Abstract
Primary hepatocytes treated with nonesterified PUFA have been used as a model for analyzing the inhibitory effects of dietary polyunsaturated fats on lipogenic gene expression. Although nonesterified fatty acids play an important signaling role in starvation, they do not completely recapitulate the mechanism of dietary fat presentation to the liver, which is delivered via chylomicron remnants. To test the effect of remnant TG on lipogenic enzyme expression, chylomicron remnants were generated from the lymph of rats intubated with either safflower oil or lard. The remnants were added to the medium of primary rat hepatocytes in culture and the accumulation of mRNA for genes involved in carbohydrate and lipid metabolism was measured. Both PUFA-enriched remnants and nonesterified PUFA inhibited the expression and maturation of sterol response element binding protein-1c (SREBP-1c) and the expression of lipogenic genes regulated by this transcription factor. These remnants also inhibited the expression of glucose-6-phosphate dehydrogenase (G6PD), a gene regulated at post-transcriptional steps. In contrast, PUFA-enriched remnants did not inhibit the accumulation of mRNA for malic enzyme, glucokinase, and l-pyruvate kinase, whereas nonesterified fatty acids caused a decrease in these mRNA. These genes are regulated independently of SREBP-1c. SFA-enriched remnants did not inhibit lipogenic gene expression, which is consistent with a lack of inhibition of lipogenesis by dietary saturated fats. Thus, the inhibitory action of dietary polyunsaturated fats on lipogenesis involves a direct action of chylomicron remnants on the liver.
Introduction
PUFA are bioactive food components that can affect the risk of cardiovascular disease (1–3). An intracellular mechanism involved in the protective effect of dietary PUFA is a decrease in the hepatic expression of the lipogenic enzymes resulting in a reduction in the production of TG and VLDL (4–6). These inhibitory actions are unique to (n-6) PUFA and (n-3) PUFA; SFA and MUFA do not inhibit de novo lipogenesis (7, 8). In previous studies, the mechanisms by which dietary PUFA inhibit the expression of lipogenic enzymes have been investigated by incubating primary rat hepatocytes with albumin-bound PUFA (9, 10). This model assumes that the increase in intracellular fatty acid concentration caused by nonesterified fatty acids results in the same changes in liver metabolism as are caused by dietary fat. The model is also limited in investigating the different abilities of PUFA compared with SFA to modulate lipogenesis, because incubation of primary rat hepatocytes with albumin-bound palmitate causes a rapid depletion in cellular ATP concentrations and stimulates apoptosis (10, 11).
Dietary TG presents to the liver in chylomicron remnants, which contain 15–35% of the dietary TG originally packaged in chylomicrons; these particles are cleared by the liver using receptor-mediated endocytosis (12, 13). In contrast, nonesterified fatty acids bound to serum albumin dramatically increase in concentration during starvation and uncontrolled diabetes. Within the liver sinusoids, the nonesterified fatty acids dissociate from albumin and enter hepatocytes by a mechanism thought to involve specific fatty acid transporters (14, 15). There is little or no information comparing the regulation of metabolic processes by chylomicron remnants compared with nonesterified fatty acids.
A growing body of evidence suggests that cells contain distinct pools of fatty acids and these pools have different functions (16–21). The mode of uptake of lipid into the hepatocyte may also dictate its regulatory potential. The goals of these experiments were first to determine whether PUFA delivered to hepatocytes as chylomicron remnant TG inhibit the expression of lipogenic genes. Second, chylomicron remnants enriched in saturated fat were tested for their potential to inhibit the expression of lipogenic and glycolytic genes.
Methods
Animal care and cell culture.
Animal experiments were conducted in conformity with the Public Health Service policy on Humane Care and Use of Laboratory Animals. The Institutional Animal Care and Use Committee of the Division of Laboratory Animal Resources at West Virginia University approved all experimental procedures. Hepatocytes were isolated from male Sprague-Dawley rat (150–200 g) livers by a modification of the method of Seglen (22), as described previously (23). Hepatocytes (3 × 106) were plated onto 60-mm collagen-coated plates in Hi/Wo/Ba medium [Waymouth MB752/1 (27.5 mmol/L glucose), 20 mmol/L HEPES, pH 7.4, 0.5 mmol/L serine, 0.5 mmol/L alanine, 0.2% BSA] plus 5% newborn calf serum. Two hours postisolation, hepatocytes were washed twice with serum-free media and incubated overnight with serum-free media and 0.3 mg Matrigel/plate (BD PharMingen). Treatments as indicated in the figure legends were added to the hepatocytes in fresh serum-free media, without Matrigel, after 20 h in culture (time zero). Fatty acids (Nu-Check Prep) were prepared in complex with BSA at a 1:4 molar ratio (BSA:fatty acid) and the stock contained butylated-hydroxytoluene 0.1%. Hepatocytes not receiving the fatty acid:BSA complex were treated with serum-free media containing an equivalent amount of BSA. All media contained supplemental α-tocopherol (5 mg/L of medium). The medium was changed every 12 h. Treatments were as follows: insulin (80 nmol/L), fatty acids (175 μmol/L), chylomicrons (100 μg TG/plate), and chylomicron remnants (100 μg TG/plate). In all cases, n refers to the number of separate rat hepatocyte experiments.
