Abstract
The effect of bulk-phase pH on the apparent affinity (Kdapp) of purified wild-type lactose permease (LacY) for sugars was studied. Kdapp values were determined by ligand-induced changes in the fluorescence of either of two covalently bound fluorescent reporters positioned away from the sugar-binding site. Kdapp for three different galactopyranosides was determined over a pH range from 5.5 to 11. A remarkably high pKa of ≈10.5 was obtained for all sugars. Kinetic data for thiodigalactoside binding measured from pH 6 to 10 show that decreased affinity for sugar at alkaline pH is due specifically to increased reverse rate. A similar effect was also observed with nitrophenylgalactoside by using a direct binding assay. Because affinity for sugar remains constant from pH 5.5 to pH 9.0, it follows that LacY is fully protonated with respect to sugar binding under physiological conditions of pH. The results are consistent with the conclusion that LacY is protonated before sugar binding during lactose/H+ symport in either direction across the membrane.
Keywords: lactose permease, membrane transporters, pH titrations, proton translocation, substrate affinity
The lactose permease of Escherichia coli (LacY), a paradigm for the major facilitator superfamily (MFS) of membrane transport proteins (1, 2), translocates a galactosidic sugar and an H+ across the cytoplasmic membrane (3, 4). This coupled translocation mechanism enables LacY to use free energy stored in an electrochemical H+ gradient (Δμ̄H+) to drive sugar accumulation against a concentration gradient (5–7).
X-ray crystal structures of LacY (8–10), all of which are in an inward-facing conformation, and a wealth of biochemical/biophysical data (7, 11–16) provide evidence for an alternating-access mechanism of action. By this means, upon sugar binding or imposition of a Δμ̄H+) (interior negative and/or alkaline), the inward-facing cavity closes with opening of an outward-facing cavity, thereby allowing alternative accessibility of the sugar-binding site, which is at the apex of the cavity in the middle of the molecule, to either side of the membrane. A similar model has been proposed for the glycerol phosphate/phosphate antiporter GlpT, a related MFS protein (17) and the ABC transporter Sav 1866 (18). The alternating-access model involves a global conformational change, which is consistent with the highly dynamic nature of LacY (7, 13, 14, 16, 19–23).
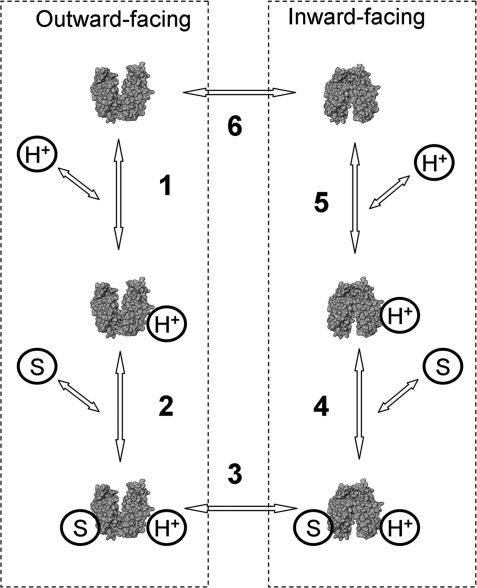
A simple kinetic model for lactose/H+ symport (Fig. 1) has been proposed based on extensive studies of partial reactions (efflux, equilibrium exchange, and entrance counterflow) catalyzed by LacY and site-directed mutants defective in the symport mechanism (see ref. 5). Wild-type LacY is tightly coupled with respect to sugar and H+ translocation and does not translocate H+ without substrate or vice versa. The effect of ambient pH on Δμ̄H+-driven symport is bell-shaped with an optimum at approximately pH 7.5 (24, 25), whereas the partial reactions exhibit different dependences on pH. For example, rates of lactose/H+ symport down a concentration gradient (efflux) are sigmoidal with respect to pH. The rates are very slow at pH 5.5–7.5 and increase sharply between pH 8.0 and 9.5. Moreover, between pH 5.5 and 7.5, efflux is inhibited 3- to 4-fold by D2O, whereas equilibrium exchange and Δμ̄H+-driven symport are unaffected, indicating that the deprotonation is probably rate-limiting for downhill lactose/H+ efflux (26). The rate of equilibrium exchange, a reaction that does not involve H+ translocation, is independent of pH from pH 5.5 to 10 (26). Furthermore, sugar affinity on either side of the membrane is independent of Δμ̄H+ (11). Nevertheless, the observation that affinity for β-d-galactopyranosyl-1-thio-β-d-galactopyranoside (TDG) decreases at the alkaline pH (27), as well as numerous other observations (for review, see refs. 5 and 7), argues that protonation may precede sugar binding.
