Abstract
An important problem in realizing personalized medicine is the development of methods for identifying disease subtypes using quantitative proteomics. Recently we found that bronchoalveolar lavage (BAL) cytokine patterns contain information about dynamic lung responsiveness. In this study, we examined physiological data from 1,048 subjects enrolled in the US Severe Asthma Research Program (SARP) to identify four largely separable, quantitative intermediate phenotypes. Upper extremes in the study population were identified for eosinophil‐ or neutrophil‐predominant inflammation, bronchodilation in response to albuterol treatment, or methacholine sensitivity. We evaluated four different statistical (“machine”) learning methods to predict each intermediate phenotype using BAL cytokine measurements on a 76 subject subset. Comparison of these models using area under the ROC curve and overall classification accuracy indicated that logistic regression and multivariate adaptive regression splines produced the most accurate methods to predict intermediate asthma phenotypes. These robust classification methods will aid future translational studies in asthma targeted at specific intermediate phenotypes. Clin Trans Sci 2010; Volume 3: 147–157
Keywords: asthma, logistic regression, multivariate regression splines, quantitative phenotypes, personalized medicine
Introduction
Asthma is a clinical syndrome characterized by recurrent episodes of symptomatic airflow obstruction and airways hyper‐reactivity to nonspecific stimuli. 1 Typically, this diagnosis is made on a constellation of signs, symptoms and the presence of reversible airflow obstruction. Despite these common clinical features, individuals with asthma show markedly heterogeneous cellular types of mucosal inflammation, 2 , 3 with sensitivity to different triggers, such as respiratory viral infections, 4 manifesting various degrees of airway remodeling,5 and occurring in response to distinct environmental/occupational exposures. 1 It is therefore likely that this syndrome is the result of different pathophysiological processes, explaining its variable clinical courses and response to therapy. 6
The development of novel methods for objectively subtyping asthma will aid in its management and intervention‐based clinical research. One approach to this problem involves the identification of quantifiable intermediate phenotypes, and developing discriminating and robust biochemical tests that can be used to detect their presence. 7 , 8 Here, data from multidimensional profiling of proteins in airway fluids is filtered and used to model quantitative physiological traits using statistical (“machine”) learning methods. However, which statistical learning method performs best for multidimensional proteomics data is not known because these techniques make different assumptions about the relationship of the features (proteins) to the outcome (phenotype).
The long‐term goal of our study is to improve the early detection and management of “severe”, or “glucocorticoid‐resistant” asthma, a syndrome that shows a relative defect in response to inhaled glucocorticoids. 9 , 10 Severe asthmatics represent 5 to 7% of the overall asthmatic population, yet account for 40 to 50% of the health costs of asthma, and incur significant morbidity, reduced lung function, and decrements in quality of life measures. 11 , 12 Additionally, some severe asthmatics are characterized by either neutrophil‐predominant inflammation or increased tissue eosinophils by endobronchial biopsy. 13 , 14 These latter patients have increased near‐fatal events, especially those with early onset disease, and are associated with airway remodeling, indicated by increased sub‐basement membrane thickening. 15 Although recent studies have shown that, as a group, glucocorticoid resistant asthmatics have reduced FEV1 and greater frequency of pneumonia (suggesting an impairment of innate immune defenses perhaps relating to high doses of inhaled glucocorticoids 16 ) than those with mild asthma, there is significant overlap in these measures that prevent their application in discriminating between these asthmatic subtypes. As a result, there is no reliable method for early recognition of treatment‐resistant subtypes, a finding that could significantly impact clinical management.
Recently we used multiplex cytokine assays to profile cytokine secretion patterns in the bronchoalveolar lavage (BAL) of 84 representative subjects with mild‐moderate and severe asthma. Our preliminary analysis indicated that molecular fingerprints of asthmatic subtypes could be identified using unsupervised clustering and decision tree analyses. 17 Preliminary analysis of the features required to identify a group enriched in severe asthmatics indicated that at least 10 cytokines were required for this classification; fewer would result in inaccurate grouping by this method. These findings suggested that BAL cytokine values could be used to predict key clinical characteristics, such as dynamic lung responsiveness and type of inflammation, but the best method for doing so was not known.
