 | — |
| 7910674 | — | — | −1.32 | 0.032 | — |
| 7929954 | — | — | −1.23 | 0.040 | — |
| 8045802 | — | — | −1.33 | 0.046 | — |
| 7973820 | — | — | −1.25 | 0.049 | — |
| Up-regulated genes | |||||
| 8026339 | Small nuclear ribonucleoprotein polypeptide G | SNRPG | 1.21 | 0.038 | RNA binding; protein binding |
| 8161774 | Transient receptor potential cation channel, subfamily M, member 6 | TRPM6 | 1.05 | 0.690 | Nucleotide binding; receptor activity; ion channel activity; calcium channel activity; calcium ion binding; ATP binding; zinc ion binding; kinase activity; transferase activity |
| 7988713 | Transient receptor potential cation channel, subfamily M, member 7 | TRPM7 | 1.07 | 0.338 | Nucleotide binding; actin binding; ion channel activity; calcium channel activity; calcium ion binding; protein binding; ATP binding; zinc ion binding; kinase activity; transferase activity; myosin binding |
| 7963406 | Keratin 6B | KRT6B | 1.24 | 0.003 | Structural constituent of cytoskeleton |
| 8088908 | Hypothetical LOC728060 | LOC728060 | 1.23 | 0.003 | — |
| 8040547 | Hypothetical LOC646049 | LOC646049 | 1.30 | 0.004 | — |
| 7984985 | Golgi autoantigen golgin subfamily a, 6 | GOLGA6 | 1.21 | 0.028 | — |
| 7984259 | RNA, U5B small nuclear 1 | RNU5B-1 | 1.26 | 0.045 | — |
| 8040336 | — | — | 1.21 | 0.004 | — |
| 7935056 | — | — | 1.23 | 0.008 | — |
| 7962916 | — | — | 1.21 | 0.011 | — |
| 8095299 | — | — | 1.61 | 0.012 | — |
| 7921031 | — | — | 1.26 | 0.016 | — |
| 8145770 | — | — | 1.21 | 0.018 | — |
| 8008965 | — | — | 1.24 | 0.020 | — |
| 7917468 | — | — | 1.31 | 0.022 | — |
| 7972016 | — | — | 1.42 | 0.022 | — |
| 7985039 | — | — | 1.29 | 0.024 | — |
| 7910377 | — | — | 1.29 | 0.024 | — |
| 8111358 | — | — | 1.23 | 0.030 | — |
| 7925790 | — | — | 1.20 | 0.030 | — |
| 8000930 | — | — | 1.24 | 0.037 | — |
| 8016980 | — | — | 1.21 | 0.038 | — |
| 8103847 | — | — | 1.20 | 0.050 | — |
MHC, major histocompatibility complex; —, gene name or function is unknown. The list of differentially expressed genes was generated by using ANOVA.
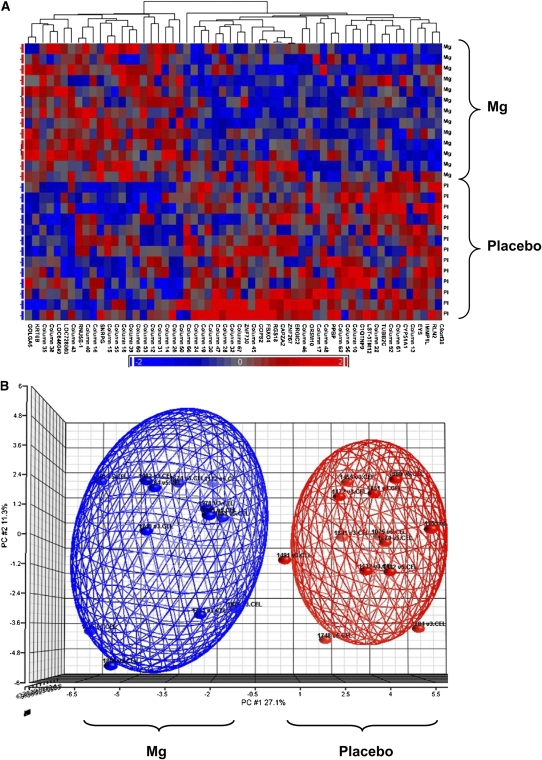
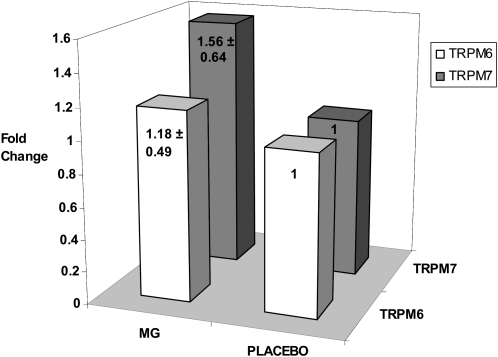
Analyses of clusters and principal components identified 2 distinct expression patterns for this set of 58 genes in response to magnesium treatment compared with in response to placebo treatment, which suggested that individuals within each treatment exhibited similar global expression profiles (Figure 2). On the basis of preliminary differences identified in the microarray analyses and our prior work (18) that suggested a possible interaction between dietary magnesium intake and TRPM6 and TRPM7 genes, we further examined the expression amounts of TRPM6 and TRPM7 by using a quantitative polymerase chain reaction and showed that both genes were up-regulated after magnesium treatment compared with after placebo treatment (average: ≈1.4 fold differences) (Figure 3).