Protein isolation and Western-blot analysis.
Hepatocytes (2 plates/treatment) were pooled and harvested after 30 min of treatment. Cell lysates were prepared as described by Hansmannel et al. (24). The cell lysates (20–40 μg protein) were subjected to Western analysis (25) using SREBP-1c antibody (Santa Cruz). Chemiluminescence was detected by ECL plus (Amersham) and imaged using the Typhoon 9410 (GE Healthcare).
Isolation of total RNA and qRT-PCR.
Hepatocytes (2 plates/treatment) were pooled and harvested after 24 h, with the treatments indicated in the figure legends. Total RNA was isolated using Tri-Reagent (Ambion) according to the manufacturer’s instructions. RNA (150 ng) was DNase I-treated and the amount of mRNA for G6PD, SREBP-1c, fatty acid synthase (FAS),6 acetyl-CoA carboxylase-1 (ACC-1), ATP-citrate lyase (ATP-CL), stearoyl-CoA desaturase 1 (SCD), S14, glucokinase (GK), L-pyruvate kinase (LPK), and malic enzyme (ME) in each sample was determined in duplicate by qPCR (BioRad iCycler iQ) analysis by one step RT-PCR using Quantitect SYBR green or probe kits (QIAGEN) according to the manufacturer’s instructions. Sequences for primers and probe are in Supplemental Table 1. The relative amount of mRNA was calculated using the comparative threshold cycle method and expressed relative to the control, cyclophilin B mRNA.
Preparation of chylomicron remnants.
Lymph fistula rats with duodenal and intestinal lymph duct fistulae (26) were infused (3 mL/h) with a lipid emulsion containing safflower oil or melted lard (0.36 g/animal) and 19 mmol/L sodium taurocholate in PBS (pH 6.4). Safflower oil contains ~78% linoleic acid and is unique among the vegetable oils in that it contains nearly all (n-6) PUFA (27). Lard contains 40% SFA, 48% MUFA, and 14% PUFA, primarily 18:2(n-6) (27). Lymph was collected for 6 h. Whole chylomicrons and remnants were isolated as previously described (28). Chylomicron TG was digested postheparin plasma (29) as a source of lipoprotein lipase and apolipoprotein (apo) E. Remnants were purified by density centrifugation (28). The amount of TG in the remnants was determined using the Triglyceride and Free Glycerol kit (Sigma-Aldrich); the amount of remnants added is expressed relative to the amount of TG in the remnants. Remnant TG (100 μg) added to a plate of hepatocytes containing 3 mL of medium results in an amount of fatty acid equivalent to 115 μmol/L.
Measurement of fatty acid synthesis.
The rate of de novo fatty acid synthesis was measured using tritiated water (30). Cells were incubated with 7.4 GBq/L 3H2O during the last 3 h of a 24-h treatment. The cells from 3 plates/treatment were pooled.
Statistics
Statistics were performed using GraphPad Prism (version 4.0). Overall significance was determined by 1-way ANOVA; multiple comparisons were made using Dunnett’s post-test if the overall P-value after ANOVA was P < 0.05. All comparisons were made to the control, which was hepatocytes incubated with insulin alone. Hepatocytes not receiving treatment (no addition) or PUFA-enriched chylomicron remnants in the absence of insulin were not compared, because these were included for reference with respect to the insulin induction or as a control for the effect of the addition of remnants per se, respectively.
Results
Composition of chylomicron remnants.
The fatty acid content of the prepared chylomicrons and chylomicron remnants was determined by GC (Table 1). Minimal composition changes were detected by conversion of the chylomicrons to the chylomicron remnants. Chylomicron remnants derived from rats receiving safflower oil (PUFA-enriched remnants) contained 57.7% PUFA. Remnants from rats receiving lard (SFA-enriched remnants) contained twice the saturated fat content compared with PUFA-enriched chylomicron remnants (45.2 vs. 21.2%) and only 21.3% PUFA, which were predominantly (n-6) fatty acids. The presence of SFA in PUFA-enriched remnants and at percentages greater than that found in safflower oil likely represented fatty acids that were synthesized de novo by the intestine or were absorbed from bile phospholipid.
TABLE 1.
Fatty acid composition of chylomicrons and chylomicron remnants prepared from rats intubated with safflower oil or lard
| Fatty acid1 | Safflower oil | Lard | ||
| CM2 | CR | CM | CR | |
| % total fatty acids | ||||
| Palmitate | 12.6 | 14.4 | 28.6 | 31.6 |
| Stearate | 4.6 | ND | 13.4 | 16.2 |
| Oleate | 13.7 | 16.1 | 29.5 | 35.1 |
| Linoleate | 65.1 | 69.5 | 23.1 | 17.1 |
| Arachidonate | 4.0 | ND | 5.4 | ND |
Each value is the mean, = 2 rats except lard CR, n = 1. Variation between the 2 values was <5.0%. Fatty acids present at <1.0% are not represented.
CM, chylomicron; CR, chylomicron remnant; ND, not detected.
G6PD is inhibited by PUFA-enriched remnants.
