Abstract
Histone H1 is commonly used to assay kinase activity in vitro. As many promising targeted therapies affect kinase activity of specific enzymes involved in cancer transformation, H1 phosphorylation can serve as potential pharmacodynamic marker for drug activity within the cell. In this report we utilized a phosphoproteomic workflow to characterize histone H1 phosphorylation changes associated with two targeted therapies in the Kasumi-1 Acute Myeloid Leukemia (AML) cell line. The phosphoproteomic workflow was first validated with standard casein phosphoproteins and then applied to the direct analysis of histone H1 from Kasumi-1 nuclear lysates. Ten H1 phosphorylation sites were identified on the H1 variants, H1.2, H1.3, H1.4, H1.5 and H1.x. Liquid chromatography mass spectrometry profiling of intact H1s demonstrated global dephosphorylation of H1.5 associated with therapy by the cyclin dependent kinase inhibitor, flavopiridol, and the Hsp90 inhibitor, 17AAG (17-(Allylamino)-17-demethoxygeldanamycin). In contrast, independent treatments with a nucleotide analog, proteosome inhibitor and histone deacetylase inhibitor did not exhibit decreased H1.5 phosphorylation. The data presented herein demonstrate that potential of histones to assess the cellular response of reagents that have direct and indirect effects on kinase activity that alters histone phosphorylation. As such, this approach may be a highly informative marker for response to targeted therapies influencing histone phosphorylation.
Keywords: Histone, Acute Myeloid Leukemia, Chemotherapy, Phosphorylation
INTRODUCTION
Higher-order chromatin structure is assembled and stabilized by linker histone H1 variants that bind to linker DNA [1, 2]. Human histone H1 has multiple sequence variants that include: H1.0, H1.1, H1.2, H1.3, H1.4, H1.5, H1.t and H1.x. The variants H1.2, H1.3, H1.4 and H1.5 exist in all human cells whereas H1.x and H1.t are tissue specific and H1.1 is found in thymus, testis, spleen, lymphocytic and neuronal cells [3]. The phosphorylation of linker histone H1 has been associated with the regulation of oncogene expression, DNA damage repair and chromatin decondensation [4–6]. For example, aberrant chromosomal condensation was induced by dephosphorylation of H1 in FM3A cells [7, 8]. However, chromosomal decondensation was linked to reduced phosphorylation of H1 after exposure of BHK cells to the topoisomerase II inhibitor VM26 or the protein kinase inhibitor staurosporine [9, 10]. Recently, the translocation of histone H1 to the cytoplasm has been implicated in signaling apoptosis [11–15]. Given the critical role of H1 isoforms in chromatin stability and their potential as chemotherapy targets, a greater understanding of the H1 phosphoisoform diversity and their response to therapeutic agents is warranted.
Antibodies directed against specific H1 phosphorylation sites are limited or have broad specificity. Mass spectrometry has no such limitation and can be used to determine global phosphorylation changes as well as site-specific changes [3, 16–20]. However, the application of MS to phosphoproteomic analysis still presents many challenges [21]. First, the low abundance of phosphorylation in vivo limits the direct application of mass spectrometry based technique due to limitations in practical dynamic range. Second, poor ionization efficiency for phosphopeptides and interferences from non-phosphopeptides can result in signal suppression in positive ion mode. Phosphoprotein and phosphopeptide enrichment have been used to overcome the limitation of mass spectrometry’s dynamic range by eliminating the chemical interferences due to non-phosphorylated peptides. Selective enrichment of phosphopeptides can be accomplished from any of the following techniques: immunoaffinity chromatography [22], immobilized metal affinity chromatography (IMAC) [23–25], metal oxide chemisorbtion [26–28] and strong cation exchange (SCX) chromatography [29]. Additionally, chemical derivatization of phosphorylated residues through β-elimination [30–33] or phosphatase treatment [25, 34–36] has been successfully used to enhance the ionization of phosphopetides prior to positive ion MS analysis [37–40].
Finally, phosphoserine and phosphothreonine-peptides are prone to loss of the phosphoric acid upon low-energy collision-induced dissociation during tandem mass spectrometric experiments [29]. The neutral loss of 98 Da for phosphoserine and phosphotheronine and 80 Da for phosphotyrosine in positive ion MS/MS can serve as a characteristic signature for phosphopeptides. Data-dependent neutral loss (DDNL) MS3 experiments are commonly used to overcome low abundant backbone cleavage resulting from the easy neutral loss [41]. The resulting interleaved set of MS2 and MS3 data from DDNL experiments present a unique challenge to automated database search programs. Alignment of the MS2 and MS3 data into a single data set has been demonstrated to improve the overall confidence of the peptide matches returned from database search programs [42–44]. An alternative approach is to exploit the hierarchical nature of the MS3 experimental data when performing the final peptide identification as previously demonstrated with metabolite identification [6] and phosphopeptide data [45].
Histone H1 is commonly used to assay kinase activity in vitro. As many promising chemotherapy reagents affect kinase activity, H1 phosphorylation can serve as potential marker for drug activity within the cell. The use of mass spectrometry to assay H1 phosphorylation allows for a new approach to preclinical assessment of H1 kinase activity during therapy for both model and primary cells. In this report we describe a proteomic workflow that combines liquid chromatography mass spectrometry, metal oxide (ZrO2/TiO2) phosphopeptide enrichment, DDNL MS3 and a hierarchical MS3 data analysis strategy to characterize histone H1 phosphorylation changes associated with flavopiridol and 17AAG (17-(Allylamino)-17-demethoxygeldanamycin) therapy in the model Acute Myeloid Leukemia (AML) cell line, Kasumi-1. Flavopiridol is a flavonoid derived from the indigenous plant from India, Dysoxylum binectariferum. It inhibits cyclin-dependent kinases CDKs 1, 2, and 4 at the G1/S and G2/M boundaries [46]. It has been reported to induce apoptosis in Chronic Lymphocytic Leukemia (CLL) via activation of caspase-3 [47]. 17AAG is an ansamycin antibiotic. It is an inhibitor to Hsp90 (Heat Shock Proteins 90). Hsp90 is responsible for the proper folding of a variety mutated and over-expressed proteins in cancer cells resulting in cell death [48] and regulates cell cycle associated proteins including CDKs [49–51]. Using the methods described above, we determined histone H1 phosphorylation sites and global H1 phosphorylation changes associated with drug therapy. Ten phosphorylation sites for the Acute Myeloid Leukemia (AML) model cell line, Kasumi-1, were identified and both drugs were shown to decrease histone H1 phosphorylation.
EXPERIMENTAL
SAMPLE PREPARATION
Bovine milk α-casein and β-casein (Sigma, St. Louis, MO) were prepared in 25 mM ammonium bicarbonate buffer to a final concentration of 25 µM. The protein mixture was digested by modified sequencing grade trypsin (Promega, Madison, WI) for 1h at 37°C at an enzyme/substrate ratio of 1:50. The digests were then dried in a vacuum concentrator (Eppendorf, Hamberg, Germany), and reconstituted with HPLC grade water containing 0.1% formic acid to a concentration of 1 µg/µL before enrichment.
Kasumi-1 cells were grown at 37 °C in a humidified atmosphere containing 5% CO2 in RPMI 1640 media supplemented with 20% heat-inactivated fetal bovine serum (FBS) (Invitrogen), 100 U/ml penicillin, and 100 µg/ml streptomycin (BioWhittaker, Walkersville, MD). Fifty million Kasumi-1 cells were treated with 17 AAG (600 nM), 1 mM valproic acid (VPA), 60 nM bortezomib (velcade) and flavopiridol (300 nM) (Sigma-Aldrich, St. Louis, MO) for 24 h. Histones were isolated from Kasumi-1 cells as described previously [52]. Briefly, the cell pellets were washed with 10 mM Tris buffer (pH = 7.5) and re-suspended in NP-40 lysis buffer with protease inhibitor cocktail (CalBiochem, San Diego, CA), PMSF, NaF, sodium orthovanadate, and phosphatase inhibitor cocktail 1 and 2 (Sigma-Aldrich, St. Louis, MO). After centrifugation, the pellets were collected and washed with Tris buffer. Sulfuric acid was added to the pellets to extract histones. The histones were precipitated overnight by addition of 80% acetone to the supernatant. The resulting precipitate was collected, dried and dissolved in 20% ACN containing 0.05% TFA.
HISTONE SEPARATION AND PURIFICATION
The histone mixtures were characterized by LC-MS. LC-MS was performed by use of reversed-phase HPLC (Waters model 2690; Milford, MA) coupled to a MicroMass Q-TOF (Micromass; Wythenshawe, UK) mass spectrometer. Histone mixtures were separated on a 1.0 mm×150 mm C18 column (Discovery Bio wide pore C18 column, 5 µm, 300 Å, Supelco, USA). Mobile phase A contained ACN with 0.05% TFA. Mobile phase B contained HPLC water with 0.05% TFA. Starting with 20% B, the gradient increased linearly to 30% B in 2 min, from 30% B to 35% B in 8 min, from 35% B to 50% B in 20 min, from 50% B to 60% B in 5 min, from 60% B to 95% B in 1 min and stayed in 95% for 4 min at the flow rate of 25 µl/min. The column was equilibrated for 30 min before each injection and washed twice with a blank run between each analysis. LC-MS data was deconvoluted by Masslynx 4.0.