FIGURE 2.
A: Gene expression patterns for 58 genes differentially regulated after magnesium (n = 13) and placebo (n = 13) treatments with the Affymetrix microarray (Affymetrix, Santa Clara, CA). Each row represents one study participant, and differentially expressed genes are shown in columns. B: Principal components (PC) analysis, which revealed similar gene expression profiles within each treatment of 58 differentially expressed genes. Cluster and PC analyses were performed on the list of differentially expressed genes generated by using ANOVA.
FIGURE 3.
Real-time polymerase chain reaction confirmation of microarray results for TRPM6 and TRPM7 (n = 9). Mean (±SD) fold changes were calculated by comparing the differences in expression across magnesium (MG) and placebo treatments.
Differential protein expression
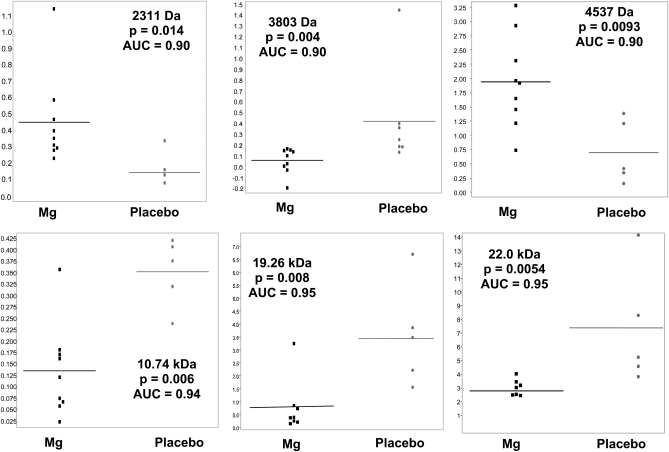
Findings from surface-enhanced laser desorption/ionization time-of-flight mass spectrometry analyses suggest clear differences in the protein expression profiles of urine collected from participants after 4 wk of magnesium supplementation compared with after 4 wk of placebo treatment. As shown in Figure 4, proteins of low molecular weights that ranged from 2311 Da to 22.0 kDa were expressed at significantly different relative intensities after 4 wk of magnesium treatment compared with after 4 wk of placebo treatment. A visible separation in the relative expression intensity of proteins of similar molecular weights was seen across treatments, which indicated that the discrimination between urine protein profiles across magnesium and placebo treatments was high. The area under the receiver operating characteristic curve ranged from 0.90 to 0.95 and provided further indication that unique proteomic signatures differentiated between magnesium and placebo treatments.
FIGURE 4.
Surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry protein expression profiles from fasting urine samples collected after 4 wk of magnesium supplementation (n = 7–9) compared with after 4 wk of placebo treatment (n = 4–7). Values shown are the relative intensity of expression of proteins at varying molecular weights. P values were calculated by using one-factor ANOVA. AUC, area under the receiver operating characteristic curve.
DISCUSSION
In this randomized crossover trial in overweight individuals, magnesium supplementation for 4 wk significantly decreased fasting concentrations of C-peptide and appeared to decrease fasting insulin concentrations. We also observed the down-regulation of genes related to metabolic and inflammatory pathways including C1QTNF9 and PPBP. Urine proteomic profiling showed a number of peptides and proteins significantly differentially expressed in response to magnesium treatment.