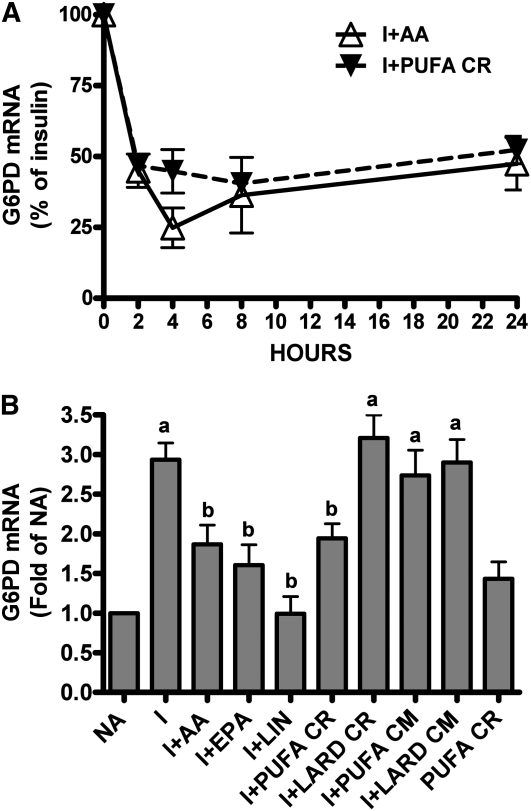
Insulin simulated G6PD expression (Fig. 1B). The addition of a maximally inhibitory concentration of PUFA-enriched remnants (Supplemental Fig. 1) decreased G6PD mRNA expression by 60% (Fig. 1A). This inhibition was observed by 4 h and continued through 24 h. Each plate of hepatocytes received 100 μg of remnant TG; if all 3 fatty acids were hydrolyzed from the glycerol backbone, this represented a dose of ~115 μmol/L fatty acid in the medium. Thus, inhibition by chylomicron remnants was achieved with a low potential concentration of fatty acids. The inhibition by the polyunsaturated-enriched remnants occurred to a similar extent and over the same time course as with the nonesterified fatty acid, arachidonic acid (AA) (Fig. 1A).
FIGURE 1.
G6PD expression in rat hepatocytes treated with PUFA-enriched chylomicron remnants or nonesterified PUFA. (A) Time course of changes in G6PD mRNA accumulation due to albumin-bound AA or PUFA-enriched chylomicron remnants (PUFA CR) in hepatocytes preincubated with insulin for 24 h. G6PD expression in cells treated with insulin was set at 100%. Each bar represents the mean ± SEM of n = 3 independent hepatocyte isolations. (B) G6PD expression after incubation with nonesterified fatty acids, chylomicrons, or remnants. Each bar represents the mean ± SEM of n = 8 independent hepatocyte isolations, with the exception of EPA and LIN, n = 5. The absolute value of no addition (NA) was 0.179 ± 0.01 corrected Ct value. Means without a common letter differ, P < 0.05. I, insulin; LIN, linoleic acid; CR, chylomicron remnants; CM, chylomicrons.
PUFA-enriched remnants inhibited G6PD mRNA accumulation to a similar extent as the nonesterified fatty acids: linoleate, arachidonate, and eicosapentaenoate (Fig. 1B). The inhibition required uptake of the remnant; whole chylomicrons that were excluded from uptake because they lack apoE did not inhibit. Remnants in the absence of insulin also did not inhibit G6PD expression (Fig. 1B), similar to the action of PUFA (25). The inhibition by chylomicron remnants was observed only with remnants derived from rats intubated with polyunsaturated fat; SFA-enriched remnants did not inhibit. This comparison could not have been made with nonesterified SFA, because palmitate causes hepatocyte death via apoptosis (10, 11).
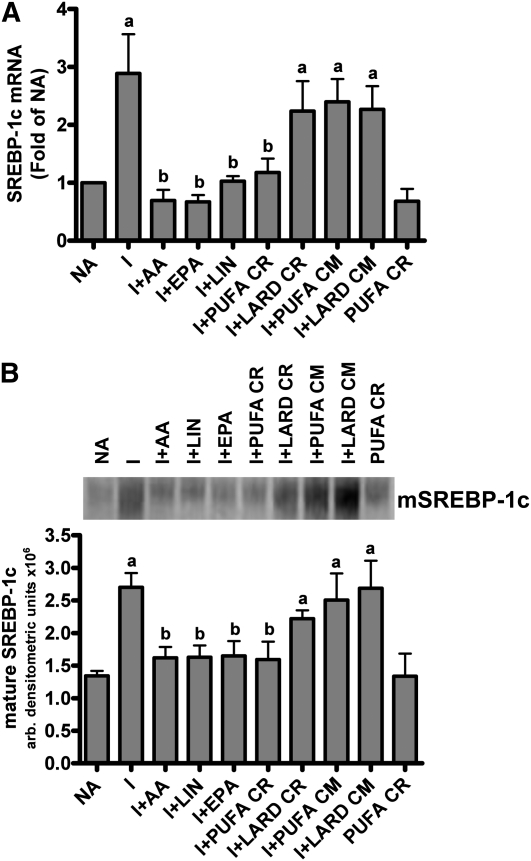
SREBP-1c#x2013 and SREBP-1c#x2013regulated genes are inhibited by PUFA-enriched chylomicron remnants.