In this study, we applied a heuristic analysis of the phenotypic parameters of the US Severe Asthma Research Program (SARP) dataset to determine quantitative intermediate phenotypes of asthma. We defined subgroups as extremes in BAL cellularity (eosinophils and neutrophils), enhanced bronchodilator response (“bronchodilators”), and methacholine sensitivity (“hyper‐responders”). Inter‐group relationships determined by network analysis showed these four groups to be largely distinct. We next sought to identify optimal statistical learning approaches that best predicted these intermediate phenotypes from BAL cytokines using logistic regression (LR), multivariate adaptive regression splines (MARS), classification and regression trees (CART), and random forest (RF) classifiers. Despite distinct assumptions about the relationship between features (cytokines) and outcomes (intermediate phenotypes), both LR and MARS approaches were comparable in accuracy and Receiver Operating Curve (ROC) characteristics and both outperformed CART and RF. These results suggest the optimal statistical learning approaches for using multidimensional protein profiling to phenotypes in investigation of airways disease.
Methods
Study population
In the U.S. SARP, enrollees are categorized as mild‐moderate (“nonsevere”) or “severe” asthma, on the basis of standardized definitions in the manual of procedures (MOP) developed from an American Thoracic Society (ATS) workshop. 10 , 18 All enrollees have history, physical examination, spirometry, bronchodilator reversibility, allergy skin testing, and methacholine challenge testing. Nonsevere asthmatics have lung function that can be normalized using standard doses of inhaled glucocorticoids, with or without long‐acting beta‐agonists or leukotriene modifiers. Severe asthmatics are defined according to ATS consensus for refractory asthma. 10 These patients are characterized by abnormal lung function in the face of aggressive inhaled glucocorticoid therapy and at least one additional control agent. For pulmonary function testing, baseline FEV1 testing required a 4–6 hour withhold of short acting bronchodilators and a 10–12 hour hold for long acting bronchodilators. Hankinson predicted values (with race correction) were utilized to obtain “percent predicted” values. 19 For dynamic testing, subjects performed spirometry before and after 4 puffs (90 μg/puff) of albuterol. Methacholine sensitivity was measured as previously described; 20 subjects with a FEV1 <55% were excluded for safety reasons. All studies were approved by the local institutional review boards and all subjects gave informed consent.
Bronchoalveolar lavage (BAL) and analysis
Bronchoscopy and BAL from 84 randomly selected patients representative of the SARP age and gender distribution was performed according to the SARP MOP [infusion of 2 aliquots of 50 mL each of 0.9% NaCl]. BAL cellular differential was measured by cytospin preparation cells subsequently stained with H&E. At least 300 cells were counted for differential analysis. Total cell neutrophil and eosinophils were determined by the product of the differential and the total cell count. Cytokine measurements were drawn from a previous analysis of 25 human cytokines measured in duplicate BAL samples from 84 subjects using multiplex immunoassays. 17 BAL fluid concentrations were analyzed as raw concentrations without normalization to total protein, albumin, or other marker. This strategy is consistent with the recommendation of the Bronchoalveolar Lavage Cooperative Study Group, 21 and with the analytic approach of the US SARP. 20
Statistical analysis
We determined the optimal cut‐off for intermediate phenotypes through an ROC curve analysis on the sensitivity‐specificity approach by ten‐fold cross‐validation. 22 Here, different cut‐offs for each outcome were selected and for each cut‐off, the following parameters were calculated: sensitivity, specificity, positive, and negative predictive values, overall performance of [sensitivity ± specificity] and the distance with respect to the square root of [(1 – sensitivity)2± (1 – specificity)2] in an ROC curve. The cut‐off producing the most accurate model was used in this study. ANOVA with multiple comparisons, t‐test for proportions and Kruskall‐Wallis tests were performed using SAS, version 9.1.3 (SAS, Inc., Cary, NC, USA) and SPSS, Release 11.0.1 (SPSS, Inc., Chicago, IL, USA). P values were adjusted using the Benjamini and Hochberg’s false discovery rate (FDR) method. The permutation and bootstrap procedures were used for estimating FDR in the smaller neutrophil class and the values were similar.