These findings lend support to the hypothesis that dietary magnesium plays a beneficial role in the regulation of insulin and glucose homeostasis. Some (13–15) but not all (16, 17) randomized trials indicated that magnesium supplementation can improve dyslipidemia and lower fasting insulin and glucose concentrations. A meta-analysis of 9 randomized trials in diabetes patients reported that 4–16 wk of magnesium supplementation was effective in reducing fasting glucose concentrations and raising HDL cholesterol concentrations but reported no effects on total cholesterol, LDL cholesterol, or triglyceride concentrations (19). In the current trial, C-peptide concentrations decreased significantly after magnesium treatment, which suggested a reduction in pancreatic insulin secretion that may have resulted from an improvement in insulin sensitivity and a subsequent lowered demand on the pancreas. We also observed a biologically consistent, although nonsignificant, decrease in fasting insulin concentrations after magnesium treatment. Decreased intracellular Mg2+ concentrations have been associated with impairment in insulin action and glucose uptake in insulin-sensitive tissues such as skeletal muscle tissue (20), heart muscles (21), and adipocytes (22), and several metabolic studies suggest that magnesium supplementation could improve insulin-induced glucose uptake (13, 14). In vitro studies also suggested that the balance of extra- and intracellular Mg2+ in pancreatic β cells may be important in regulating the secretion of insulin directly (12, 23).
In contrast to observational studies that linked higher dietary magnesium intakes to lower concentrations of biomarkers of systemic inflammation (8, 24), we observed a significant increase in IL-6 concentrations after magnesium treatment. IL-6 is a proinflammatory cytokine secreted by macrophages and T cells and is the major mediator of the acute-phase response (25). Because elevated IL-6 concentrations may provide an early indicator of acute inflammation, the observed increase in IL-6 concentrations may reflect a variation because of unmeasured underlying acute infection disproportionately present in the magnesium treatment by chance, although we do not have recorded measurements of transitory illness to confirm this explanation. Very few randomized trials have examined the effects of magnesium supplementation on systemic inflammation in overweight individuals (24), and our findings suggest a need for further investigation in larger trials.
Changes in the expression of several genes consistent with metabolic and inflammatory pathways were detected. C1QTNF9, a gene that may play a role in insulin and glucose metabolism, was down-regulated in the magnesium treatment relative to in the placebo treatment. The protein encoded by the C1QTNF9 gene is expressed in the adipose tissue and is a paralog of adiponectin (26), a protein hormone with insulin sensitizing and antiinflammatory properties. PPBP, a platelet derived growth factor in the CXC chemokine family and a potent chemoattractant and activator of neutrophils (27), was also down-regulated after magnesium treatment. PPBP mRNA has also been shown to be down-regulated by glucocorticoids in monocytes and platelets (27). Thus, further investigation into the mechanism of action of these genes in response to magnesium supplementation is warranted.
TRPM6 and TRPM7, 2 members of the “transient receptor potential” family of cation channels, play an essential role in magnesium homeostasis (28–30) and may be important for glucose and insulin homeostasis. Coding-region variants recently identified in the TRPM6 gene may interact with dietary magnesium intake in determining the risk of type 2 diabetes (18). Based on this prior work, we investigated the effects of magnesium supplementation on TRPM6 and TRPM7 genes and showed that both were differentially up-regulated in the magnesium treatment compared with in the placebo treatment at an average 1.4-fold change, which supported the theory that these genes may interact with dietary magnesium intake and ultimately affect metabolic functioning.
Although a number of the other genes identified as differentially expressed in this trial are unknown, our exploratory findings indicated a systemic effect of magnesium supplementation at the level of gene expression. This is consistent with our findings that showed a distinct protein profile in urine collected after treatment with magnesium compared with after treatment with the placebo. In healthy individuals, 70% of the urinary proteome originates from the kidney and urinary tract, and 30% represents plasma proteins filtered by the glomerulus (31). We conducted this exploratory analysis of the urinary proteome to investigate the way magnesium might affect the expression of systemic and renal proteins related to metabolic and inflammatory disease. Our findings were suggestive of measurable physiologic changes in the urinary proteome after treatment with magnesium for 4 wk, which warrants further investigation into these changes and identification of the proteins involved.
Several limitations need to be kept in mind when interpreting findings of this trial. First, the small size and short time frame limited our power in detecting changes because of the magnesium treatment. Second, we made an explicit assumption that a washout period of 4 wk was adequate in clearing the system of magnesium, which was a reasonable assumption because of the moderately low dose of magnesium administered in the study. Third, magnesium citrate was administered in our study because of its superior bioavailability over other formulations (32), but the inclusion of citrate may have led to systemic changes in the acid-base balance that may have been partially responsible for the differential regulation of the organic anion transporter LST-3b as well as changes in urine protein profiles. Our microarray and proteomics analyses were exploratory in nature and limited with respect to the specificity of pathways and proteins identified; however, our findings provide evidence to support the effects of short-term magnesium supplementation on global gene expression and proteomic profiling consistent with metabolic pathways. Finally, an inherent limitation of our systems biology approach combined with the small size of the trial was that not all potentially important findings reached statistical significance at the conventional α = 0.05 level; however, the scientific value in stimulating further research in this area is nonetheless present.