Regulation of G6PD expression by dietary fat occurs exclusively at a post-transcriptional step (31). We next asked if the inhibitory effect of PUFA-enriched chylomicron remnants would extend to lipogenic genes inhibited at transcriptional steps and, in particular, genes induced by SREBP-1c. The addition of insulin increased SREBP-1c mRNA expression by 3-fold compared with no addition (Fig. 2A). Both nonesterified fatty acids and PUFA-enriched chylomicron remnants inhibited the insulin induction of SREBP-1c mRNA by 60% or more. SFA-enriched chylomicron remnants had no effect on SREBP-1c mRNA expression. Treatment with nascent chylomicrons or remnants in the absence of insulin also did not affect SREBP-1c mRNA abundance. Accompanying the decrease in SREBP-1c expression, the release of active or mature SREBP-1c was inhibited by both nonesterified PUFA and PUFA-enriched remnants (Fig. 2B). Treatment with SFA-enriched chylomicron remnants or nascent chylomicrons did not inhibit mature SREBP-1c protein formation. Therefore, PUFA-enriched chylomicron remnants mimicked the effect of dietary PUFA on SREBP-1c expression and generation of the active protein.
FIGURE 2.
(A) SREBP-1c expression after incubation of rat hepatocytes with nonesterified fatty acids, chylomicrons, or remnants. Each bar represents the mean ± SEM, n = 8 (NA, INS, AA), 3 (EPA, LIN, LARD CR, PUFA CM, LARD CM), or 6 (PUFA CR). The absolute value of NA was 0.0281 ± 0.004 corrected Ct value. (B) A representative Western blot of mature SREBP-1c. Each bar represents the means ± SEM, n = 3 independent hepatocyte isolations. Means without a common letter differ, P < 0.05. LIN, Linoleic acid; CR, chylomicron remnants; CM, chylomicrons.
FAS, ACC-1, SCD-1, ATP-CL, and S14 are SREBP-1c target genes. Changes in SREBP-1c activity should therefore coordinately regulate these genes. Insulin increased the expression of these genes (Table 2). Expression of these genes was stimulated only with insulin; therefore, the increase in expression was less than reports in which the medium was supplemented with additional hormones such as glucocorticoids and/or thyroid hormone (6, 32). PUFA-enriched chylomicron remnants significantly inhibited the expression of these SREBP-1c–dependent genes and the magnitude of this decrease was the same as that observed with the nonesterified fatty acids (Table 2). The inhibition of these genes occurred in parallel with the changes in the amount of mature SREBP-1c and SREBP-1c mRNA. SFA-enriched chylomicron remnants had no effect.
TABLE 2.
Expression of the SREBP-1c regulated genes in rat hepatocytes after incubation with nonesterified fatty acids, chylomicrons, or remnants1
| mRNA |
|||||
| Treatment | FAS | ACC-1 | ATP-CL | SCD-1 | S14 |
| Fold of no addition2 | |||||
| I | 2.46 ± 0.33a | 1.40 ± 0.23a | 2.87 ± 0.47a | 3.20 ± 0.72a | 5.27 ± 0.67a |
| I + AA | 0.74 ± 0.14b | 0.76 ± 0.07b | 0.98 ± 0.26b | 1.3 ± 0.27b | 0.92 ± 0.20b |
| I + EPA | 1.12 ± 0.16b | 0.75 ± 0.15b | 0.98 ± 0.13b | 1.09 ± 0.24b | 1.11 ± 0.36b |
| I + LIN3 | 1.48 ± 0.30b | 0.70 ± 0.10b | 1.30 ± 0.29b | 1.30 ± 0.32b | 1.68 ± 0.58b |
| I + PUFA CR | 1.44 ± 0.14b | 0.68 ± 0.13b | 1.28 ± 0.23b | 1.58 ± 0.27b | 2.22 ± 0.44b |
| I + LARD CR | 2.16 ± 0.25 | 1.04 ± 0.09 | 2.17 ± 0.30 | 2.47 ± 0.63 | 3.84 ± 0.84 |
| I + PUFA CM | 2.20 ± 1.19 | 1.06 ± 0.10 | 2.02 ± 0.37 | 2.74 ± 0.42 | 4.30 ± 0.78 |
| I + LARD CM | 2.76 ± 0.27 | 1.31 ± 0.19 | 2.45 ± 0.35 | 2.65 ± 0.46 | 3.55 ± 0.69 |
| PUFA CR4 | 1.26 ± 0.22 | 1.14 ± 0.23 | 1.05 ± 0.21 | 1.32 ± 0.37 | 0.98 ± 0.18 |
Data are means ± SEM, = 5–8. Means in a column with superscripts without a common letter differ, P < 0.05.
The no addition values were set to 1.0 and the relative Ct values were 0.048 ± 0.01 (FAS), 0.0218 ± 0.002 (ACC-1), 0.107 ± 0.04 (ATP-CL), 0.0536 ± 0.005 (SCD), and 0.054 ± 0.01 (S14).
LIN, linoleate.
Reference sample.
SREBP-1c independent genes are not inhibited by chylomicron remnants.