Logistic regression (LR)
LR classification is a parametric method for dichotomous dependent variable prediction using a linear combination of independent variables. LR modeling was performed on log‐transformed cytokine concentrations by evaluating the misclassification error (the sum of the false positive and false negative error rates) by changing the predictive probability cut‐off value from 0.4 to 0.6 in 0.1 increments (SAS, Version 9.1.3). Cytokines whose values were undetectable in >50 of subjects were excluded from modeling because they did not improve accuracy or model performance. These cytokines included IL‐1 β, IL‐7, IL‐10, IL‐12p40, IL‐13, IFN‐α, and GM‐CSF. The final models used a cut‐off value of 0.5, a value that minimized the misclassification error. Best subsets prediction regression using Akaike Information Criterion (AIC) was used in variable selection. AIC is asymptotically equivalent to the cross validation criterion. 23 Regression coefficients of the statistically significant cytokines (p= 0.05) included in the regression analysis were used in predicting the corresponding clinical outcome. Calculation accuracy is calculated for each of the models. An ROC curve was used to estimate the model performance.
Multivariate adaptive regression splines (MARS)
MARS is a nonparametric method that uses piecewise linear spline functions (basis functions) as predictors. The basis functions are combinations of independent variables, and so this method allows detection of feature interactions and performs well with complex data structures. 24 For this analysis, we varied the number of basis functions from 1 to 3 times the number of features, allowing two‐way interactions selecting the optimal model as having the lowest test mean square error (Salford Systems, Inc, San Diego, CA, USA).
Classification and regression trees (CART) and random forests (RF)
CART is an iterative nonlinear discrimination method that splits the sample into smaller nodes, generating a binary tree (Salford Systems, Inc). For each independent variable, the best split is determined by a rule that produces a daughter node consisting of one phenotype. Symmetric “gini” rule was used for eosinophil and neutrophil classes, and the “two‐ing” rule was used for bronchodilator and hyper‐responder classes. The process is iteratively repeated until a binary tree is produced, and the tree is then reduced by pruning. RF generates an ensemble classifier consisting of 500 decision trees. Each decision tree is produced from the training set with replacement. The RF predicts the class that is the mode of the class’s output by individual trees (Salford Systems, Inc).
Results
The clinical features of 461 severe versus 587 mild‐moderate (“nonsevere”) asthmatics for the currently enrolled SARP study population are summarized in Table 1 . Analysis of these data indicate that in the severe asthmatics in this study population, women predominate over men by a ratio of 1.6:1, consistent with earlier analyses of these subjects. 20 Relative to nonsevere asthmatics, severe asthmatics had significant reductions in forced expiratory volume in 1 sec (FEV1), a feature also consistent with the operational definition of severe asthma. Specifically, severe asthmatics had an FEV1 (percent predicted) of 65 ± 22.81% versus 85.26 ± 17.86% for subjects with nonsevere disease (p < 0.01). These differences remained significant for both genders. However, severe and nonsevere asthmatics did not differ by age of onset or serum IgE ( Table 1 ).
Table 1.
Study subject characteristics. Mean and standard deviation of study population demographics. * indicates p < 0.01 in pairwise comparison between mild‐moderate and severe asthmatics (ATS consensus criteria).