In conclusion, findings from this randomized crossover trial indicated that magnesium supplementation for 4 wk may improve insulin and glucose homeostasis in overweight or obese individuals. Systemic changes in gene and protein expression also provided further leads that should be investigated in future studies of large populations.
Acknowledgments
We express appreciation to all study participants for their enthusiastic commitment to this study. We also express gratitude to the staff at the UCLA General Clinical Research Center for their support with conducting this study and Najib Aziz for his assistance with laboratory procedures for the biochemical assays.
The authors' responsibilities were as follows—SL: designed the research; SAC, JS, and YY: collected the data; XL, JL, YY, and AB: provided essential reagents and materials; SAC, XL, JL, and YY: analyzed the data; SAC, YS, and SL: wrote the manuscript; and all authors: read and approved the final manuscript. General Mills played no role in the design, implementation, analysis, or interpretation of the data. None of the authors had a conflict of interest.
REFERENCES
- 1.Paolisso G, Barbagallo M. Hypertension, diabetes mellitus, and insulin resistance: the role of intracellular magnesium. Am J Hypertens 1997;10:346–55 [DOI] [PubMed] [Google Scholar]
- 2.Song Y, Sesso HD, Manson JE, Cook NR, Buring JE, Liu S. Dietary magnesium intake and risk of incident hypertension among middle-aged and older US women in a 10-year follow-up study. Am J Cardiol 2006;98:1616–21 [DOI] [PubMed] [Google Scholar]
- 3.Song Y, Ridker PM, Manson JE, Cook NR, Buring JE, Liu S. Magnesium intake, C-reactive protein, and the prevalence of metabolic syndrome in middle-aged and older U.S. women. Diabetes Care 2005;28:1438–44 [DOI] [PubMed] [Google Scholar]
- 4.Song Y, Manson JE, Buring JE, Liu S. Dietary magnesium intake in relation to plasma insulin levels and risk of type 2 diabetes in women. Diabetes Care 2004;27:59–65 [DOI] [PubMed] [Google Scholar]
- 5.He K, Liu K, Daviglus ML, et al. Magnesium intake and incidence of metabolic syndrome among young adults. Circulation 2006;113:1675–82 [DOI] [PubMed] [Google Scholar]
- 6.Al-Delaimy WK, Rimm EB, Willett WC, Stampfer MJ, Hu FB. Magnesium intake and risk of coronary heart disease among men. J Am Coll Nutr 2004;23:63–70 [DOI] [PubMed] [Google Scholar]
- 7.Chacko SA, Song Y, Nathan L, et al. Relations of dietary magnesium intake to biomarkers of inflammation and endothelial dysfunction in an ethnically diverse cohort of postmenopausal women. Diabetes Care 2010;33:304–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Y, Li TY, van Dam RM, Manson JE, Hu FB. Magnesium intake and plasma concentrations of markers of systemic inflammation and endothelial dysfunction in women. Am J Clin Nutr 2007;85:1068–74 [DOI] [PubMed] [Google Scholar]
- 9.Gueux E, Rayssiguier Y. The effect of magnesium deficiency on glucose stimulated insulin secretion in rats. Horm Metab Res 1983;15:594–7 [DOI] [PubMed] [Google Scholar]
- 10.Matsunobu S, Terashima Y, Senshu T, Sano H, Itoh H. Insulin secretion and glucose uptake in hypomagnesemic sheep fed a low magnesium, high potassium diet. J Nutr Biochem 1990;1:167–71 [DOI] [PubMed] [Google Scholar]
- 11.Malpuech-Brugere C, Nowacki W, Daveau M, et al. Inflammatory response following acute magnesium deficiency in the rat. Biochim Biophys Acta 2000;1501:91–8 [DOI] [PubMed] [Google Scholar]
- 12.Ishizuka J, Bold RJ, Townsend CM, Jr, Thompson JC. In vitro relationship between magnesium and insulin secretion. Magnes Res 1994;7:17–22 [PubMed] [Google Scholar]
- 13.Guerrero-Romero F, Tamez-Perez HE, Gonzalez-Gonzalez G, et al. Oral magnesium supplementation improves insulin sensitivity in non-diabetic subjects with insulin resistance. A double-blind placebo-controlled randomized trial. Diabetes Metab 2004;30:253–8 [DOI] [PubMed] [Google Scholar]