Fig. 1.
Kinetic scheme for lactose/H+ transport cycle. Six steps include protonation/deprotonation of LacY (1 and 5), substrate binding/release (2 and 4), and conformational changes (3 and 6). Steps 2, 3, and 4 are involved in equilibrium exchange and entrance counterflow that proceed without deprotonation (5, 7).
In the present work, we investigate the effect of pH on substrate binding to LacY. Galactoside binding is readily detected and quantified by using V331C LacY labeled with 2-(4′-maleimidylanilino)naphthalene-6-sulfonic acid (MIANS), which places the reporter away from the sugar-binding site (Fig. 2A), and in this position, the fluorophore is sensitive to long-range conformational changes. This approach has been used to measure affinity for TDG (28), for detection of sugar binding to LacY mutants (29, 30), and for stopped-flow kinetic studies (31). It is noteworthy that binding is measured indirectly, but the fluorescence changes observed are the immediate result of sugar binding. Therefore, reference is made here to apparent affinity (Kdapp) only. We used purified V331C LacY labeled with either MIANS or N-(7-dimethylamino-4-methylcoumarin-3-yl)maleimide (DACM) to study the effect of pH on Kdapp. The findings demonstrate that sugar binding exhibits an unusually high apparent pKa of ≈10.5, thereby indicating that protonation is essential for sugar binding under physiological conditions.
Fig. 2.
Effect of sugar binding on fluorescence of DACM- or MIANS-labeled LacY. (A) Overall structure of LacY [Protein Data Bank (PDB) ID code 1pv7] with TDG occupying the sugar-binding site in the middle of the molecule at the apex of the hydrophilic cavity opened to the cytoplasmic side. The black sphere represents Cα atom of position 331 where the V331C mutation was introduced. LacY structure is displayed by using Pymol 0.97 (DeLano Scientific). (B and C) Structures of fluorophores and emission spectra of V331C LacY labeled with DACM or MIANS, respectively. Solid line, no sugar added or 10 mM sucrose; broken line, 10 mM TDG added. The spectra were recorded at 0.4 μM protein in 50 mM NaPi (pH 7.5), 0.02% DDM with excitation wavelengths of 397 nm or 330 nm for DACM- or MIANS-labeled LacY, respectively.
Results
TDG Binding.
In the presence of galactopyranosides specifically, MIANS-labeled V331C LacY displays a decrease in fluorescence intensity (28, 30–32). Several other fluorophores were tested, and DACM-labeled V331C LacY was also found to be sensitive to sugar binding. Saturating concentrations of TDG alter the fluorescence of DACM- or MIANS-labeled LacY in inverse fashion (Fig. 2). Thus, TDG binding to DACM-labeled V331C LacY causes an increase in DACM fluorescence with a clear blue shift in the spectrum (Fig. 2B), whereas MIANS fluorescence decreases under the same conditions with no shift (Fig. 2C). The inverse response of the fluorophores is likely caused by different interactions with the local environment induced by sugar binding. Although both fluorophores report a sugar-induced conformational change (31), the reporters may sample different local environments because DACM is more hydrophobic relative to negatively charged MIANS. In contrast, large maleimide-based fluorophores such as different Alexa dyes, rhodamine, and Oregon Green, as well as smaller fluorophores like acrylodan and pyrene derivatives or surprisingly another courmarin derivative [7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin] do not act as reporters for sugar binding with V331C LacY.