We next examined the expression of genes (GK, LPK, ME) whose expression can be inhibited by dietary polyunsaturated fat but whose regulation does not involve SREBP-1c (32–34). Expression of each of these genes was enhanced by insulin and significantly inhibited by albumin-bound PUFA (Table 3). In contrast, PUFA-enriched chylomicron remnants did not significantly inhibit the accumulation of these mRNA compared with treatment with insulin alone.
TABLE 3.
Expression of the SREBP-1c–independent genes in rat hepatocytes after incubation with nonesterified fatty acids, chylomicrons, or remnants1
| mRNA |
|||
| Treatment | GK | LPK | ME |
| Fold of no addition | |||
| I | 5.28 ± 0.83a2 | 2.38 ± 0.32a | 2.58 ± 0.40a |
| I + AA | 1.59 ± 0.20b | 1.24 ± 0.19b | 1.04 ± 0.37b |
| I + EPA | 0.74 ± 0.26b | 1.17 ± 0.29b | 0.92 ± 0.34b |
| I + LIN2 | 0.82 ± 0.24b | 0.92 ± 0.18b | 0.99 ± 0.21b |
| I + PUFA CR | 3.44 ± 0.59 | 1.88 ± 0.43 | 1.80 ± 0.53 |
| I + LARD CR | 3.35 ± 0.54 | 1.73 ± 0.19 | 1.66 ± 0.53 |
| I + PUFA CM | 3.79 ± 0.42 | 1.99 ± 0.26 | 1.63 ± 0.38 |
| I + LARD CM | 3.86 ± 1.17 | 1.77 ± 0.16 | 1.92 ± 0.24 |
| PUFA CR3 | 0.43 ± 0.17 | 1.14 ± 0.14 | 1.04 ± 0.32 |
Data are the means ± SEM, = 5–8 independent hepatocyte isolations. Means without a common letter differ, P < 0.05. The no addition values were set to 1. The relative Ct values for these samples were 0.053 ± 0.001 (GK), 0.198 ± 0.04 (LPK), and 0.274 ± 0.03 (ME).
LIN, linoleate.
PUFA CR without insulin was a reference sample.
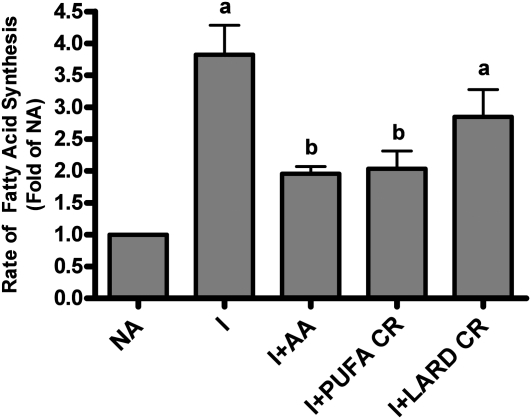
Polyunsaturated- but not SFA-enriched remnants inhibit lipogenic rate.
The coordinate decrease in the expression of lipogenic enzymes suggested that the overall rate of fatty acid biosynthesis should be similarly regulated. Insulin enhanced the rate of de novo fatty acid synthesis by 3.5-fold (Fig. 3). Addition of either nonesterified AA or PUFA-enriched chylomicron remnants inhibited the lipogenic rate by 50%. Treatment of hepatocytes with saturated fat-enriched chylomicron remnants did not significantly inhibit the insulin stimulation of fatty acid synthesis.
FIGURE 3.
The rate of de novo fatty acid synthesis in rat hepatocytes after incubation with nonesterified fatty acids or chylomicron remnants. Each bar represents the means ± SEM, n = 3 independent hepatocyte isolations. The absolute value of no addition (NA) was 5.4 ± 1 nmol 3H2O incorporated/(h⋅mg protein). Means without a common letter differ, P < 0.05.
Discussion
Primary rat hepatocytes contain the necessary proteins involved in uptake of the remnant particles from the medium (35–37). These include hepatic lipase (38, 39), LDL receptor (40, 41), and LDL receptor-related protein 1 (42) and have been used as a model to study uptake of lipoproteins, including chylomicron remnants (37, 41). ApoE, necessary for hepatic uptake, is acquired by the remnants during the incubation with post-heparin plasma. Chylomicron remnants enriched in PUFA inhibited the expression of several lipogenic genes. This inhibition was specific to remnants enriched in PUFA and was not observed with remnants derived from rats fed lard. Selective inhibition of lipogenic rate and lipogenic gene expression by PUFA- as opposed to SFA-enriched chylomicron remnants is consistent with the pattern of regulation by dietary fat observed in animals (5, 8). This differential effect suggests that the inhibition of these genes involves a direct intracellular action of dietary PUFA as opposed to inhibition secondary to extrahepatic regulation of other humoral factors.
Uptake of the remnant fatty acid can occur by both hepatic lipase hydrolysis on the extracellular surface and lysosomal digestion of the internalized remnant particle (13, 37). Because hepatic lipase digestion of chylomicron remnants will increase nonesterified concentrations of fatty acids in the medium, the inhibitory effect of remnants on lipogenesis might be secondary to generation of nonesterified fatty acids at the cell surface; however, the medium used in these studies contains BSA at concentrations that are demonstrated to reduce FFA uptake (19). Thus, the action of remnants observed in the present studies likely involves internalization of the intact TG.