- 14.Paolisso G, Sgambato S, Gambardella A, et al. Daily magnesium supplements improve glucose handling in elderly subjects. Am J Clin Nutr 1992;55:1161–7 [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen HS, Aurup P, Goldstein K, et al. Influence of magnesium substitution therapy on blood lipid composition in patients with ischemic heart disease. A double-blind, placebo controlled study. Arch Intern Med 1989;149:1050–3 [PubMed] [Google Scholar]
- 16.Purvis JR, Cummings DM, Landsman P, et al. Effect of oral magnesium supplementation on selected cardiovascular risk factors in non-insulin-dependent diabetics. Arch Fam Med 1994;3:503–8 [DOI] [PubMed] [Google Scholar]
- 17.Marken PA, Weart CW, Carson DS, Gums JG, Lopes-Virella MF. Effects of magnesium oxide on the lipid profile of healthy volunteers. Atherosclerosis 1989;77:37–42 [DOI] [PubMed] [Google Scholar]
- 18.Song Y, Hsu YH, Niu T, Manson JE, Buring JE, Liu S. Common genetic variants of the ion channel transient receptor potential membrane melastatin 6 and 7 (TRPM6 and TRPM7), magnesium intake, and risk of type 2 diabetes in women. BMC Med Genet 2009;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song Y, He K, Levitan EB, Manson JE, Liu S. Effects of oral magnesium supplementation on glycaemic control in Type 2 diabetes: a meta-analysis of randomized double-blind controlled trials. Diabet Med 2006;23:1050–6 [DOI] [PubMed] [Google Scholar]
- 20.Naitoh T, Kobayashi S, Kimura I, Kimura M. Intracellular Ca2+ and Mg2+ regulation for insulin-stimulated glucose uptake into mouse diaphragm muscles. Jpn J Pharmacol 1991;56:241–4 [DOI] [PubMed] [Google Scholar]
- 21.Paxton R, Ye L. Regulation of heart insulin receptor tyrosine kinase activity by magnesium and spermine. Mol Cell Biochem 2005;277:7–17 [DOI] [PubMed] [Google Scholar]
- 22.Nadler J, Scott S. Evidence that pioglitazone increases intracellular free magnesium concentration in freshly isolated rat adipocytes. Biochem Biophys Res Commun 1994;202:416–21 [DOI] [PubMed] [Google Scholar]
- 23.Murakami M, Ishizuka J, Sumi S, et al. Role of extracellular magnesium in insulin secretion from rat insulinoma cells. Proc Soc Exp Biol Med 1992;200:490–4 [DOI] [PubMed] [Google Scholar]
- 24.Almoznino-Sarafian D, Berman S, Mor A, et al. Magnesium and C-reactive protein in heart failure: an anti-inflammatory effect of magnesium administration? Eur J Nutr 2007;46:230–7 [DOI] [PubMed] [Google Scholar]
- 25.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med 1999;340:448–54 [DOI] [PubMed] [Google Scholar]
- 26.Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, et al. Identification and characterization of CTRP9, a novel secreted glycoprotein, from adipose tissue that reduces serum glucose in mice and forms heterotrimers with adiponectin. FASEB J 2009;23:241–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Gedaily A, Schoedon G, Schneemann M, Schaffner A. Constitutive and regulated expression of platelet basic protein in human monocytes. J Leukoc Biol 2004;75:495–503 [DOI] [PubMed] [Google Scholar]
- 28.Voets T, Nilius B, Hoefs S, et al. TRPM6 forms the Mg2+ influx channel involved in intestinal and renal Mg2+ absorption. J Biol Chem 2004;279:19–25 [DOI] [PubMed] [Google Scholar]
- 29.Montell C. The TRP superfamily of cation channels. Sci STKE 2005;2005:re3. [DOI] [PubMed] [Google Scholar]
- 30.Schlingmann KP, Weber S, Peters M, et al. Hypomagnesemia with secondary hypocalcemia is caused by mutations in TRPM6, a new member of the TRPM gene family. Nat Genet 2002;31:166–70 [DOI] [PubMed] [Google Scholar]
- 31.Thongboonkerd V, Malasit P. Renal and urinary proteomics: current applications and challenges. Proteomics 2005;5:1033–42 [DOI] [PubMed] [Google Scholar]
- 32.Walker AF, Marakis G, Christie S, Byng M. Mg citrate found more bioavailable than other Mg preparations in a randomised, double-blind study. Magnes Res 2003;16:183–91 [PubMed] [Google Scholar]