Histone extracts from Kasumi-1 cells (1×107) were purified on a 4.6 mm×25 mm, 5 µm C18 discovery column (Sigma-Aldrich, St. Louis, MO) by use of the same 40 min gradient as before. The proteins eluting at 14.04–14.91 min (H1.5), 17.74–18.90 min (H1.2, H1.3 and H1.4) were collected and vacuum dried. The H1 extract was reconstituted in 25 mM ammonium bicarbonate and digested with trypsin at ratio of 100:1 (protein: enzyme) at 37°C for 1 h. H1 tryptic digests were dried and dissolved in 50% acetonitrile with 0.3% trifluoroacetic acid (TFA) before phosphopeptide enrichment.
PHOSPHOPEPTIDE ENRICHMENT
Zirconium dioxide and titanium dioxide coated Nutips (Glygen Corp, Columbia MD) were used to enrich the phosphopeptides from casein [26] and histone H1 digests. The loading buffer was made up of 0.3% trifluoroacetic acid (TFA) in 50% acetonitrile. Nutips (ZrO2 and TiO2) were washed with HPLC grade water and equilibrated with the loading buffer before loading samples. Casein peptides and histone H1 peptides mixture were reconstituted in loading buffer, and loaded onto equilibrated Nutips. Unbound peptides were washed with either 0.1% formic acid or 50% acetonitrile with 0.3% TFA; bound phosphopeptides were eluted off the tip with 1% ammonium hydroxide (pH 10.5). Eluted peptides were dried, reconstituted in 0.1% formic acid and subjected to data dependent neutral loss (DDNL) MS3.
LIQUID CHROMATOGRAPHY - TANDEM MASS SPECTROMETRY
Experiments were carried out on the following instruments: LTQ-FT mass spectrometer (Thermo Finnigan, San Jose, CA) and LTQ-Orbitrap mass spectrometer (Thermo Finnigan, San Jose, CA). Nano-RPLC separations were performed on a Dionex Ultimate U3000 HPLC (Dionex, Sunnyvale, CA) with a 5 cm × 75 µm Pico Frit C18 column (New Objective, Woburn MA) directly connected to a New Objectives nanospray emitter (15 µm, New Objectives, Woburn, MA). A 2 µl sample volume was injected and then eluted by use of a gradient (5% – 50% B, 0–28 min, 50% – 75% B, 28–32 min, 75% – 90% B, 32–38 min, 90% - 90% B, 43–48 min) of mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in ACN) at a overall flow rate of 0.3 µl/min. The heated capillary temperature and electrospray voltage were set at 150 °C and 1.5 kV, respectively. All data were acquired in positive ion mode. Data-dependent neutral loss MS3 acquisition was used for all experiments. In these experiments full MS scans (m/z 300–2000) were followed by subsequent MS2 scans on the top five most abundant peptide ions using a normalized collision energy of 35%. When a neutral loss of 98, 49 or 32.7 (H3PO4 for +1, +2 and +3 charge states) was observed the MS3 scan was triggered to isolate and fragment the corresponding neutral loss product ions from the preceding MS2 scan.
DATA ANALYSIS
Caseins DDNL neutral loss MS3 data sets were analyzed by use of MassMatrix [53, 54], Mascot [55], X!Tandem [56] and a hierarchical MS3 search strategy [45]. Histone MS data were searched by use of MassMatrix using the hierarchical MS3 search strategy [45, 56]. Casein tandem MS data were searched against a protein database containing the sequences for bovine α casein S1, S2, β casein and decoy sequences comprised of the reversed casein sequence database. Phosphorylation (serine and threonine residues) and sodium adducts (aspartic acid and glutamic acid) were included as variable modifications. Three missed cleavages were allowed. Trypsin was selected as the enzyme for digestion which will cleave the proteins after lysine and arginine. The mass tolerance is 10 ppm for precursor ion search, 1 Da for product ion.
Histone H1 data were searched against the NCBInr histones and/or NCBInr human protein database (2009-05-26) appended with the equivalent reversed database as decoy sequences. Acetylation on the N terminus, acetylation of lysine, phosphorylation (serine and threonine residues), methylation of lysine and arginine, formylation of lysine and sodium adducts (aspartic acid and glutamic acid) were included as variable modifications. Three missed cleavages were allowed. Trypsin was selected as the enzyme for digestion. The mass tolerance is 0.02 Da for precursor ion search, 1 Da for product ion search. Additionally, the mass tolerance is 5 ppm for precursor ion when the data were searched by several search engines for comparison. All the raw data were converted to .mzData using BioWorks (Thermo Fisher, San Jose, CA) and converted to mzXML. Then a Perl file was used to combine these two files together to generate a third “fixed mzXML” file which was used as the final file for searching the results. False positive rates were calculated as described by Elias et al [57].
REAL TIME REVERSE TRANSCRIPTION-POLYMERASE CHAIN REACTION (RT-PCR) ASSAYS
Quantitative RT–PCR was used to quantify Mcl-1-1 expression (primers available upon request) after flavopiridol treatment. Untreated cells were used as a negative control. RT-PCR was done using 2 µg of total RNA extracted with Trizol reagent (Invitrogen, Carlsbad, CA) and reverse transcribed by Moloney murine leukemia virus reverse transcriptase (Invitrogen). The comparative cycle threshold (CT) method was used to determine target gene expression levels relative to an internal control 18S.
RESULTS AND DISCUSSION
VALIDATION OF PHOSPHOPROTEOMIC WORKFLOW AND HIERARCHICAL DATA ANALYSIS
A component of this work is the novel hierarchical data analyses of DDNL MS3 data from enriched phosphopeptides [45]. Before carrying out the workflow on AML cells we first validated the workflow with commercially available phosphoproteins. Casein phosphorylation has been extensively characterized and was thus selected as a control to validate the proteomic platform [23, 26, 27, 34, 35, 40, 45, 58–60]. Zirconium dioxide and titanium dioxide solid phase extraction were used in parallel to enrich phosphopeptides from casein tryptic digests [61]. Polyoxyanions including phosphate, sulfate and carboxylate show high binding selectivity towards ZrO2 and TiO2 [62]. It was anticipated that ZrO2 would show a higher selectivity towards the phosphate group because it is a stronger Lewis acid than other polyoxy anions [63]. Initially, 0.1 % formic acid was used to wash the sample after loading to remove nonspecific binding peptides [26]. However, the 0.1% formic acid solution did not adequately eliminate nonspecific binding in all samples. Several alternative elution conditions were then evaluated. Larsen et al. reported the use of 50%–80% ACN and 20 mg/mL 2,5-dihydroxybenzoic acid in 0.1% TFA as loading and washing solutions for enhanced selectivity and improved MALDI TOF analysis [27]. While they reported exciting results using these conditions, the presence of the DHBA would be problematic for LC-MS/MS. Ishihama et al. reported the use of 50% acetonitrile with 0.3% TFA as an effective loading buffer for IMAC [25]. They concluded that this loading buffer will effectively neutralize acidic residues whilst minimizing nonspecific hydrophobic binding between peptides and IMAC columns. We found that this later condition (50% acetonitrile with 0.3% TFA) was also optimal as the loading and washing buffers for the zirconium dioxide and titanium dioxide solid phase extraction of phosphopeptides. To elute the phosphopeptides, the column was washed with 0.1% ammonia (pH 10.5) [26]. We observed that ZrO2 showed stronger binding affinity than TiO2 toward singly phosphorylated peptides as previously reported [64]. The total number of the phosphopeptides identified after ZrO2 enrichment was greater compared with TiO2 enrichment. We identified 234 and 149 phosphopeptides from α casein sample after ZrO2 and TiO2 enrichment respectively, while the number of phosphopeptides identified from β casein was 212 and 126, respectively using the same approach. Therefore, ZrO2 shows higher binding affinity to phosphate groups than TiO2 according to the MS spectral counting results [65, 66].