A common feature of these lipogenic genes is that their expression is stimulated by insulin and this stimulation involves SREBP-1c (5). In this regard, both nonesterified PUFA and the PUFA-enriched remnants only inhibit in the presence of insulin ( (25). This interaction with insulin is key in the inhibitory action of remnants, because remnants enriched in PUFA do not affect SREBP-1 expression in response to liver X-receptor agonists (43). Thus, inhibition of FAS, ACC-1, ATP-CL, SCD-1, and S14 appears to be secondary to the decrease in SREBP-1c. G6PD has been a useful prototype for studying regulation by fatty acids, because its expression is stimulated by insulin and the insulin stimulation is inhibited by PUFA in the absence of other hormones, such as glucocorticoids or thyroid hormone, which could confound the interpretation of the results. G6PD, an enzyme is regulated at a post-transcriptional step (23, 31) as well as indirectly by SREBP-1c (44) was also inhibited by the PUFA-enriched remnants and in an insulin-dependent manner. Previous data from our laboratory demonstrate that nonesterified PUFA inhibit insulin signal transduction within hepatocytes (25). PUFA-enriched remnants may act in a similar manner or the inhibition of G6PD expression may also be secondary to reduced SREBP-1c activity.
In contrast to key lipogenic enzymes regulation of LPK, GK and ME does not involve SREBP-1c, but involves carbohydrate response element binding protein (ChREBP) in the case of LPK, hepatocyte nuclear factor-4 for GK, and ME remains incompletely characterized (32–34). Accumulations of mRNA of GK, LPK, and ME were not inhibited by the PUFA-enriched remnants with or without insulin. Yet dietary polyunsaturated fat does inhibit the expression of these genes in the livers of intact animals (9, 45). Although increased concentrations of remnant lipid may be necessary to inhibit these genes in primary hepatocytes, these genes are as inhibited by the same amount of dietary polyunsaturated fat as the SREBP-1c–dependent genes. Regulation of GK, LPK, and ME may involve other humoral factors whose levels change with dietary fat feeding. In this regard, bile acid release is stimulated by fat in the intestine and can decrease activity of malic enzyme and glucokinase (46). FGF-19, a growth factor produced by the intestine, inhibits expression of pyruvate kinase and glucokinase (47). Additional gut-derived hormones, such as glucagon like peptide-1, are implicated in the regulation of hepatic TG synthesis (48). Factors such as these may play an important role in signaling dietary status to the liver and are candidates for polyunsaturated fat-derived, signals regulating metabolic genes.
The data presented here support the conclusion that diet-derived lipoproteins are capable of inhibiting lipogenic gene expression in liver. Although some effects of dietary fat can be modeled by the addition of nonesterified PUFA, they may not reflect the mechanisms of regulation used by dietary lipid. The aim of future work is to decipher the different intracellular signals generated by nonesterified fatty acids and chylomicron remnants and in particular how these signals regulate gene expression.
Supplementary Material
Acknowledgments
We thank Dr. Callee M. Walsh for experimental advice and for critical reviews of the manuscript and Dr. George Kelley for help with the statistics. A.K., H.C., and Y.Q. conducted research. A.K., H.C., and L.S. wrote the manuscript. A.K., P.T., and L.S. analyzed data and designed experiments. All authors read and approved the final manuscript.
Footnotes
Supported by NIH grant DK46897 (L.M.S.), NIH T32 HL090610 (A.B.K.), and the Mouse Metabolic Phenotyping Center at University of Cincinnati no. DK059630 (P.T.).
Supplemental Table 1 and Supplemental Figure 1 are available with the online posting of this paper at jn.nutrition.org.
Abbreviations used: AA, arachidonic acid; apo, apolipoprotein; LIN, linoleate.