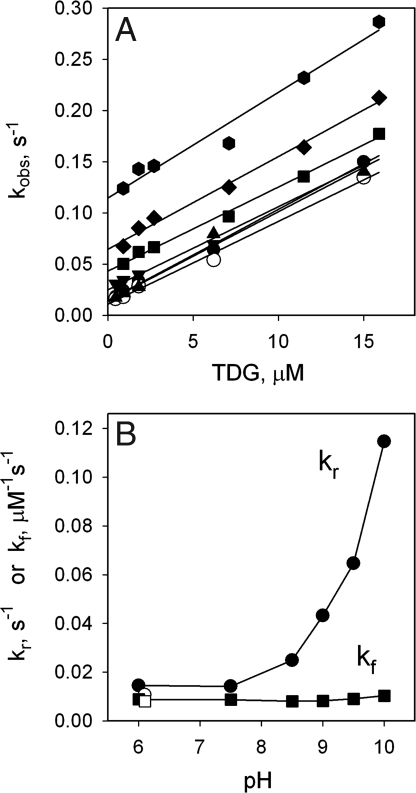
TDG affinity measured with DACM- or MIANS-labeled LacY (Fig. 3A; 21.4 and 1.2 μM, respectively) is significantly higher than the affinity of unlabeled LacY, which is characterized by a Kdapp in the low mM range [ref. 32 and supporting information (SI) Fig. S1 A and B]. Replacement of Val-331 with Cys itself does not affect Δμ̄H+-driven lactose/H+ symport (33) or alter affinity for sugar (see Fig. S1C). Thus, the covalently bound fluorophore is responsible for the change in affinity. Analysis of kinetic parameters for TDG binding to DACM- or MIANS-labeled LacY (Fig. 3 B and C) demonstrates that kreverse (kr) for MIANS-labeled LacY is ≈100 times lower than that for DACM-labeled LacY, whereas kforward (kf) is ≈3 times lower, resulting in a much lower Kdapp for MIANS-labeled LacY relative to DACM-labeled LacY.
Fig. 3.
TDG binding to DACM-labeled or MIANS-labeled V331C LacY. (A) Dependence of fluorescence on TDG concentration as percentage of initial emission level before TDG addition for each fluorophore. Titration traces were recorded as a function of time at 0.4 μM protein with excitation and emission wavelengths of 397 and 440 nm for DACM-labeled or 330 and 415 nm for MIANS-labeled LacY, respectively. Solid lines are hyperbolic fits to the data with estimated Kdapp values of 21.4 ± 0.4 and 1.2 ± 0.1 μM for DACM- and MIANS-labeled LacY, respectively. ●, DACM; ▾, MIANS. (B) Kinetics of TDG binding to DACM-labeled LacY. Concentration dependence of the rates (kobs) of the fluorescence changes was measured at 0.4 μM protein in 50 mM NaPi (pH 7.5), 0.02% DDM. Data were collected after rapid mixing of equal volumes of TDG and labeled protein by stopped-flow. Final concentrations of TDG are shown. Rates were estimated from single exponential fitting to time traces. Each point is an average of five to seven measurements. Linear fit to the data is shown as a solid line. The intercept with the y axis is kr (1.0 ± 0.1 s−1), and the slope is kf (28.66 ± 0.02 mM−1s−1). Estimated Kdapp (kr/kf) is 34.9 ± 3.5 μM. (C) Kinetics of TDG binding to MIANS-labeled LacY. Measurements were done as described above. Data were collected by stopped-flow at TDG concentrations from 0.05 to 4 mM and from time traces at TDG concentrations from 0.45 to 15 μM. Estimated parameters are kr = 0.011 ± 0.001 s−1; kf = 10.81 ± 0.03 mM−1s−1; and Kdapp = 1.0 ± 0.1 μM.
pH Dependence of TDG Binding.
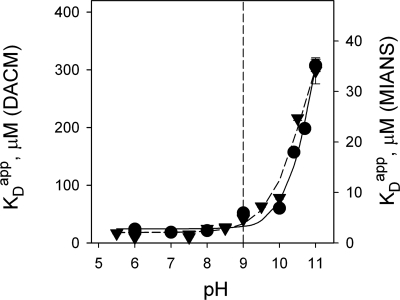
The emission spectra of V331C LacY labeled with either DACM or MIANS do not change significantly from pH 5.5 to 11.0 (see Fig. S2). However, specific sugar binding markedly affects fluorescence and allows titration measurements over a wide pH range (see Figs. S2 and S3). Values for Kdapp estimated from TDG concentration dependence at each pH (see Fig. S4) are plotted as a function of pH in Fig. 4. Remarkably, no significant change in Kdapp is observed for LacY labeled with either fluorophore in the pH range 5.5–9.0, whereas a sharp increase in Kdapp (i.e., decreased affinity) is observed between pH 9.5 and 11. The estimated pKa value is at least 10.5 for protein labeled with either DACM or MIANS (for details, see Fig. S5). Extremely high pH results in a decrease in fluorescence with time. An experiment testing protein stability at alkaline pH shows that MIANS-labeled LacY after preincubation at pH 11 for 12 min has the same affinity for TDG at pH 6 as the control sample, although the amplitude of the sugar-binding effect is smaller (see Figs. S3B and S4). Therefore, LacY is not rapidly denatured at high pH. The data demonstrate that the affinity of LacY for TDG depends on H+ concentration and that over the physiological range of pH, LacY is likely protonated.