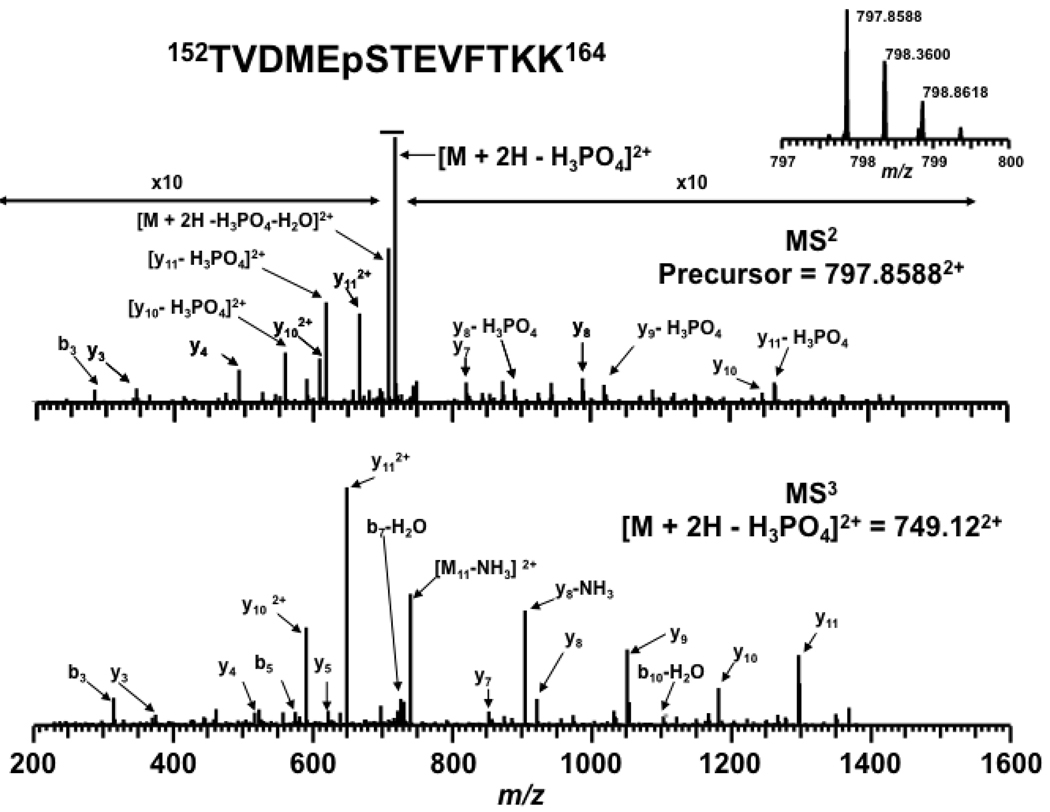
The neutral loss of the side-chain phosphate may dominate product ion formation and can serve as a signature ion for phosphopeptides. Unfortunately, when the neutral loss product ion dominates the MS2 spectrum it may be difficult to identify the phosphopeptide and locate the phosphorylation site. This problem has been largely alleviated by the use of data-dependent neutral loss MS3 experiments [25, 41, 43, 67–70]. In DDNL MS3 experiments the dominant NL product ion is selected and fragmented by CID yielding a higher abundance of product ions from peptide backbone fragmentation. An example is shown in Figure 1 where the top spectrum is from the MS2 and the bottom is for the MS3 of the neutral loss product ions (−98 Da). The MS2 data show the expected NL as well as a low abundance of peptide backbone cleavages. The MS2 and MS3 data are then used to identify the peptide and locate the site of phosphorylation. The MS2 and MS3 data are often analyzed independently and the results merged to form a master peptide list [44]. However, in our workflow we evaluated the MS2 and MS3 data in series through the use of a hierarchical database search algorithm [45]. In this approach the MS2 data is first analyzed to obtain a list of peptide matches based on the molecular weight of the precursor ion as well as peptide backbone cleavages if present. The MS3 spectrum generated from the DDNL experiment is then evaluated against all the possible neutral loss products obtained from sequences returned in the MS2 analysis. The constraints on the MS3 database search from the MS2 search results limits the number of potential false positive candidates resulting in overall improved confidence in peptide matches. The direct analysis of the casein tryptic digests using phosphopeptide enrichment, nano-ESI-LC-MS3 combined with hierarchical MS2/MS3 database search resulted in the identification of 70 unique casein peptides including 26 unique casein phosphopeptides (Table 1) with no matches returned from the decoy database. Two phosphorylation sites were identified by our workflow which were not reported [26, 27, 40, 60, 71, 72]. One is phosphorylation at T56 from the peptide 48FQpSEEQQQpTEDELQDK63 of β casein [26, 27, 40] (Table 1 and Supplementary Figure 1). Another one is phosphorylation of Ser 44 from the peptide 39NMAINPpSKENLCSTFCK55 of α casein S2. We also detected several peptides with multiple phosphorylations that undergo multiple neutral losses resulting in poor quality tandem MS spectra [59, 60, 73] (Table 1 and Supplementary Figure 2).
Figure 1.
Identification of phosphorylation sites using hierarchical MS2/MS3 data analysis. (Top) MS2 spectrum of the doubly charged precursor ion at m/z of 797.86 Da. (Bottom) MS3 spectrum of neutral loss product ion at m/z 749.12. The peptide was identified as α-casein S2 phosphopeptide 152TVDMEpSTEVFTKK164. The MS2 spectrum is magnified to accentuate the lower abundant y ions and b ions.
Table 1.
Bovine milk casein phosphopeptides identified by hierarchical data analysis.
| Casein Isoform |
Peptide Sequence | # of Phos |
Expt. Mass | Calc. Mass | Method | Ref. | |
|---|---|---|---|---|---|---|---|
| α-S1 |
58DIGpSESTEDQAMEDIK73 or 58DIGSEpSTEDQAMEDIK73 |
1 | 1847.7281 | (+2) | 1847.7223 | Zr, Ti | [26, 27, 40, 60] |
| 58DIGpSEpSTEDQAMEDIK73 | 2 | 1927.6937 | (+2) | 1927.6916 | Zr, Ti | [26, 60, 71, 72] | |
| 1927.6919 | (+3) | 1927.6916 | Zr, Ti | ||||
| 58DIGpSEpSpTEDQAMEDIK73 | 3 | 2008.6677 | (+2) | 2007.6580 | Zr | [26] | |
| 2029.6459 | (+2) | 2029.6399 | Zr, Ti | ||||
| 52VNELpSK57 | 1 | 769.34972 | (+2) | 769.34915 | Zr | [27], [40, 60] | |
| 50EKVNELpSK57 | 1 | 1026.4879 | (+2) | 1026.4868 | Zr | [27], [40, 60] | |
| 1048.4691 | (+2) | 1048.4687b | Zr | [27, 40, 60] | |||
| 50VNELSKDIGpSEpSTEDQAMEDIK57 | 2 | 2599.0676 | (+3) | 2598.0566a | Zr, Ti | [27, 40, 60] | |
| 50VNELpSKDIGpSEpSTEDQAMEDIK57 | 3 | 2679.0326 | (+3) | 2678.0229a | Zr, Ti | [27, 60] | |
| 2679.0315 | (+2) | 2678.0229a | Zr, Ti | ||||
| 121VPQLEIVPNpSAEER134 | 1 | 1682.7761 | (+3) | 1682.7775b | Zr, Ti | [26, 27, 40, 58, 60, 71, 72] | |
| 1660.7955 | (+2) | 1660.7942 | Zr, Ti | ||||
| 119YKVPQLEIVPNpSAEER134 | 1 | 1951.9575 | (+2) | 1951.9525 | Zr, Ti | [26, 27, 40, 60, 71, 72] | |
| 1951.9544 | (+3) | 1951.9525 | Zr, Ti | ||||
| 118KYKVPQLEIVPNpSAEER134 | 1 | 2102.0270 | (+3) | 2102.0295b | Ti | [27], [26, 60] | |
| 74QMEAEpSISpSpSEEIVPNpSVEQK94 | 4 | 2662.9280 | (+3) | 2662.9286b | Zr, Ti | [27], [26, 60] | |
| 74QMEAEpSIpSpSpSEEIVPNpSVEQK94 | 5 | 2743.9004 | (+3) | 2742.8950b | Zr | [27], [26, 60, 71, 72] | |
| α-S2 | 17NTMEHVpSpSpSEESIIpSQETYKQEK39 | 4 | 3005.1161 | (+3) | 3004.1010a | Zr, Ti | [27], [26, 60, 71, 72] |
| 39NMAINPpSKENLCSTFCK55 | 1 | 1979.8247 | (+3) | 1979.8425 | Zr | - | |
| 61NANEEEYSIGpSpSpSEEpSAEVATEEVK85 | 4 | 3008.59 | (+2) | 3008.0295c | Zr | [26, 60, 72] | |
| 61NANEEEYSIGpSpSpSEEpSAEVATEEVK85 | 4 | 3031.0155 | (+2) | 3030.0115a,b | Ti | [26, 60, 71, 72] | |
| 3030.0242 | (+3) | 3030.0116b | Zr, Ti | ||||
| 152KTVDMEpSTEVFTK164 | 1 | 1722.8033 | (+3) | 1722.8020 | Zr, Ti | [26, 60, 71] | |
| 153TVDMEpSTEVFTKK165 | 1 | 1594.7097 | (+2) | 1594.7071 | Zr, Ti | [27, 60, 71] | |
| 153TVDMEpSTEVFTK164 | 1 | 1466.6127 | (+2) | 1466.6121 | Zr, Ti | [27, 60, 71] | |
| 141EQLpSTpSEENSK151 | 2 | 1539.6015 | (+2) | 1539.5976 | Zr | [27], [26, 60, 72] | |
| β | 48FQpSEEQQQTEDELQDK 63 | 1 | 2061.8303 | (+2) | 2061.8285 | Zr, Ti | [26, 27, 40, 60, 72] |
| 2061.8300 | (+3) | 2061.8285 | Zr, Ti | ||||
| 48FQpSEEQQQpTEDELQDK 63 | 2 | 2142.7993 | (+2) | 2141.7940a | Zr | - | |
| 45IEKFQpSEEQQQTEDELQDK63 | 1 | 2433.0548 | (+3) | 2432.0505a | Zr, Ti | [27], [26, 60] | |
| 44KIEKFQpSEEQQQTEDELQDK63 | 1 | 2561.1506 | (+3) | 2560.1445a | Ti | [27], [26, 60, 72] | |
Isotopic Precursor
[M+Na]+
A detailed comparison of hierarchical MS3 search results [45] and parallel search results obtained with Mascot [55] and X!Tandem [56] for high and low mass accuracy data confirmed the improvement in sensitivity and specificity. Overall, hierarchical data analysis significantly reduced the number of false positives matching resulting in improved sensitivity and specificity. For sake of comparison, peptides identified in other reports are also denoted in Table 1 [26, 27, 40, 60, 71, 72].