| Phenotype | Characteristic | Males (n= 385; 36.7%) | Females (n= 663; 63.26%) | All subjects |
|---|---|---|---|---|
| Mild‐Moderate (n= 587) | n = 207 (35.2%) | n = 380 (64.7%) | n = 587 (100%) | |
| Age of onset | 10.87 ± 12.41 | 13.8 ± 12.5 | 12.83 ± 12.6 | |
| FEV1 (%) | 81 ± 18.8 * | 87.57 ± 16.85 * | 85.26 ± 17.86 * | |
| IgE (log) | 2.29 ± 0.6 | 1.97 ± 0.7 | 2.09 ± 0.67 | |
| Severe (n= 461) | 178 (38.6%) | 283 (61.3%) | N = 461 (100%) | |
| Age of onset | 12.83 ± 15.5 | 14.7 ± 14.66 | 14 ± 15 | |
| FEV1 (%) | 63.3 ± 21.27 | 66.21 ± 23.71 | 65 ± 22.81 | |
| IgE (log) | 2.27 ± 0.61 | 2.05 ± 0.8 | 2.14 ± 0.73 |
Previously, we applied shrunken centroids analysis to determine the minimum number of cytokines that accurately classified the syndrome of severe asthma. 17 From this analysis, we concluded that at least 10 cytokines were required for accurate grouping. We therefore sought to simplify the problem by defining quantitative, intermediate phenotypes of asthma severity. Our approach is to identify extremes of selected quantitatiave phenotypes spectra to maximize our ability to discover cytokines that predict them. The strategy for our modeling approach is schematically diagrammed in Figure 1 .
Figure 1.
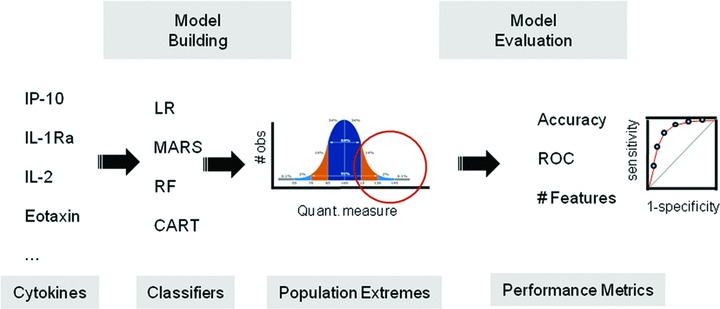
Study Overview. Shown is a schematic diagram of the approach in this study to identify the most reliable statistical learning methods (“classifiers”) that map the relationship between BAL cytokines and population extremes of intermediate phenotypes in asthma.
To identify quantitative intermediate phenotypic traits in the study population we used a heuristic approach focusing on the distribution of BAL cellular components. We first used absolute eosinophil and neutrophil counts because airway eosinophils correlate with clinical severity of asthma, 25 , 26 , 27 and neutrophilic inflammation has been described in fatal 15 and severe asthma. 15 We examined the FEV1 response to β agonist‐induced bronchodilation because FEV1 response may be linked to responsiveness to glucocorticoids, and our previous statistical analysis indicated a greater FEV1 response in severe asthmatics ( Table 1 ). 28 Finally, we identified extremes in methacholine sensitivity, because this is an independent index of airway responsiveness, a correlate of airway inflammation, and is an objective measurement of therapeutic response. 29
We plotted population histograms for each phenotype to identify subclasses that represent the most extreme outliers in the measurements. For example, data for eosinophil counts was available for 165 subjects in the SARP study population; this population histogram indicated that eosinophil numbers were distributed in a non‐Gaussian distribution, with most BAL eosinophils being clustered around 0 ( Figure 2A ). Using an absolute eosinophil count of greater than 0.2 × 106 eosinophils in the BAL sample, 26 of the 165 subjects (15.8%) were arbitrarily designated as the “high eosinophil” class. For neutrophils, the majority of subjects had no neutrophils; using a cut‐off of greater than 1 × 106 neutrophils per BAL sample, 11 of 228 subjects (4.8%), where neutrophil counts were available, were identified as the “high neutrophil” class ( Figure 2B ). Analysis of the study population FEV1 response to albuterol showed that the majority of subjects had 0 to 20% change in percent corrected FEV1; 196 of 925 subjects (21.8%) who had albuterol responses measured had responses of greater than 20% of their baseline FEV1; these patients were identified as “bronchodilators” ( Figure 2C ). Finally, a wide distribution of sensitivity to methacholine was identified in the population; PC20 values of <0.5 mg/ml were used to identify the “hyper‐responder” class, representing 189 of 959 subjects (19.7%, Figure 2D ).