Fig. 4.
Effect of pH on Kdapp for TDG with DACM-labeled or MIANS-labeled V331C LacY. Time traces from TDG titration experiments were recorded at indicated ambient pH values with excitation and emission wavelengths at 397 and 440 nm for DACM- or 330 and 415 nm for MIANS-labeled LacY, respectively. Data collection and fitting details were as described in Fig. 3A (see also Figs. S3–S5). Kdapp values estimated from titrations at each pH were plotted versus H+ concentration and fitted with a hyperbolic equation (see Fig. S5); they are presented as a function of pH. Estimated pKa values are 10.9 and 10.7 for DACM-labeled (●) and MIANS-labeled (▾) V331C LacY.
Kinetics of Sugar Binding.
Rates of sugar-induced changes in MIANS fluorescence were measured from pH 6 to 10, and the data exhibit a linear dependence of observed rate on TDG concentration from 0.5 to 15.0 μM at all pH values tested (Fig. 5A). The slope, which represents kf, does not depend on pH, but intercept with the y axis (kr) is clearly dependent on pH. The estimated values of the kinetic parameters from Fig. 5A are plotted as a function of pH in Fig. 5B. Strikingly, kf is constant as a function of pH, whereas kr is constant from pH 6.0 to 7.5 and increases sharply above pH 9.0. Hence, the effect of pH on apparent affinity for sugar is mediated by an increase in kr at high pH. Data from control experiments with protein preincubated at pH 11 and then shifted back to pH 6.1 are also presented (open symbols) and practically indistinguishable from the data for untreated LacY. Furthermore, Kdapp values determined from kinetic data (Kdapp = kf/kr) are practically identical to those obtained from the titration studies and can be superimposed when plotted as a function of pH (see Fig. S6).
Fig. 5.
Kinetics of TDG binding to MIANS-labeled V331C LacY as a function of pH. (A) TDG concentration dependence of the rate of fluorescence change (kobs) at given ambient pH values. Details of data collection and fitting are described in Fig. 3. Solid lines are linear fits to each dataset at pH 6 (●), 7.5 (▴), 8.5 (▾), 9.0 (■), 9.5 (♦), and 10.0 (⬣). ○, control experiment at pH 6.1 with labeled protein preincubated at pH 11.0 for 12 min. (B) Kinetic parameters of kr (●) and kf (■) estimated from A for each dataset are plotted as a function of pH. ○, □, kf and kr, respectively, for protein preincubated at pH 11.0 for 12 min and returned to pH 6.1 (in addition, see Fig. S6 for comparison of Kdapp values calculated from kinetic parameters and obtained from titration data).
A similar effect of pH on the kinetic parameters was observed with 4-nitrophenyl-α-d-galactopyranoside (NPG) by using a direct binding method based on Trp→NPG FRET (31). Rates of binding to unlabeled LacY were measured by stopped-flow at pH 5.5, 7.5, and 9.5. Kinetic parameters estimated from the NPG concentration dependence of kobs are presented in Table 1. The calculated kon and koff values are at least 3 orders of magnitude larger, but the effect of pH is very similar to that observed with TDG (Fig. 5). Alkaline pH decreases the affinity of LacY for NPG by increasing koff with little effect on kon. Unfortunately, it is not possible to measure rapid kinetics of NPG binding at higher pH accurately because of increased rates and decreased amplitudes of the fluorescence changes.
Table 1.