HISTONE H1 PHOSPHORYLATION
Histone H1 has long been used to assess kinase activity in vitro. By examining the changes in H1 phosphorylation using mass spectrometry we can investigate kinase inhibition within model and primary cells. Such an approach would be beneficial for preclinical assessment of new targeted therapies on model and primary cells treated ex vivo. Histone H1s were first fractionated by RP-HPLC. The collected factions were digested with trypsin. ZrO2 solid phase extraction was used to enrich H1 phosphopeptides for reasons described above. We identified 4 phosphorylation sites on H1.5, 4 phosphorylation sites on H1.4, 2 phosphorylation sites on H1.3, 2 phosphorylation sites on H1.2 and 1 phosphorylation site from H1.x (Table 2). Most of the CDK dependent phosphorylation sites occur within the C-terminal tails of H1 at residues T138, T155 of H1.1; T146, S173, T154 of H1.2; T147, T155, S189, of H1.3; T146, T154, S172, S187 of H1.4; and T138, T155, S173, S189 of H1.5 [3, 74–77]. N-terminal phosphorylation on the tails is also CDK dependent: S31 of H1.X; T31 of H1.2; T18 of H1.3 and H1.4; and S18 of H1.5 [3, 76, 77]. Non-CDK phosphorylation sites have also been reported and include S2 of H1.1, S2, S36 and T165 of H1.2; S36 and T180 of H1.3; S2, T4, S27, and S36 of H1.4, and S2, T4 and T11 of H1.5 [3, 76]. The data from our phosphoproteomic workflow of H1 phosphopeptides corroborated many of the previously reported sites of H1 phosphorylation (Table 2 and Supplementary Figure 3). Ser 2 of H1.x was identified as a novel phosphorylation sites by our workflow. Although there was no MS3 data for this peptide (Ac2pSVELEEALPVTTAEGMAK19), the unmodified MS spectra in the same experiment confirmed the identification (Supplementary Figure 3b).
Table 2.
Histone phosphorylation sites detected in Kasumi-1 cells identified by hierarchical data analysis.
| H1 isoform | Peptide Sequence | Exp. Mass | calc. Mass | Ref | |
|---|---|---|---|---|---|
| H1.2,H1.3,H1.4 | 34KApSGPPVSELITK46 | 1406.7270 | (+3) | 1406.7291 | [3, 75, 76] |
| 34KApSGPPVSELITK46 | 1406.7272 | (+2) | 1406.7291 | ||
| 33RKApSGPPVSELITK46 | 1562.8277 | (+3) | 1562.8302 | ||
| H1.2 |
2SEpTAPAAPAAAPPAEK17 or 2pSETAPAAPAAAPPAEK17 |
1558.7200 | (+2) | 1558.7149 | [3, 76] |
| H1.3 | Ac2SETAPLAPTIPAPAEKpTPVK21 | 2140.0970 | (+2) | 2140.0937 | [3] |
| Ac2SETAPLAPTIPAPAEKpTPVKK22 | 2268.1981 | (+3) | 2268.1887 | ||
| Ac2SETAPLAPTIPAPAEKpTPVKKK23 | 2396.3062 | (+3) | 2396.2837 | ||
| H1.4 | Ac2SETAPAAPAAPAPAEKpTPVK21 | 2026.0060 | (+2) | 2025.9893 | [3, 75, 76] |
|
Ac2SEpTAPAAPAAPAPAEKpTPVK21 or Ac2pSETAPAAPAAPAPAEKpTPVK21 |
2063.9547 | (+2) | 2063.9450 | ||
| Ac2SETAPAAPAAPAPAEKpTPVKK22 | 2154.0865 | (+2) | 2154.0842 | ||
| Ac2SETAPAAPAAPAPAEKpTPVKKK23 | 2282.1954 | (+3) | 2282.1792 | ||
| Ac2SEpTAPAAPAAPAPAEKpTPVKKK23 | 2362.1631 | (+3) | 2362.1455 | ||
| 2SETAPAAPAAPAPAEKpTPVKK22 | 2112.0893 | (+3) | 2112.0737 | ||
| 2SETAPAAPAAPAPAEKpTPVKKK23 | 2240.1874 | (+3) | 2240.1686 | ||
| H1.5 |
Ac2pSETAPAETATPAPVEK17 or Ac2SEpTAPAETATPAPVEK17 |
1720.7782 | (+2) | 1720.7677 | [3] |
| Ac2SETAPAETATPAPVEKpSPAK21 | 2103.9844 | (+2) | 2103.9846 | [3, 77] | |
| 2104.9895 | (+3) | 2104.9880a | |||
| 2SETAPAETATPAPVEKpSPAKK22 | 2190.0738 | (+3) | 2190.0690 | ||
| 2SETAPAETATPAPVEKpSPAK21 | 2061.9744 | (+2) | 2061.9740 | ||
| Ac2SETAPAETApTPAPVEKpSPAK21 | 2183.9683 | (+2) | 2183.9509 | ||
|
Ac2SEpTAPAETATPAPVEKpSPAK21or Ac2SETAPAETATPAPVEKpSPAK21 |
2183.9515 | (+2) | 2183.9509 | ||
|
Ac2SETAPAETATPAPVEKpSPAKK22 or Ac2SETAPAETApTPAPVEKSPAKK22 |
2232.0800 | (+2) | 2232.0795 | ||
| Ac2SETAPAETATPAPVEKpSPAKKK23 | 2360.1731 | (+3) | 2360.1754 | ||
|
2SETAPAEpTATPAPVEKpSPAKK22 or 2SEpTAPAETATPAPVEKpSPAKK22 |
2270.0358 | (+4) | 2270.0353 | [3] | |
|
Ac2pSETAPAETATPAPVEKpSPAKK22 or Ac2SEpTAPAETATPAPVEKpSPAKK22 |
2312.0614 | (+3) | 2312.0459 | ||
|
Ac2pSETAPAETATPAPVEKpSPAKKK23 or Ac2SEpTAPAETATPAPVEKpSPAKKK23 |
2440.1457 | (+3) | 2440.1408 | ||