Literature Cited
- 1.Hegsted DM, McGandy RB, Myers ML, Stare FJ. Quantitative effects of dietary fat on serum cholesterol in man. Am J Clin Nutr. 1965;17:281–95 [DOI] [PubMed] [Google Scholar]
- 2.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–52 [DOI] [PubMed] [Google Scholar]
- 3.Hu FB, Manson JE, Willett WC. Types of dietary fat and risk of coronary heart disease: a critical review. J Am Coll Nutr. 2001;20:5–19 [DOI] [PubMed] [Google Scholar]
- 4.Hill R, Linazasoro JM, Chevallier F, Chaikoff IL. Regulation of hepatic lipogenesis: the influence of dietary fats. J Biol Chem. 1958;233:305–10 [PubMed] [Google Scholar]
- 5.Jump DB, Clarke SD. Regulation of gene expression by dietary fat. Annu Rev Nutr. 1999;19:63–90 [DOI] [PubMed] [Google Scholar]
- 6.Ntambi JM. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J Lipid Res. 1999;40:1549–58 [PubMed] [Google Scholar]
- 7.Bartley JC, Abraham S. Hepatic lipogenesis in fasted, re-fed rats and mice: response to dietary fats of differing fatty acid composition. Biochim Biophys Acta. 1972;280:258–66 [DOI] [PubMed] [Google Scholar]
- 8.Clarke SD, Romsos DR, Leveille GA. Specific inhibition of hepatic fatty acid synthesis exerted by dietary linoleate and linolenate in essential fatty acid adequate rats. Lipids. 1976;11:485–90 [DOI] [PubMed] [Google Scholar]
- 9.Jump DB, Clarke SD, Thelen A, Liimatta M. Coordinate regulation of glycolytic and lipogenic gene expression by polyunsaturated fatty acids. J Lipid Res. 1994;35:1076–84 [PubMed] [Google Scholar]
- 10.Salati LM, Clarke SD. Fatty acid inhibition of hormonal induction of acetyl-coenzyme A carboxylase in hepatocyte monolayers. Arch Biochem Biophys. 1986;246:82–9 [DOI] [PubMed] [Google Scholar]
- 11.Wei Y, Wang D, Topczewski F, Pagliassotti MJ. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab. 2006;291:E275–81 [DOI] [PubMed] [Google Scholar]
- 12.Redgrave TG. Formation of cholesteryl ester-rich particulate lipid during metabolism of chylomicrons. J Clin Invest. 1970;49:465–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji ZS, Dichek HL, Miranda RD, Mahley RW. Heparan sulfate proteoglycans participate in hepatic lipaseand apolipoprotein E-mediated binding and uptake of plasma lipoproteins, including high density lipoproteins. J Biol Chem. 1997;272:31285–92 [DOI] [PubMed] [Google Scholar]
- 14.Stahl A, Gimeno RE, Tartaglia LA, Lodish HF. Fatty acid transport proteins: a current view of a growing family. Trends Endocrinol Metab. 2001;12:266–73 [DOI] [PubMed] [Google Scholar]
- 15.Stremmel W. Mechanism of hepatic fatty acid uptake. J Hepatol. 1989;9:374–82 [DOI] [PubMed] [Google Scholar]
- 16.Coleman RA, Lewin TM, Muoio DM. Physiological and nutritional regulation of enzymes of triacylglycerol synthesis. Annu Rev Nutr. 2000;20:77–103 [DOI] [PubMed] [Google Scholar]
- 17.Doege H, Baillie RA, Ortegon AM, Tsang B, Wu Q, Punreddy S, Hirsch D, Watson N, Gimeno RE, et al. Targeted deletion of FATP5 reveals multiple functions in liver metabolism: alterations in hepatic lipid homeostasis. Gastroenterology. 2006;130:1245–58 [DOI] [PubMed] [Google Scholar]
- 18.Fulgencio JP, Kohl C, Girard J, Pegorier JP. Troglitazone inhibits fatty acid oxidation and esterification, and gluconeogenesis in isolated hepatocytes from starved rats. Diabetes. 1996;45:1556–62 [DOI] [PubMed] [Google Scholar]
- 19.Ruby MA, Goldenson B, Orasanu G, Johnston TP, Plutzky J, Krauss RM. VLDL hydrolysis by LpL activates PPAR-{alpha} through generation of unbound fatty acids. J Lipid Res. 2010;51:2275–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanderson LM, Degenhardt T, Koppen A, Kalkhoven E, Desvergne B, Muller M, Kersten S. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) but not PPARalpha serves as a plasma free fatty acid sensor in liver. Mol Cell Biol. 2009;29:6257–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YL, Hernandez-Ono A, Ko C, Yasunaga K, Huang LS, Ginsberg HN. Regulation of hepatic apolipoprotein B-lipoprotein assembly and secretion by the availability of fatty acids. I. Differential response to the delivery of fatty acids via albumin or remnant-like emulsion particles. J Biol Chem. 2004;279:19362–74 [DOI] [PubMed] [Google Scholar]
- 22.Seglen PO. Preparation of rat liver cells. 3. Enzymatic requirements for tissue dispersion. Exp Cell Res. 1973;82:391–8 [DOI] [PubMed] [Google Scholar]
- 23.Stabile LP, Klautky SA, Minor SM, Salati LM. Polyunsaturated fatty acids inhibit the expression of the glucose-6-phosphate dehydrogenase gene in primary rat hepatocytes by a nuclear posttranscriptional mechanism. J Lipid Res. 1998;39:1951–63 [PubMed] [Google Scholar]
- 24.Hansmannel F, Mordier S, Iynedjian PB. Insulin induction of glucokinase and fatty acid synthase in hepatocytes: analysis of the roles of sterol-regulatory-element-binding protein-1c and liver X receptor. Biochem J. 2006;399:275–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talukdar I, Szeszel-Fedorowicz W, Salati LM. Arachidonic acid inhibits the insulin induction of glucose-6-phosphate dehydrogenase via p38 MAP kinase. J Biol Chem. 2005;280:40660–7 [DOI] [PubMed] [Google Scholar]