Kinetic paramerters for NPG binding to LacY measured by stopped-flow at different pH using direct sugar binding method based on Trp→NPG FRET (31)
| pH | koff, s−1 | kon, M−1 s−1 | Kdapp, μM |
|---|---|---|---|
| 5.5 | 35 | 0.50 × 106 | 70 |
| 7.5 | 18 | 0.43 × 106 | 42 |
| 9.5 | 114 | 0.50 × 106 | 228 |
pH Dependence of Lactose or Melibiose Binding.
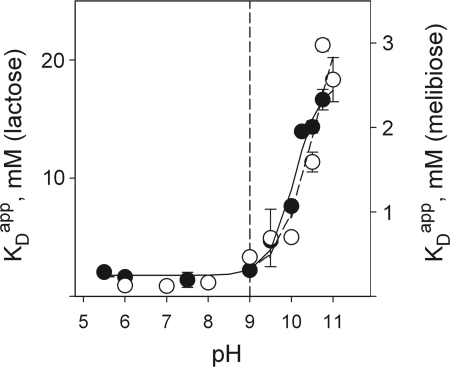
The increase in sugar affinity observed for MIANS-labeled V331C LacY allows measurement of the effect of pH on binding of natural substrates that bind with low affinity. For example, a Kdapp of ≈10 mM was estimated for lactose by protection of Cys-148 against alkylation at pH 7.4 (34). Changes in fluorescence of MIANS-labeled V331C LacY were measured with lactose or melibiose at concentrations ranging from 0.05 to 30 mM at different pH values (see Fig. S7), and Kdapp values were plotted as a function of pH (Fig. 6). The data exhibit marked similarity to those obtained for TDG (Fig. 4) despite the difference in affinity for the two disaccharides (Kdapp values for lactose and melibiose at neutral pH are ≈1.5 mM and 0.14 mM, respectively). There is no significant change in Kdapp for either lactose or melibiose from pH 5.5 to 9.0, and a sharp increase in Kdapp (i.e., decreased affinity) is observed between pH 9.5 and 11; pKa values are estimated to be 10.1 and 10.5 for lactose and melibiose, respectively.
Fig. 6.
Effect of pH on Kdapp for lactose or melibiose with MIANS-labeled V331C LacY. Experimental manipulations, data collection, and curve fitting were carried out as described in Fig. 4 (in addition, see Fig. S7). Estimated pKa values are 10.1 and 10.5 for lactose (●) and melibiose (○), respectively.
Discussion
In the kinetic model originally proposed for lactose/H+ symport (35) and supported by many findings (for reviews, see refs. 5 and 7), LacY binds a H+ before sugar on either side of the membrane to initiate translocation in either direction across the membrane (Fig. 1). Numerous studies demonstrate that sugar binding at neutral pH induces a conformational change with LacY in the membrane (12, 15) or with purified protein in detergent (13, 14, 28). Furthermore, rates of NPG binding measured by stopped-flow demonstrate that rapid sugar binding is followed by a slower conformational change detected with covalently bound MIANS as a reporter (31). This technique is used here to study the effect of pH on sugar binding to test the assertion that protonation is a prerequisite for sugar binding.
V331C LacY labeled with fluorescent reporter exhibits the same change in Kdapp as a function of ambient pH with TDG, lactose, and melibiose (Figs. 4 and 6) as well as unlabeled LacY with NPG (Table 1). In each instance, Kdapp is constant from pH 5.5 to 9.0 and increases dramatically from pH 9.5 to 11.0. Kinetic studies provide a clear explanation for the decrease in affinity at alkaline pH. Sugar binds with a forward rate that is independent of pH; however, the reverse rate increases dramatically at high pH (Fig. 5 and Table 1). Both rates are complex kinetic functions that include conformational changes. The pKa values approximate 10.5 as a lower limit because a complete sigmoidal dependence of Kdapp on pH cannot be obtained because the protein denatures too rapidly above pH 11.0. Because the Kdapp is constant over the physiological range of pH and increases only above pH 9.0, the data indicate that H+ binding is required for sugar binding. It has also been shown (J. Vazquez-Ibar, S. Schuldiner, and H.R.K., unpublished data) that addition of TDG to a concentrated solution of purified, detergent-solubilized LacY induces no change in pH. In contrast, a positive control with the antiporter EmrE under identical conditions releases 1 H+ per mol of EmrE upon addition of tetraphenylphosphonium (36). Because LacY exhibits no change in pH upon addition of TDG under conditions where EmrE releases an H+ upon substrate binding, the findings are consistent with the argument that LacY is already protonated before sugar binding.