| H1.x | Ac2pSVELEEALPVTTAEGMAK19 | 1996.9308 | (+2) | 1996.9185 | N/A |
H1 PHOSPHORYLATION DURING DRUG TREATMENT
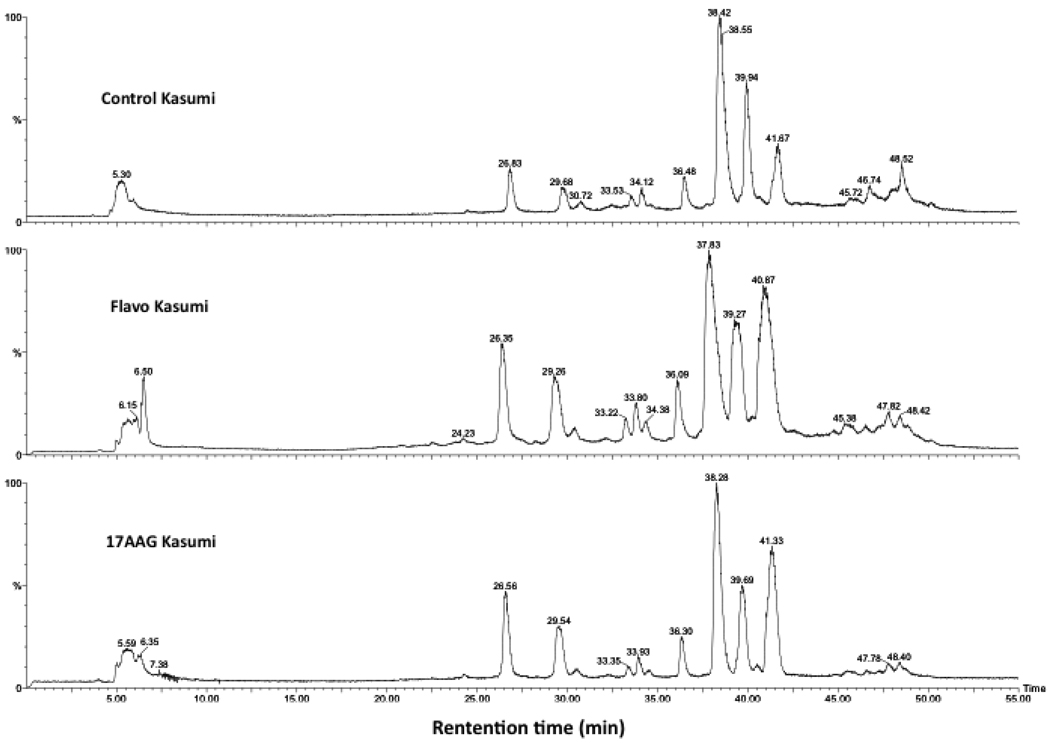
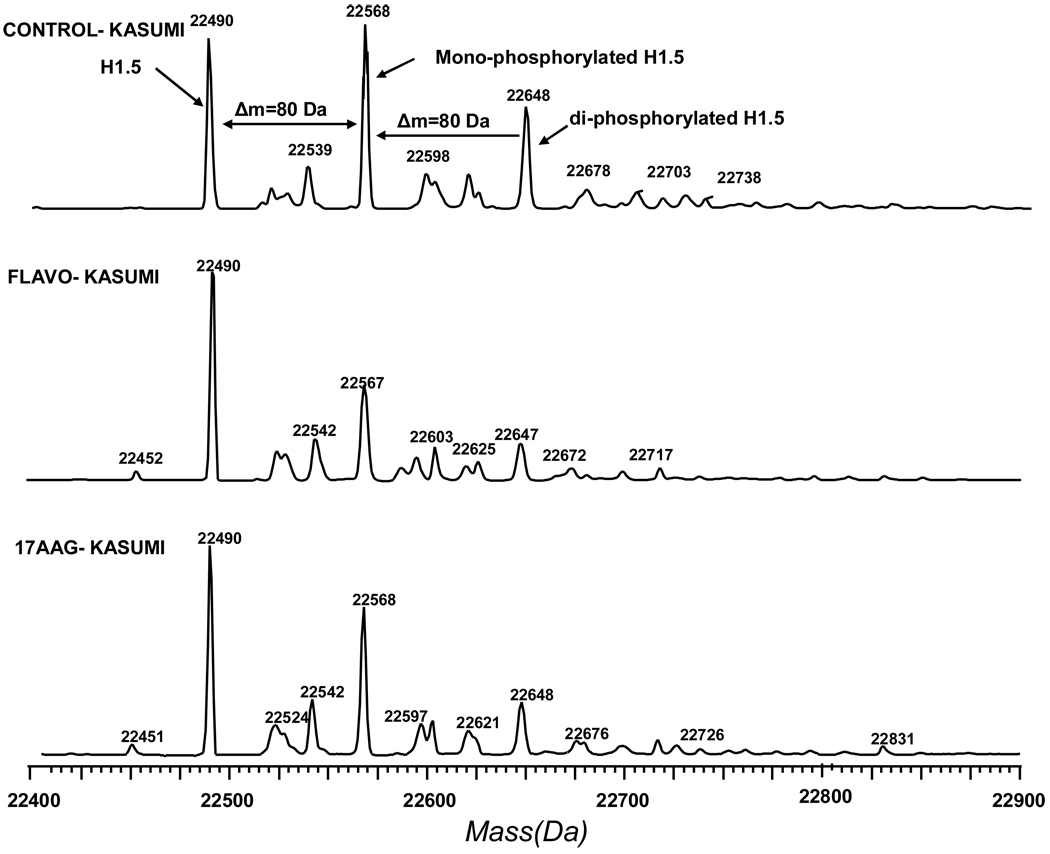
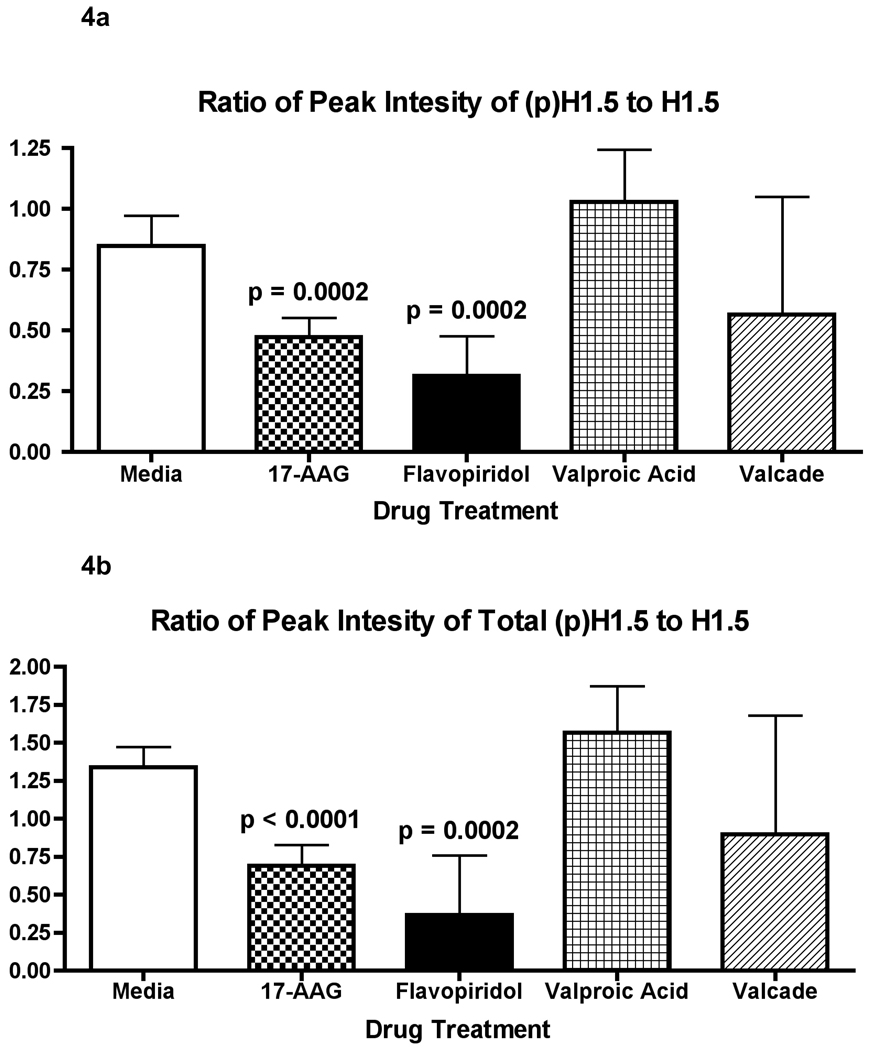
We next applied the LC-MS histone profiling approach to assess histone phosphorylation changes in the acute myeloid leukemia model cell line (Kasumi-1) associated with chemotherapy. Acid-extracted histones from Kasumi-1 cells were profiled by LC-MS. The histones were well separated and each peak was deconvoluted by MaxEnt algorithm in MassLynx 4.0. The first peak, eluted from the column at a retention time greater than 20 minutes, was identified as N-terminally acetylated H1.5 (Mw = 22,491 Da). The second peak, eluting two to three minutes later than the H1.5 peak, was a mixture of N-terminally acetylated H1.4 (Mw = 21,774 Da) and H1.3 (Mw = 22,257 Da) (Figure 2 and Supplementary Figure 4). To examine the effect of drug treatment on histone H1, we treated Kasumi-1 cells with flavopiridol and 17AAG for 24 h. For these drugs, we observed that the phosphorylation of H1.5 decreased dramatically (Figure 3 and Supplementary Figure 5). To further demonstrate the decrease in phosphorylation induced by flavopiridol and 17AAG, a bar chart was constructed showing the relative abundance of monophosphorylated H1.5 to unphosphorylated H1.5 in each drug treatment (Figure 4a). A significant decrease in the peak abundance ratio was observed when compared with the untreated Kasumi-1 control (N=3–4 biological replicates per treatment). Furthermore, a similar chart was constructed showing the ratio of the total phosphorylated H1.5 peak abundances to the unphosphorylated H1.5 peak abundance for each drug treatment (Figure 4b). These data demonstrate the significant change in H1.5 phosphorylation in response to flavopiridol and 17AAG treatments in vitro.
Figure 2.
HPLC separation of histones derived from Kasumi-1 cells. Top: histones from Kasumi-1 cells. Middle and Bottom: histones from Kasumi-1 cells treated by flavopiridol and 17 AAG for 24 h, respectively
Figure 3.
Deconvoluted mass spectra of H1.5. Top: Mass distribution of Histone H1.5 from control Kasumi-1 cells, the first peak at 22,490 Da is the mass peak of unmodified H1.5, the peak at 22,567 Da is mono-phosphorylated H1.5, the peak at 22,648 Da is diphosphorylated H1.5; Middle & Bottom: Mass distribution of histone H1.5 from Kasumi-1 cells treated with Flavopiridol and 17 AAG, respectively. Mono- and di-phosphorylated H1.5 peak magnititude of the cells treated by Flavopiridol and 17 AAG was decreased compared to the mass spectra of the control sample.
Figure 4.
Bar chart depicting the changes in the monophosphorylated (top) and total phosphorylated (bottom) H1.5 peak abundance in response to each drug treatment. The charts show the significant decrease in the peak ratios for the flavopiridol and 17AAG drug treatments when compared to the untreated Kasumi control as determined by a student’s t-test (N=3–4 for each sample). The other drug treatment’s abundance ratios were not significantly different from the Kasumi control abundance ratio. The error bars show the 95% confidence interval for the abundance ratios.
17 AAG has been reported to induce apoptosis of AML cells including Kasumi-1 cells by inhibiting Hsp90 [78–83]. Hsp90 is strong binder of histones and their kinases. Early reports suggested that HSP90 is autophosphorylated in the presence of histones and in turn phosphorylates the histones inducing chromatin condensation [84, 85]. The most plausible explanation for this activity is that early Hsp90 cellular isolates also contained bound kinases. CDKs are known substrates of Hsp90 and the inhibition of Hsp90 by 17AAG has been show to disrupt their activity [49–51]. Our data show that treatment with 17AAG resulted in a loss of H1.5 phosphorylation supporting a decrease in kinase activity induced by this therapy.