Figure 2.

Class distributions and definitions. Shown are population histograms of the intermediate phenotypes selected for modeling. (A) Frequency for total BAL eosinophil count (in millions) in the study population. The population cut‐off for “high eosinophils” is indicated. (B) Frequency of neutrophil count (in millions). (C) Bronchodilators in response to b‐agonist. Plotted on the x‐axis is percent change FEV1 in response to albuterol inhalation. (D) Methacholine hyper‐responders. X‐axis is PC20 methacholine. The cut‐offs used to define eosinophil rich, neutrophil rich, bronchodilator response, and methacholine hyper‐sensitive classes are shown.
We next examined the distribution of these intermediate phenotypes in the syndromes of severe and nonsevere asthma using set analysis. We noted that the 26 members of the high eosinophil class were evenly distributed between nonsevere (12) and severe asthma syndromes (14, Figure 3A ), whereas 8 of the 11 members in high neutrophil class were surprisingly distributed in the nonsevere asthmatic syndromes ( Figure 3B ). Comparing the proportions of specific BAL cell types in the severe versus nonsevere asthma syndromes indicated that there was no significant difference in the proportions of severe asthmatics with high eosinophils versus the proportion of nonsevere asthmatics with high eosinophils.
Figure 3.
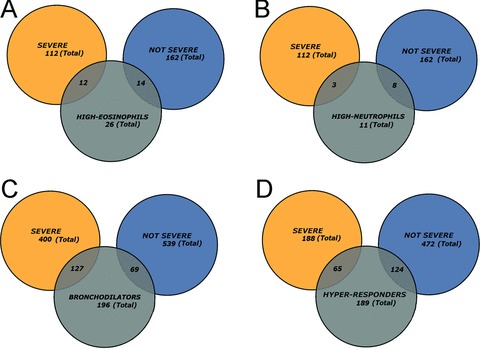
Inter‐relationship of asthmatic phenotypes with SARP classification. Venn diagram analysis of phenotypes with clinical asthmatic groups. Shown is the intersection for various groups. (A) High BAL eosinophils (eosinophils); (B) High BAL neutrophils (neutrophils); (C) Bronchodilation in response to 4 puffs of albuterol (bronchodilators); (D) Methacholine “hyper‐responder” class.
Conversely the bronchodilator class was primarily distributed in the severe asthma syndrome, with 127 of the 196 bronchodilators being severe asthmatics ( Figure 3C ); in fact, the proportion of bronchodilators in the severe group was significantly different from that of the nonsevere group (p < 0.0001, proportional t‐test). This finding is consistent with our previous comparison of the severe versus nonsevere asthmatics where the severe asthmatics as a group had a lower FEV1 but a greater bronchodilatory response. 17 Finally, 124 of the 189 methacholine hyper‐responder class was primarily distributed in the nonsevere asthmatics. This proportion of hyper‐responders in the nonsevere diagnosis was also statistically significant (p= 0.03).
This preliminary analysis indicated that the intermediate phenotypes have different segregation patterns in severe and nonsevere asthma. For example, bronchodilators are largely contained in the syndrome of severe asthma, whereas the hyper‐responders were largely nonsevere asthmatics. These intermediate phenotypes, then, may represent indicators of distinct pathophysiological processes. To more fully understand the inter‐relationships of these phenotypes, class memberships for all phenotypes were analyzed. Strikingly, membership of these classes were largely distinct, with no subject being contained in all four classes ( Figure 4 ). Of the 189 subjects classified as hyper‐responders, the greatest overlap was with the bronchodilator class, where 47 subjects were shared ( Figure 4 ). In addition, 6 hyper‐responders were also in the high neutrophil class. Of the 196 bronchodilators, apart from the 47 shared members with the hyper‐responder class, 6 were shared with the high eosinophil class. Of those 26 classified as having the high eosinophil phenotype, 11 were members of the hyper‐responders, 6 were also members of the bronchodilator class, and 4 were also in the high neutrophil class. Only 2 subjects were jointly in three groups, representing the hyper‐responder, neutrophil, and eosinophil classes. We note that the bronchodilators and high neutrophil groups were completely disjoint, with no shared members ( Figure 4 ). Based on this analysis, we concluded that these cellular and physiological phenotypes are relatively distinct groups.