So what becomes protonated? Tyr-236 (helix VII), Glu-269 (helix VIII), Arg-302 (helix IX), His-322 (helix X), and Glu-325 (helix X) are the residues in LacY that play a central role in H+ translocation (for review, see ref. 7). As shown by x-ray crystallography (8–10), these residues are in the C-terminal 6-helix bundle and positioned in the approximate middle of the molecule across from the sugar-binding site. The distances between the OH group of Tyr-236 and Arg-302, His-322, or Glu-325 are estimated to be 3–4 Å. It is also noteworthy that remarkable conservation of these residues has been found in many other members of the MFS (37, 38). Although the high pKa could be caused by Arg-302 or Tyr-236, it is unlikely that any single residue is responsible for the unique pH dependence of sugar binding. Moreover, several single site-directed mutants in side chains involved in H+ translocation (for example, Y236A, H322A, and R302A) display a shift in pKa of ≈2 pH units in the acidic direction. In contrast, E325Q LacY maintains high affinity for sugar that remains constant from pH 5.5 to 11.0 (unpublished observations). Therefore, the observed affinity change at alkaline pH results specifically from H+ interaction with a site that directly alters affinity for sugar.
H+ binding and translocation across membranes are thought to involve movement of H+ through pathways that run perpendicular to the plane of the membrane and parallel to the transmembrane helices. However, this situation does not appear to be the case for LacY where the residues involved in H+ translocation run perpendicular to the transmembrane helices and parallel to the plane of the membrane (Fig. 7). Therefore, as an alternative, we suggest that these residues form a coordination site for one or two buried water molecules that act as a hydronium ion intermediate(s) during H+ binding and release. In other words, water may act as a cofactor for H+ translocation, a mechanism suggested originally by Boyer (39) and applied recently to F1Fo by von Ballmoos and Dimroth (40). By this means, H+ translocation might occur much like sugar translocation, involving alternating access of both binding sites to either side of the membrane. Such a scenario would at least explain the long-standing problem of how sugar gradient-driven H+ translocation can occur in either direction across the membrane with the same residues.
Fig. 7.
Configuration of residues involved in H+ translocation in LacY located in C-terminal 6-helix bundle (PDB ID code 1pv7) presented as green sticks. Bound TDG is shown as spheres. (A) Viewed parallel to the membrane. (B) Viewed along the membrane normal from the cytoplasmic side. LacY structure is displayed by using Pymol 0.97 (DeLano Scientific).
Experimental Procedures
Materials.
Oligonucleotides were synthesized by Integrated DNA Technologies. Restriction enzymes were purchased from New England Biolabs. The QuikChange II kit was from Stratagene. All sugars were purchased from Sigma. Fluorophores were obtained from Molecular Probes/Invitrogen. Talon superflow resin was purchased from BD Clontech. All other materials were of reagent grade obtained from commercial sources.
Construction of Mutants and LacY Purification.
Construction of mutants and purification of LacY were carried out as described in ref. 14. Purified proteins (10–15 mg/ml) in 50 mM NaPi, 0.01% dodecyl-β-d-maltopyranoside (DDM) (pH 7.5) were frozen in liquid nitrogen and stored at −80°C until use.
Labeling with Fluorophores.
Purified V331C LacY (40–50 μM) was labeled with an equimolar concentration of fluorophore in 50 mM NaPi (pH 7.0), 0.02% DDM in the presence of 15 mM TDG to protect Cys-148 against alkylation (13, 14). Reactions were carried out for 15 min at room temperature in the dark. TDG and unreacted label were removed by buffer exchange on an Amicon Ultra-15 concentrator with a 30-kDa cutoff (Millipore). The extent of labeling estimated from absorption spectra was 0.7–0.8 mol/mol of protein. Control experiments with wild-type LacY exhibited essentially no labeling by MIANS under the same conditions.
Fluorescence Measurements.