- 26.Tso P, Karlstad MD, Bistrian BR, DeMichele SJ. Intestinal digestion, absorption, and transport of structured triglycerides and cholesterol in rats. Am J Physiol. 1995;268:G568–77 [DOI] [PubMed] [Google Scholar]
- 27.Ziegler EE, Filer LJ, Jr, editor Present knowledge in nutrition. 7th ed Washington, DC: ILSI Press; 1996 [Google Scholar]
- 28.Floren CH, Nilsson A. Degradation of chylomicron remnant cholesteryl ester by rat hepatocyte monolayers. Inhibition by chloroquine and colchicine. Biochem Biophys Res Commun. 1977;74:520–8 [DOI] [PubMed] [Google Scholar]
- 29.Nilsson A. Effects of anti-microtubular agents and cycloheximide on the metabolism of chylomicron cholesteryl esters by hepatocyte suspensions. Biochem J. 1977;162:367–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowenstein JM, Brunengraber H, Wadke M. Measurement of rates of lipogenesis with deuterated and tritiated water. Methods Enzymol. 1975;35:279–87 [DOI] [PubMed] [Google Scholar]
- 31.Stabile LP, Hodge DL, Klautky SA, Salati LM. Posttranscriptional regulation of glucose-6-phosphate dehydrogenase by dietary polyunsaturated fat. Arch Biochem Biophys. 1996;332:269–79 [DOI] [PubMed] [Google Scholar]
- 32.Stoeckman AK, Towle HC. The role of SREBP-1c in nutritional regulation of lipogenic enzyme gene expression. J Biol Chem. 2002;277:27029–35 [DOI] [PubMed] [Google Scholar]
- 33.Dozin B, Rall JE, Nikodem VM. Tissue-specific control of rat malic enzyme activity and messenger RNA levels by a high carbohydrate diet. Proc Natl Acad Sci USA. 1986;83:4705–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth U, Curth K, Unterman TG, Kietzmann T. The transcription factors HIF-1 and HNF-4 and the coactivator p300 are involved in insulin-regulated glucokinase gene expression via the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2004;279:2623–31 [DOI] [PubMed] [Google Scholar]
- 35.Floren CH, Nilsson A. Uptake and degradation of iodine-labelled chylomicron remnant particles by monolayers of rat hepatocytes. Biochem J. 1978;174:827–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert MS, Avella MA, Botham KM, Mayes PA. Comparison of short- and long-term effects of different dietary fats on the hepatic uptake and metabolism of chylomicron remnants in rats. Br J Nutr. 1998;79:203–11 [DOI] [PubMed] [Google Scholar]
- 37.Nagata Y, Chen J, Cooper AD. Role of low density lipoprotein receptor-dependent and -independent sites in binding and uptake of chylomicron remnants in rat liver. J Biol Chem. 1988;263:15151–8 [PubMed] [Google Scholar]
- 38.Leitersdorf E, Stein O, Stein Y. Synthesis and secretion of triacylglycerol lipase by cultured rat hepatocytes. Biochim Biophys Acta. 1984;794:261–8 [DOI] [PubMed] [Google Scholar]
- 39.Sundaram GS, Shakir KM, Barnes G, Margolis S. Release of phospholipase A and triglyceride lipase from rat liver. J Biol Chem. 1978;253:7703–10 [PubMed] [Google Scholar]
- 40.Qin W, Infante J, Wang SR, Infante R. Regulation of HMG-CoA reductase, apoprotein-B and LDL receptor gene expression by the hypocholesterolemic drugs simvastatin and ciprofibrate in Hep G2, human and rat hepatocytes. Biochim Biophys Acta. 1992;1127:57–66 [DOI] [PubMed] [Google Scholar]
- 41.Salter AM, Saxton J, Brindley DN. Characterization of the binding of human low-density lipoprotein to primary monolayer cultures of rat hepatocytes. Biochem J. 1986;240:549–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laatsch A, Merkel M, Talmud PJ, Grewal T, Beisiegel U, Heeren J. Insulin stimulates hepatic low density lipoprotein receptor-related protein 1 (LRP1) to increase postprandial lipoprotein clearance. Atherosclerosis. 2009;204:105–11 [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Soldado I, Avella M, Botham KM. Suppression of VLDL secretion by cultured hepatocytes incubated with chylomicron remnants enriched in n-3 polyunsaturated fatty acids is regulated by hepatic nuclear factor-4alpha. Biochim Biophys Acta. 2009,1791:1181–9 [DOI] [PubMed] [Google Scholar]
- 44.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herzberg GR, Janmohamed N. Regulation of hepatic lipogenesis by dietary maize oil or tripalmitin in the meal-fed mouse. Br J Nutr. 1980;43:571–9 [DOI] [PubMed] [Google Scholar]
- 46.Tsai AC, Dyer IA. Influence of dietary cholesterol and cholic acid on liver carbohydrate metabolism enzymes in rats. J Nutr. 1973;103:93–101 [DOI] [PubMed] [Google Scholar]
- 47.Bhatnagar S, Damron HA, Hillgartner FB. Fibroblast growth factor-19, a novel factor that inhibits hepatic fatty acid synthesis. J Biol Chem. 2009;284:10023–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parlevliet ET, Schroder-van der Elst JP, Corssmit EP, Picha K, O'Neil K, Stojanovic-Susulic V, Ort T, Havekes LM, Romijn JA, et al. CNTO736, a novel glucagon-like peptide-1 receptor agonist, ameliorates insulin resistance and inhibits very low-density lipoprotein production in high-fat-fed mice. J Pharmacol Exp Ther. 2009;328:240–8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.