Figure 4.
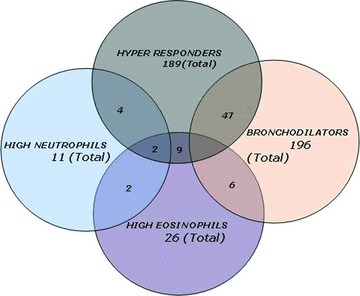
Inter‐relationship of asthmatic phenotypes. Shown is a Venn diagram of the membership for all classes.
Cytokine patterns associated with asthma phenotypes
We next sought to determine if we could relate cytokine concentrations in the BAL fluid to the subjects classified within these relatively distinct asthma phenotypes. For this purpose, a previously determined data set of 25 cytokines in BAL fluid from 84 randomly selected subjects was measured by multiplex ELISAs. 17 This group included 41 severe and 43 nonsevere asthmatics representative of the overall SARP population in terms of gender, differences in FEV1 reversal, and age of onset (Supporting information, Table SI, Figure S1, and Figure S2). To determine whether the phenotypes were associated with different patterns of cytokine expression, BAL cytokine concentrations were compared between members of the class versus those not in the class (e.g., high eosinophils vs. the remainder of the population) using a ranked (nonparametric) test. A nonparametric t‐test was used because the distribution of cytokine measurements were skewed, and not normally distributed. The FDR of Benjamini and Hochberg was used to adjust the p values to reduce the effect of multiple hypothesis testing.
We noted that BAL IL‐2 was different between “high eosinophil” and “low” eosinophil classes at the p < 0.05 cut‐off ( Table 2 ). Conversely, in the comparison between the subjects with high‐ versus low‐neutrophil classes, IP‐10, IL‐7, and GM‐CSF were significantly different. A different pattern of cytokines was significant between the bronchodilators with IL‐1Ra, IL‐4, TNF, Eotaxin, and GM‐CSF being significant. For the methacholine hyper‐responders, the cytokines IL‐1Ra, IL‐5, IL‐15, MIG, Eotaxin, MCP‐1, and GM‐CSF were significant. Of these, only IL‐4 (for the bronchodilators) and IL‐1Ra, MIG (hyper‐responders) reached the p < 0.01 level of significance. Together these data suggest that the asthma phenotypes are associated with distinct BAL cytokine patterns.
Table 2.
Comparison of cytokine expression for four dichotomous phenotypes. Shown is a class‐by‐class comparison for each BAL cytokine. Values shown are median cytokine concentrations for each class. *P value is nonparametric pair‐wise test adjusted for FDR.