Fluorescence was measured at room temperature in steady-state or stopped-flow modes by using an SLM-Aminco 8100 spectrofluorometer modified by OLIS as described in ref. 31 with excitation and emission wavelengths as indicated. Titration traces were recorded for 10–20 min with sequential additions of 5–10 μl of concentrated sugars to 2 ml of protein solution (0.4 μM) in 0.02% DDM and 50 mM buffers with overlapping pH ranges. Buffers used were: citrate-Pi for pH 5.5–7.0; NaPi for pH 7.0–8.0; sodium borate for pH 8.0–9.5; sodium glycine for pH 9.0–10.5, and Na-CAPS for pH 9.5–11.0. Ttitration data were corrected for dilution and fluorescence drift at high pH. Data analysis was done by using SigmaPlot 10 (Systat Software). Stopped-flow time traces were fitted by using OLIS GlobalWorks software.
Supplementary Material
Acknowledgments.
We are indebted to Shimon Schuldiner (Hebrew University of Jerusalem, Israel) for providing purified EmrE, and we thank Junichi Sugihara for excellent technical assistance. This work was supported in part by National Institutes of Health Grants DK51131, DK069463, GM073210 and GM074929 and National Science Foundation Grant 0450970 (to H.R.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803577105/DCSupplemental.
References
- 1.Saier MH, Jr, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 2.Saier MH., Jr Families of transmembrane sugar transport proteins. Mol Microbiol. 2000;35:699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 3.West IC. Lactose transport coupled to proton movements in Escherichia coli. Biochem Biophys Res Commun. 1970;41:655–661. doi: 10.1016/0006-291x(70)90063-x. [DOI] [PubMed] [Google Scholar]
- 4.West IC, Mitchell P. Stoichiometry of lactose-H+ symport across the plasma membrane of Escherichia coli. Biochem J. 1973;132:587–592. doi: 10.1042/bj1320587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaback HR, Sahin-Toth M, Weinglass AB. The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 6.Kaback HR. Structure and mechanism of the lactose permease. C R Biol. 2005;328:557–567. doi: 10.1016/j.crvi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 7.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 9.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H(+) symport in LacY. EMBO J. 2006;25:1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci USA. 2007;104:15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan L, Kaback HR. Binding affinity of lactose permease is not altered by the H+ electrochemical gradient. Proc Natl Acad Sci USA. 2004;101:12148–12152. doi: 10.1073/pnas.0404936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaback HR, et al. Site-directed alkylation and the alternating access model for LacY. Proc Natl Acad Sci USA. 2007;104:491–494. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumdar DS, et al. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc Natl Acad Sci USA. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci USA. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie Y, Ermolova N, Kaback HR. Site-directed alkylation of LacY: Effect of the proton electrochemical gradient. J Mol Biol. 2007;374:356–364. doi: 10.1016/j.jmb.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, Guan L, Freites JA, Kaback HR. Opening and closing of the periplasmic gate in lactose permease. Proc Natl Acad Sci USA. 2008;105:3774–3778. doi: 10.1073/pnas.0800825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol 3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 18.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 19.le Coutre J, Narasimhan LR, Patel CK, Kaback HR. The lipid bilayer determines helical tilt angle and function in lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1997;94:10167–10171. doi: 10.1073/pnas.94.19.10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Fourier transform infrared spectroscopy reveals a rigid α-helical assembly for the tetrameric Streptomyces lividans K+ channel. Proc Natl Acad Sci USA. 1998;95:6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patzlaff JS, Moeller JA, Barry BA, Brooker RJ. Fourier transform infrared analysis of purified lactose permease: A monodisperse lactose permease preparation is stably folded, α-helical, and highly accessible to deuterium exchange. Biochemistry. 1998;37:15363–15375. doi: 10.1021/bi981142x. [DOI] [PubMed] [Google Scholar]