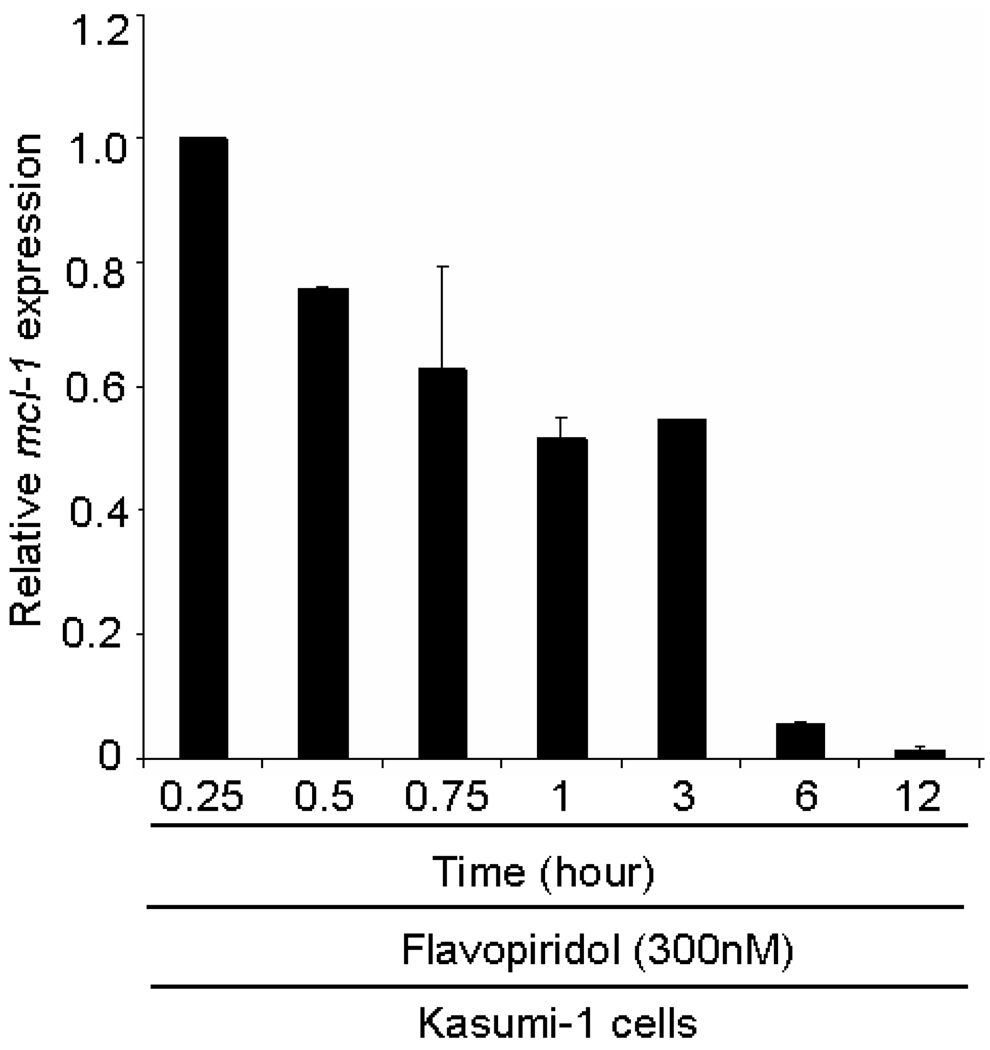
Flavopiridol, the first active cyclin-dependent kinase inhibitor, has been reported to induce apoptosis in CML (chronic myelogenous leukemia) and AML cells [86–89]. Flavopiridol treatment can lead to apoptosis via a mechanism that has been associated with down-regulation of a key antiapoptotic protein Mcl-1 in lung carcinoma cells [90]. This down-regulation can lead to release of the histone kinase Cdc2 / Cdk1. However, flavopiridol is also a potent Cdk inhibitor. We examined the expression of Mcl-1 by RT-PCR in Kasumi-1 cells (300 nM flavopiridol treatment from 15 min to 12 h). Three replicates were obtained and the percent expression of Mcl-1 relative to the control was plotted versus the time of treatment with flavopiridol. As shown in Figure 5, the Mcl-1 expression decreased significantly in a time-responsive manner and Mcl-1 expression was barely detectable after 12 hour-treatment with flavopiridol. Despite the decrease in Mcl-1, our data show that flavopiridol was effective at inhibiting phosphorylation of H1.5.
Figure 5.
Quantitative RT–PCR showed that flavopiridol treatment reduces the Mcl-1 expression in Kasumi-1 cells.
The decrease of H1.2 and H1.5 phosphorylation are reported to be markers for early onset of apoptosis. Kratzmeier et al. described the rapid dephosphorylation of certain histone subtypes shortly after induction of apoptosis and before the onset of internucleosomal cleavage [91]. The phosphorylation of linker histone H1 has been associated with the regulation of oncogene expression, DNA damage repair and chromatin decondensation [4–6]. The role of H1 phosphorylation is postulated to modulate DNA recognition and/or DNA cleavage by the caspase 3-mediated activation of caspase activated DNase. However, we postulated that treatment of 17AAG and flavopiridol result in dephosphorylation of H1.5 principally through inhibition of kinase signaling pathways. Because these two drugs are examples of direct and indirect inhibition of CDK activity, we also examined two additional drugs with different primary mechanisms of action. These drugs, velcade [92] and valproic acid [93] act via proteome inhibition and histone deacetylase inhibition. While each of these drugs has been shown to induce apoptosis or inhibit proliferation in myeloid cells, none of these drugs showed significant changes in H1.5 phosphorylation when compared to the untreated Kasumi-1 control (see Supplementary Figures 5 and 6) [93–98].
CONCLUSIONS
To facilitate the characterization of phosphorylation sites and improve the reliability of phosphopeptides identification we employed a proteomic workflow that combined phosphopeptide enrichment, data-dependent neutral loss MS3 and hierarchical data analysis. The workflow was first validated against standard casein phosphoproteins and then applied to Histone H1 phosphoproteins from Kasumi-1 cells. Known phosphorylation sites of caseins and histone H1 variants were successfully identified as well as several novel sites of phosphorylations. Furthermore, by examining the histone phosphorylation profiles in Kasumi-1 cancer cell lines using LC-MS we were able to monitor modulation of histone H1 phosphorylation associated with flavopiridol and 17AAG chemotherapy. The data presented herein demonstrate the histones may be a potent marker to assess the cellular response of therapy for reagents that have direct and indirect effects on kinase activity.
Supplementary Material
ACKNOWLEDGEMENTS
The study was funded by the Ohio State University, the National Institutes of Health (CA107106, RR023647, CA101956), the V Foundation (AACR Translational Cancer Research Grant) and the Leukemia & Lymphoma Society (SCOR).
REFERENCES
- 1.Thoma F, Koller T. Influence of histone H1 on chromatin structure. Cell. 1977;12:101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- 2.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wisniewski JR, Zougman A, Kruger S, Mann M. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics. 2007;6:72–87. doi: 10.1074/mcp.M600255-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Roth SY, Allis CD. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem Sci. 1992;17:93–98. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- 5.Herrera RE, Chen F, Weinberg RA. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc Natl Acad Sci U S A. 1996;93:11510–11515. doi: 10.1073/pnas.93.21.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopfgartner AG, Sleno L, Loftus N, Warrander J, Ashton S. A statistical based approach in metabolite identification by high mass accuracy MSn analysis. 55th ASMS Conference on MAss Spectrometry and Allied Topics; 2007. p. 34. Oral. [Google Scholar]
- 7.Matsumoto Y, Yasuda H, Mita S, Marunouchi T, Yamada M. Evidence for the involvement of H1 histone phosphorylation in chromosome condensation. Nature. 1980;284:181–183. doi: 10.1038/284181a0. [DOI] [PubMed] [Google Scholar]
- 8.Yasuda H, Matsumoto Y, Mita S, Marunouchi T, Yamada M. A mouse temperature-sensitive mutant defective in H1 histone phosphorylation is defective in deoxyribonucleic acid synthesis and chromosome condensation. Biochemistry. 1981;20:4414–4419. doi: 10.1021/bi00518a028. [DOI] [PubMed] [Google Scholar]
- 9.Roberge M, Th'ng J, Hamaguchi J, Bradbury EM. The topoisomerase II inhibitor VM-26 induces marked changes in histone H1 kinase activity, histones H1 and H3 phosphorylation, and chromosome condensation in G2 phase and mitotic BHK cells. J Cell Biol. 1990;111:1753–1762. doi: 10.1083/jcb.111.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Th'ng JP, Guo XW, Swank RA, Crissman HA, Bradbury EM. Inhibition of histone phosphorylation by staurosporine leads to chromosome decondensation. J Biol Chem. 1994;269:9568–9573. [PubMed] [Google Scholar]
- 11.Okamura H, Yoshida K, Amorim BR, Haneji T. Histone H1.2 is translocated to mitochondria and associates with Bak in bleomycin-induced apoptotic cells. J Cell Biochem. 2008;103:1488–1496. doi: 10.1002/jcb.21537. [DOI] [PubMed] [Google Scholar]
- 12.Gine E, Crespo M, Muntanola A, Calpe E, et al. Induction of histone H1.2 cytosolic release in chronic lymphocytic leukemia cells after genotoxic and non-genotoxic treatment. Haematologica. 2008;93:75–82. doi: 10.3324/haematol.11546. [DOI] [PubMed] [Google Scholar]
- 13.Zong WX. Histone, H1.2: another housekeeping protein that kills. Cancer Biol Ther. 2004;3:42–43. doi: 10.4161/cbt.3.1.738. [DOI] [PubMed] [Google Scholar]
- 14.Konishi A, Shimizu S, Hirota J, Takao T, et al. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 2003;114:673–688. doi: 10.1016/s0092-8674(03)00719-0. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie DA, Vousden KH. The secret life of histones. Cell. 2003;114:655–656. doi: 10.1016/s0092-8674(03)00723-2. [DOI] [PubMed] [Google Scholar]
- 16.Snijders AP, Pongdam S, Lambert SJ, Wood CM, et al. Characterization of post-translational modifications of the linker histones H1 and H5 from chicken erythrocytes using mass spectrometry. J Proteome Res. 2008;7:4326–4335. doi: 10.1021/pr800260a. [DOI] [PubMed] [Google Scholar]
- 17.Villar-Garea A, Imhof A. Fine mapping of posttranslational modifications of the linker histone H1 from Drosophila melanogaster. PLoS One. 2008;3:e1553. doi: 10.1371/journal.pone.0001553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia BA, Joshi S, Thomas CE, Chitta RK, et al. Comprehensive phosphoprotein analysis of linker histone H1 from Tetrahymena thermophila. Mol Cell Proteomics. 2006;5:1593–1609. doi: 10.1074/mcp.M600086-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Deterding LJ, Banks GC, Tomer KB, Archer TK. Understanding global changes in histone H1 phosphorylation using mass spectrometry. Methods. 2004;33:53–58. doi: 10.1016/j.ymeth.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 20.Mizzen CA. Purification and analyses of histone H1 variants and H1 posttranslational modifications. Methods Enzymol. 2004;375:278–297. doi: 10.1016/s0076-6879(03)75019-8. [DOI] [PubMed] [Google Scholar]
- 21.Mann M, Ong SE, Gronborg M, Steen H, et al. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 22.Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, et al. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc Natl Acad Sci U S A. 2000;97:179–184. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu WW, Wang G, Rattanasinganchan P, Zhang T, et al. Quantitative Phosphoproteomics: Higher Sensitivity and Multiplexing Using Two-Staged IMAC Enrichment,iTRAQ Labeling, and Linear Ion Trap/PQD. 55th ASMS Conference on MAss Spectrometry and Allied Topics; 2007. poster. [Google Scholar]