| Cytokines | Eosinophils (n= 72) | Neutrophils (n= 72) | Bronchodilators (n= 83) | Hyper‐responders (n= 68) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | P | Low | High | P | Low | High | P | Low | High | P | |
| IL‐2 | 8.69 | 2.3 | *0.041 | – | – | – | – | – | – | – | – | – |
| IP‐10 | – | – | – | 8.93 | 28.36 | *0.034 | – | – | – | |||
| IL‐7 | – | – | – | 2.113 | 15.36 | *0.042 | – | – | – | – | – | |
| GM‐CSF | – | – | – | 1.618 | 1.62 | *0.048 | 1.618 | 0.13 | *0.045 | 1.618 | 0.131 | *0.031 |
| IL‐4 | – | – | – | – | – | – | 14.62 | 15.49 | *0.007 | – | – | – |
| TNF‐a | – | – | – | – | – | – | 11.7 | 10.63 | *0.026 | – | – | – |
| IL‐5 | – | – | – | – | – | – | 2.575 | 1.91 | *0.035 | 0.74 | 1.02 | *0.013 |
| Eotaxin | – | – | – | – | – | – | 6.4 | 6.37 | *0.017 | 1.08 | 0.99 | *0.038 |
| IL‐1Ra | – | – | – | – | – | – | 272.47 | 199.15 | *0.048 | 0.22 | 1.08 | **0.000 |
| MIG | – | – | – | – | – | – | – | – | – | 0.01 | 1.22 | *0.010 |
| IL‐15 | – | – | – | – | – | – | – | – | – | 0.61 | 1.06 | *0.012 |
| MCP‐1 | – | – | – | – | – | – | – | – | – | 0.61 | 1.21 | *0.043 |
| IL‐12 | – | – | – | – | – | – | – | – | – | 4.02 | 0.087 | *0.003 |
Comparison of statistical learning methods
We next used four statistical learning approaches to model intermediate phenotypes and compared the performance for each. The specific models developed using each machine learning approach is displayed in Supporting Tables SII–SIII and Figures S3–S6; major features selected for each model are displayed in Supporting Tables SIV–SVII. Model comparison was based on overall accuracy, sensitivity, specificity and area under the Receiver Operator Characteristics (ROC) Curve (AUC). ROC curves plot the sensitivity versus 1‐specificity; a diagonal line indicates that the output is a random guess, whereas an ideal classifier with a high true positive rate and low false positive rate will be located in the upper left quadrant of the plot. 30 The AUC is a scalar value between 0.5 and 1.0, equivalent to the probability that two cases, one randomly chosen from each group, are correctly ordered by the classifier. 31
High eosinophils
The best LR model predicting the “high eosinophil” class produced an overall accuracy of 85% ( Table 3 ) and AUC of 0.84 ( Figure 5A ). Similarly, the best MARS model that best predicted “high eosinophil” class showed an overall accuracy of 85% and an AUC of 0.89, indicating that these two modeling approaches produced indistinguishable performance. By contrast, the CART and RF classifiers were substantially less accurate with a rate of 67% and 49%, respectively, and had AUCs of 0.48 and 0.40, essentially equivalent to a random guess. Inspection of the rank‐ordered cytokine features that most contributed to the comparable models showed that the cytokines Eotaxin, IL‐2, and IL‐1Ra had the highest χ 2 score statistics in the LR modeling, whereas IL‐15, MCP‐1, and IL‐6 were the cytokines with the highest variable importance in the MARS models (Supporting Table SII). These data indicated that although the prediction performance for both LR and MARS were comparable, these models were based on different cytokines.
Table 3.
Model performance comparisons. Shown is the model performance comparisons for the machine learning approaches for accuracy, AUC, sensitivity and specificity. AUC 5 area under the ROC curve; LR 5 logistic regression; MARS 5 multivariate adaptive regression splines; CART 5 classification and regression trees; RF 5 random forests.
| Phenotype | Classifiers | Accuracy (%) | AUC | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| High eosinophils | LR | 85 | 0.84 | 53 | 94 |
| MARS | 85 | 0.89 | 46 | 93 | |
| CART | 67 | 0.48 | 30 | 74 | |
| RF | 49 | 0.40 | 46 | 49 | |
| High neutrophils | LR | 97 | 0.76 | 60 | 98 |
| MARS | 93 | 0.77 | 20 | 98 | |
| CART | 83 | 0.51 | 20 | 88 | |
| RF | 66 | 0.47 | 40 | 68 | |
| Bronchodilators | LR | 76 | 0.71 | 23 | 85 |
| MARS | 88 | 0.85 | 23 | 100 | |
| CART | 75 | 0.69 | 53 | 81 | |
| RF | 61 | 0.65 | 54 | 64 | |
| Hyper‐responders | LR | 85 | 0.85 | 53 | 94 |
| MARS | 90 | 0.88 | 73 | 94 | |
| CART | 79 | 0.68 | 53 | 86 | |
| RF | 70 | 0.77 | 73 | 70 |
Figure 5.

Receiver Operating Curves of phenotype classifiers. (A) ROC of high eosinophil model for all four statistical learning methods. In these plots, the x‐axis is 1‐specificity and y‐axis is sensitivity. (B) ROC of high neutrophil models. (C) ROC of bronchodilator models. (D) ROC of methacholine hyper‐responder models. The corresponding AUCs for each curve are presented in Table 3.