- 22.Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nie Y, Smirnova I, Kasho V, Kaback HR. Energetics of ligand-induced conformational flexibility in the lactose permease of Escherichia coli. J Biol Chem. 2006;281:35779–35784. doi: 10.1074/jbc.M607232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He MM, Voss J, Hubbell WL, Kaback HR. Use of designed metal-binding sites to study helix proximity in the lactose permease of Escherichia coli. 1. Proximity of helix VII (Asp-237 and Asp-240) with helices X (Lys-319) and XI (Lys-358) Biochemistry. 1995;34:15661–15666. doi: 10.1021/bi00048a009. [DOI] [PubMed] [Google Scholar]
- 25.Frillingos S, Kaback HR. Monoclonal antibody 4B1 alters the pKa of a carboxylic acid at position 325 (helix X) of the lactose permease of Escherichia coli. Biochemistry. 1996;35:10166–10171. doi: 10.1021/bi960995r. [DOI] [PubMed] [Google Scholar]
- 26.Viitanen P, Garcia ML, Foster DL, Kaczorowski GJ, Kaback HR. Mechanism of lactose translocation in proteoliposomes reconstituted with lac carrier protein purified from Escherichia coli. 2. Deuterium solvent isotope effects. Biochemistry. 1983;22:2531–2536. doi: 10.1021/bi00279a034. [DOI] [PubMed] [Google Scholar]
- 27.Sahin-Tóth M, Karlin A, Kaback HR. Unraveling the mechanism of lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:10729–10732. doi: 10.1073/pnas.200351797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, Frillingos S, Voss J, Kaback HR. Ligand-induced conformational changes in the lactose permease of Escherichia coli: Evidence for two binding sites. Protein Sci. 1994;3:2294–2301. doi: 10.1002/pro.5560031214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesan P, Kaback HR. The substrate-binding site in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1998;95:9802–9807. doi: 10.1073/pnas.95.17.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnova IN, Kaback HR. A mutation in the lactose permease of Escherichia coli that decreases conformational flexibility and increases protein stability. Biochemistry. 2003;42:3025–3031. doi: 10.1021/bi027329c. [DOI] [PubMed] [Google Scholar]
- 31.Smirnova IN, Kasho VN, Kaback HR. Direct sugar binding to LacY measured by resonance energy transfer. Biochemistry. 2006;45:15279–15287. doi: 10.1021/bi061632m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ermolova NV, Smirnova IN, Kasho VN, Kaback HR. Interhelical packing modulates conformational flexibility in the lactose permease of Escherichia coli. Biochemistry. 2005;44:7669–7677. doi: 10.1021/bi0502801. [DOI] [PubMed] [Google Scholar]
- 33.Sahin-Tóth M, Kaback HR. Cysteine scanning mutagenesis of putative transmembrane helices IX and X in the lactose permease of Escherichia coli. Protein Sci. 1993;2:1024–1033. doi: 10.1002/pro.5560020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Kaback HR. Cysteine 148 in the lactose permease of Escherichia coli is a component of a substrate binding site. 2. Site-directed fluorescence studies. Biochemistry. 1994;33:12166–12171. doi: 10.1021/bi00206a020. [DOI] [PubMed] [Google Scholar]
- 35.Kaczorowski GJ, Robertson DE, Kaback HR. Mechanism of lactose translocation in membrane vesicles from Escherichia coli. 2. Effect of imposed Δψ, ΔpH, and ΔμH+ Biochemistry. 1979;18:3697–3704. doi: 10.1021/bi00584a010. [DOI] [PubMed] [Google Scholar]
- 36.Soskine M, Adam Y, Schuldiner S. Direct evidence for substrate-induced proton release in detergent-solubilized EmrE, a multidrug transporter. J Biol Chem. 2004;279:9951–9955. doi: 10.1074/jbc.M312853200. [DOI] [PubMed] [Google Scholar]
- 37.Kasho VN, Smirnova IN, Kaback HR. Sequence alignment and homology threading reveals prokaryotic and eukaryotic proteins similar to lactose permease. J Mol Biol. 2006;358:1060–1070. doi: 10.1016/j.jmb.2006.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vadyvaloo V, Smirnova IN, Kasho VN, Kaback HR. Conservation of residues involved in sugar/H(+) symport by the sucrose permease of Escherichia coli relative to lactose permease. J Mol Biol. 2006;358:1051–1059. doi: 10.1016/j.jmb.2006.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyer PD. Bioenergetic coupling to protonmotive force: Should we be considering hydronium ion coordination and not group protonation? Trends Biochem Sci. 1988;13:5–7. doi: 10.1016/0968-0004(88)90005-9. [DOI] [PubMed] [Google Scholar]
- 40.von Ballmoos C, Dimroth P. Two distinct proton-binding sites in the ATP synthase family. Biochemistry. 2007;46:11800–11809. doi: 10.1021/bi701083v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.