- 24.Neville DC, Rozanas CR, Price EM, Gruis DB, et al. Evidence for phosphorylation of serine 753 in CFTR using a novel metal-ion affinity resin and matrix-assisted laser desorption mass spectrometry. Protein Sci. 1997;6:2436–2445. doi: 10.1002/pro.5560061117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishihama Y, Wei FY, Aoshima K, Sato T, et al. Enhancement of the efficiency of phosphoproteomic identification by removing phosphates after phosphopeptide enrichment. J Proteome Res. 2007;6:1139–1144. doi: 10.1021/pr060452w. [DOI] [PubMed] [Google Scholar]
- 26.Kweon HK, Hakansson K. Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal Chem. 2006;78:1743–1749. doi: 10.1021/ac0522355. [DOI] [PubMed] [Google Scholar]
- 27.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 29.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark RC, Dijkstra J. Chemical modification of phosvitin: preparation of dimethylaminovitin and methylmercaptovitin and their utility for elucidation of phosvitin primary structure. Int J Biochem. 1980;11:577–585. doi: 10.1016/0020-711x(80)90268-2. [DOI] [PubMed] [Google Scholar]
- 31.Meyer HE, Hoffmann-Posorske E, Korte H, Heilmeyer LM., Jr Sequence analysis of phosphoserine-containing peptides. Modification for picomolar sensitivity. FEBS Lett. 1986;204:61–66. doi: 10.1016/0014-5793(86)81388-6. [DOI] [PubMed] [Google Scholar]
- 32.Oda Y, Nagasu T, Chait BT. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 33.Goshe MB, Conrads TP, Panisko EA, Angell NH, et al. Phosphoprotein isotope-coded affinity tag approach for isolating and quantitating phosphopeptides in proteome-wide analyses. Anal Chem. 2001;73:2578–2586. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- 34.Liao PC, Leykam J, Andrews PC, Gage DA, Allison J. An approach to locate phosphorylation sites in a phosphoprotein: mass mapping by combining specific enzymatic degradation with matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem. 1994;219:9–20. doi: 10.1006/abio.1994.1224. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Herring CJ, Romano PR, Szczepanowska J, et al. Identification of phosphorylation sites in proteins separated by polyacrylamide gel electrophoresis. Anal Chem. 1998;70:2050–2059. doi: 10.1021/ac971207m. [DOI] [PubMed] [Google Scholar]
- 36.Stensballe A, Andersen S, Jensen ON. Characterization of phosphoproteins from electrophoretic gels by nanoscale Fe(III) affinity chromatography with off-line mass spectrometry analysis. Proteomics. 2001;1:207–222. doi: 10.1002/1615-9861(200102)1:2<207::AID-PROT207>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Adamczyk M, Gebler JC, Wu J. Selective analysis of phosphopeptides within a protein mixture by chemical modification, reversible biotinylation and mass spectrometry. Rapid Commun Mass Spectrom. 2001;15:1481–1488. doi: 10.1002/rcm.394. [DOI] [PubMed] [Google Scholar]
- 38.Thompson AJ, Hart SR, Franz C, Barnouin K, et al. Characterization of protein phosphorylation by mass spectrometry using immobilized metal ion affinity chromatography with on-resin beta-elimination and Michael addition. Anal Chem. 2003;75:3232–3243. doi: 10.1021/ac034134h. [DOI] [PubMed] [Google Scholar]
- 39.Knight ZA, Schilling B, Row RH, Kenski DM, et al. Phosphospecific proteolysis for mapping sites of protein phosphorylation. Nat Biotechnol. 2003;21:1047–1054. doi: 10.1038/nbt863. [DOI] [PubMed] [Google Scholar]
- 40.Ahn YH, Ji ES, Kwon KH, Lee JY, et al. Protein phosphorylation analysis by site-specific arginine-mimic labeling in gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem. 2007;370:77–86. doi: 10.1016/j.ab.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 41.DeGnore JP, Qin J. Fragmentation of phosphopeptides in an ion trap mass spectrometer. J Am Soc Mass Spectrom. 1998;9:1175–1188. doi: 10.1016/S1044-0305(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 42.Ozlu N, Bensadek D, Patterson T, Mitchison T, et al. Robust Method for the Confident Location of Protein Phosphorylation Sites. 55th ASMS Conference on MAss Spectrometry and Allied Topics; 2007. posters. [Google Scholar]
- 43.Zabrouskov V, Senko MW, Du Y, Leduc RD, Kelleher NL. New and automated MSn approaches for top-down identification of modified proteins. J Am Soc Mass Spectrom. 2005;16:2027–2038. doi: 10.1016/j.jasms.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang X, Han G, Feng S, Jiang X, et al. Automatic Validation of Phosphopeptide Identifications by the MS2/MS3 Target-Decoy Search Strategy. J Proteome Res. 2008 doi: 10.1021/pr700675j. [DOI] [PubMed] [Google Scholar]
- 45.Xu H, Wang L, Sallans L, Freitas MA. A hierarchical MS2/MS3 database search algorithm for automated analysis of phosphopeptide tandem mass spectra. Proteomics. 2009;9:1763–1770. doi: 10.1002/pmic.200800282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senderowicz AM. Flavopiridol: the first cyclin-dependent kinase inhibitor in human clinical trials. Invest New Drugs. 1999;17:313–320. doi: 10.1023/a:1006353008903. [DOI] [PubMed] [Google Scholar]
- 47.Byrd JC, Shinn C, Waselenko JK, Fuchs EJ, et al. Flavopiridol induces apoptosis in chronic lymphocytic leukemia cells via activation of caspase-3 without evidence of bcl-2 modulation or dependence on functional p53. Blood. 1998;92:3804–3816. [PubMed] [Google Scholar]
- 48.Workman P. Altered states: selectively drugging the Hsp90 cancer chaperone. Trends Mol Med. 2004;10:47–51. doi: 10.1016/j.molmed.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Georgakis GV, Li Y, Rassidakis GZ, Martinez-Valdez H, et al. Inhibition of heat shock protein 90 function by 17-allylamino-17-demethoxy-geldanamycin in Hodgkin's lymphoma cells down-regulates Akt kinase, dephosphorylates extracellular signal-regulated kinase, and induces cell cycle arrest and cell death. Clin Cancer Res. 2006;12:584–590. doi: 10.1158/1078-0432.CCR-05-1194. [DOI] [PubMed] [Google Scholar]
- 50.Prince T, Sun L, Matts RL. Cdk2: a genuine protein kinase client of Hsp90 and Cdc37. Biochemistry. 2005;44:15287–15295. doi: 10.1021/bi051423m. [DOI] [PubMed] [Google Scholar]
- 51.Burrows F, Zhang H, Kamal A. Hsp90 activation and cell cycle regulation. Cell Cycle. 2004;3:1530–1536. doi: 10.4161/cc.3.12.1277. [DOI] [PubMed] [Google Scholar]
- 52.Ren C, Zhang L, Freitas MA, Ghoshal K, et al. Peptide mass mapping of acetylated isoforms of histone H4 from mouse lymphosarcoma cells treated with histone deacetylase (HDACs) inhibitors. J Am Soc Mass Spectrom. 2005;16:1641–1653. doi: 10.1016/j.jasms.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J Proteome Res. 2008;7:138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 56.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 57.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 58.McCormick DJ, Holmes MW, Muddiman DC, Madden BJ. Mapping sites of protein phosphorylation by mass spectrometry utilizing a chemical-enzymatic approach: characterization of products from alpha-S1 casein phosphopeptides. J Proteome Res. 2005;4:424–434. doi: 10.1021/pr049804u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee CH, McComb ME, Bromirski M, Jilkine A, et al. On-membrane digestion of beta-casein for determination of phosphorylation sites by matrix-assisted laser desorption/ionization quadrupole/time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2001;15:191–202. doi: 10.1002/1097-0231(20010215)15:3<191::AID-RCM209>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 60.Hsieh HC, Sheu C, Shi FK, Li DT. Development of a titanium dioxide nanoparticle pipette-tip for the selective enrichment of phosphorylated peptides. J Chromatogr A. 2007;1165:128–135. doi: 10.1016/j.chroma.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 61.Amplett CB. Inorganic ion exchangers. Amersterdam, The Netherlands: Elsevier; 1964. [Google Scholar]
- 62.Kraus KA, Philips HO, Carlson TA, Johnson JS. Proceedings of the second International Conference on Peaceful Uses of Atomic Energy. Geneva, Switerland: 1958. [Google Scholar]
- 63.Blackwell JA, Carr PW. Fluoride-modified zirconium oxide as a biocompatible stationary phase for high-performance liquid chromatography. J. Chromatogr., A. 1991;549:43–57. [Google Scholar]
- 64.Ballif BA, Roux PP, Gerber SA, MacKeigan JP, et al. Quantitative phosphorylation profiling of the ERK/p90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc Natl Acad Sci U S A. 2005;102:667–672. doi: 10.1073/pnas.0409143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balgley BM, Wang W, Song T, Fang X, et al. Evaluation of confidence and reproducibility in quantitative proteomics performed by a capillary isoelectric focusing-based proteomic platform coupled with a spectral counting approach. Electrophoresis. 2008;29:3047–3054. doi: 10.1002/elps.200800050. [DOI] [PubMed] [Google Scholar]
- 66.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994–999. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 67.Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci U S A. 2004;101:13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gruhler A, Olsen JV, Mohammed S, Mortensen P, et al. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 69.Macek B, Waanders LF, Olsen JV, Mann M. Top-down protein sequencing and MS3 on a hybrid linear quadrupole ion trap-orbitrap mass spectrometer. Mol Cell Proteomics. 2006;5:949–958. doi: 10.1074/mcp.T500042-MCP200. [DOI] [PubMed] [Google Scholar]
- 70.Amoresano A, Monti G, Cirulli C, Marino G. Selective detection and identification of phosphopeptides by dansyl MS/MS/MS fragmentation. Rapid Commun Mass Spectrom. 2006;20:1400–1404. doi: 10.1002/rcm.2461. [DOI] [PubMed] [Google Scholar]
- 71.Wang J, Green K, McGibbon G, McCarry B. Analysis of effect of casein phosphopeptides on zinc binding using mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:1546–1554. doi: 10.1002/rcm.2992. [DOI] [PubMed] [Google Scholar]
- 72.Zhou W, Merrick BA, Khaledi MG, Tomer KB. Detection and sequencing of phosphopeptides affinity bound to immobilized metal ion beads by matrix-assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom. 2000;11:273–282. doi: 10.1016/s1044-0305(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 73.Yang X, Wu H, Kobayashi T, Solaro RJ, van Breemen RB. Enhanced ionization of phosphorylated peptides during MALDI TOF mass spectrometry. Anal Chem. 2004;76:1532–1536. doi: 10.1021/ac035203v. [DOI] [PubMed] [Google Scholar]
- 74.Deterding LJ, Bunger MK, Banks GC, Tomer KB, Archer TK. Global changes in and characterization of specific sites of phosphorylation in mouse and human histone H1 Isoforms upon CDK inhibitor treatment using mass spectrometry. J Proteome Res. 2008;7:2368–2379. doi: 10.1021/pr700790a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parseghian MH, Henschen AH, Krieglstein KG, Hamkalo BA. A proposal for a coherent mammalian histone H1 nomenclature correlated with amino acid sequences. Protein Sci. 1994;3:575–587. doi: 10.1002/pro.5560030406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia BA, Busby SA, Barber CM, Shabanowitz J, et al. Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J Proteome Res. 2004;3:1219–1227. doi: 10.1021/pr0498887. [DOI] [PubMed] [Google Scholar]
- 77.Sarg B, Helliger W, Talasz H, Forg B, Lindner HH. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. J Biol Chem. 2006;281:6573–6580. doi: 10.1074/jbc.M508957200. [DOI] [PubMed] [Google Scholar]
- 78.Yu WJ, Rao Q, Wang M, Tian Z, et al. [The heat shock protein 90 inhibitor induces apoptosis and differentiation of Kasumi-1 and its mechanisms] Zhonghua Xue Ye Xue Za Zhi. 2005;26:728–731. [PubMed] [Google Scholar]
- 79.George P, Bali P, Annavarapu S, Scuto A, et al. Combination of the histone deacetylase inhibitor LBH589 and the hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood. 2005;105:1768–1776. doi: 10.1182/blood-2004-09-3413. [DOI] [PubMed] [Google Scholar]
- 80.George P, Bali P, Cohen P, Tao J, et al. Cotreatment with 17-allylamino-demethoxygeldanamycin and FLT-3 kinase inhibitor PKC412 is highly effective against human acute myelogenous leukemia cells with mutant FLT-3. Cancer Res. 2004;64:3645–3652. doi: 10.1158/0008-5472.CAN-04-0006. [DOI] [PubMed] [Google Scholar]
- 81.Hawkins LM, Jayanthan AA, Narendran A. Effects of 17-allylamino-17-demethoxygeldanamycin (17-AAG) on pediatric acute lymphoblastic leukemia (ALL) with respect to Bcr-Abl status and imatinib mesylate sensitivity. Pediatr Res. 2005;57:430–437. doi: 10.1203/01.PDR.0000153871.45184.19. [DOI] [PubMed] [Google Scholar]
- 82.Mesa RA, Loegering D, Powell HL, Flatten K, et al. Heat shock protein 90 inhibition sensitizes acute myelogenous leukemia cells to cytarabine. Blood. 2005;106:318–327. doi: 10.1182/blood-2004-09-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas X, Campos L, Le QH, Guyotat D. Heat shock proteins and acute leukemias. Hematology. 2005;10:225–235. doi: 10.1080/10245330500093120. [DOI] [PubMed] [Google Scholar]
- 84.Csermely P, Kajtar J, Hollosi M, Oikarinen J, Somogyi J. The 90 kDa heat shock protein (hsp90) induces the condensation of the chromatin structure. Biochem Biophys Res Commun. 1994;202:1657–1663. doi: 10.1006/bbrc.1994.2124. [DOI] [PubMed] [Google Scholar]
- 85.Park M, Yong Kang C, Krishna P. Brassica napus hsp90 can autophosphorylate and phosphorylate other protein substrates. Mol Cell Biochem. 1998;185:33–38. doi: 10.1023/a:1006884306169. [DOI] [PubMed] [Google Scholar]
- 86.Liesveld JL, Abboud CN, Lu C, McNair C, et al. Flavonoid effects on normal and leukemic cells. Leuk Res. 2003;27:517–527. doi: 10.1016/s0145-2126(02)00265-5. [DOI] [PubMed] [Google Scholar]
- 87.Rosato RR, Almenara JA, Kolla SS, Maggio SC, et al. Mechanism and functional role of XIAP and Mcl-1 down-regulation in flavopiridol/vorinostat antileukemic interactions. Molecular cancer therapeutics. 2007;6:692–702. doi: 10.1158/1535-7163.MCT-06-0562. [DOI] [PubMed] [Google Scholar]
- 88.Dasmahapatra G, Almenara JA, Grant S. Flavopiridol and histone deacetylase inhibitors promote mitochondrial injury and cell death in human leukemia cells that overexpress Bcl-2. Molecular pharmacology. 2006;69:288–298. doi: 10.1124/mol.105.016154. [DOI] [PubMed] [Google Scholar]
- 89.Hubeek I, Peters GJ, Broekhuizen AJ, Sargent J, et al. Potentiation of in vitro ara-C cytotoxicity by ribonucleotide reductase inhibitors, cyclin-dependent kinase modulators and the DNA repair inhibitor aphidicolin in paediatric acute myeloid leukaemia. British journal of haematology. 2005;131:219–222. doi: 10.1111/j.1365-2141.2005.05760.x. [DOI] [PubMed] [Google Scholar]
- 90.Ma Y, Cress WD, Haura EB. Flavopiridol-induced apoptosis is mediated through up-regulation of E2F1 and repression of Mcl-1. Mol Cancer Ther. 2003;2:73–81. [PubMed] [Google Scholar]
- 91.Kratzmeier M, Albig W, Hanecke K, Doenecke D. Rapid dephosphorylation of H1 histones after apoptosis induction. J Biol Chem. 2000;275:30478–30486. doi: 10.1074/jbc.M003956200. [DOI] [PubMed] [Google Scholar]
- 92.Colado E, Alvarez-Fernandez S, Maiso P, Martin-Sanchez J, et al. The effect of the proteasome inhibitor bortezomib on acute myeloid leukemia cells and drug resistance associated with the CD34+ immature phenotype. Haematologica. 2008;93:57–66. doi: 10.3324/haematol.11666. [DOI] [PubMed] [Google Scholar]
- 93.Liu S, Klisovic RB, Vukosavljevic T, Yu J, et al. Targeting AML1/ETO-histone deacetylase repressor complex: a novel mechanism for valproic acid-mediated gene expression and cellular differentiation in AML1/ETO-positive acute myeloid leukemia cells. J Pharmacol Exp Ther. 2007;321:953–960. doi: 10.1124/jpet.106.118406. [DOI] [PubMed] [Google Scholar]
- 94.Gomez-Bougie P, Wuilleme-Toumi S, Menoret E, Trichet V, et al. Noxa up-regulation and Mcl-1 cleavage are associated to apoptosis induction by bortezomib in multiple myeloma. Cancer Res. 2007;67:5418–5424. doi: 10.1158/0008-5472.CAN-06-4322. [DOI] [PubMed] [Google Scholar]
- 95.Matondo M, Bousquet-Dubouch MP, Gallay N, Uttenweiler-Joseph S, et al. Proteasome inhibitor-induced apoptosis in acute myeloid leukemia: A correlation with the proteasome status. Leuk Res. 2009 doi: 10.1016/j.leukres.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 96.Conticello C, Adamo L, Vicari L, Giuffrida R, et al. Antitumor activity of bortezomib alone and in combination with TRAIL in human acute myeloid leukemia. Acta Haematol. 2008;120:19–30. doi: 10.1159/000151511. [DOI] [PubMed] [Google Scholar]
- 97.Flotho C, Claus R, Batz C, Schneider M, et al. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- 98.Khan R, Schmidt-Mende J, Karimi M, Gogvadze V, et al. Hypomethylation and apoptosis in 5-azacytidine-treated myeloid cells. Exp Hematol. 2008;36:149–157. doi: 10.1016/j.exphem.2007.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.