High neutrophils
LR and MARS were similarly highly accurate for prediction of high neutrophil phenotype, producing models with 97% and 93% accuracy, and AUCs of 0.76 and 0.77, respectively, whereas lower accuracy and AUC values were produced by the CART and RF classifiers ( Table 3 , Figure 5B ). The cytokine most important for the classification was IL‐6 for both LR and MARS classifiers (Supporting Table III).
Bronchodilators
The MARS classifier was more accurate than that produced by LR for predicting the bronchodilator phenotype ( Figure 5C ), with an accuracy of 88% and AUC of 0.85 using 5 basis functions (Supplemental Table IV). Here the LR was only marginally better than CART in overall model accuracy with both outperforming RF. The cytokines with the greatest variable importance in the MARS classifier for bronchodilators were MCP‐1 and IL‐2 (Supporting Table IV).
Hyper‐responders
As with the bronchodilators, MARS produced a slightly more accurate model than did LR with 90% accuracy and an AUC value of 0.88, with LR producing an overall accuracy of 85% and AU value of 0.85 ( Table 3 , Figure 5A ). Both of these models outperformed the CART and RF. The one cytokine with the greatest χ 2 score statistic in the LR and the greatest variable importance in the MARS was IL‐1Ra (Supporting Table SVI). We note that IL‐1Ra was also highly statistically significant in the group‐wise comparison of cytokines in the hyper‐responders versus non‐hyper‐responder groups (p < 0.001, Table 2 ).
Together we interpret these data to indicate that the LR and MARS statistical learning methods consistently outperformed the CART and RF for prediction of intermediate asthma phenotypes.
Discussion
Asthma exhibits significant variation in its onset, etiology and treatment response. These subgroups cannot be distinguished based on clinical phenotypes alone, and no reliable proteomic markers have yet been identified. Because a prominent pathophysiological aspect of asthma involves mucosal inflammation, 27 we are exploring the application of multidimensional protein profiling in BAL to more objectively understand the relationship between indices of airway inflammation and heterogeneity in clinical phenotypes. For this reason, we have selected BAL for multiplex cytokine measurements because this biofluid represents a reasonably proximal sampling of direct inflammatory processes in the lung. Here, successful application of supervised learning approaches may identify robust methods of identifying subtypes of asthmatics differing in treatment response or clinical outcome.
In this study, we extend on our preliminary work that BAL cytokine measurements contain information that can be related to distinct clinical phenotypes, including cellular inflammation and dynamic airway responses. 17 A schematic diagram of how BAL cytokines, intermediate phenotypes and underlying disease processes can conceptually be mapped to the syndrome of severe asthma is shown in Figure 6 . Here, we have extended our previous work to: (1) Empirically identify distinct quantitative (intermediate) phenotypes of asthma based on 1,048 subjects enrolled in the SARP program, and (2) Evaluate the performance of four statistical learning models to identify that approach that could be best used to associate cytokine features with the intermediate phenotypes. This work, therefore, informs the first step of mapping BAL proteins to the syndrome of asthma ( Figure 6 ).
Figure 6.

BAL cytokines and intermediate phenotypes. Shown is a schematic diagram of the relationship between BAL cytokines, intermediate phenotypes indicative of disease processes, and asthma syndromes. Our study informs the best methods for mapping cytokines to distinct intermediate phenotypes using LR and MARS. As the spectrum of intermediate phenotypes are defined and their relationship to severe asthma better understood, this work may be used in clinical study design, prediction of therapeutic response, and ultimately personalized therapy.
Intermediate phenotypes in asthma‐cytokine associations
The application of sputum eosinophil measurement to guide pharmacological dosing 32 may make the application of proteomic markers of airway eosinophilia useful clinically. Because there is no universally accepted formal definition of BAL eosinophilia, our classification of the “high eosinophil” phenotype is entirely driven by the characteristics of our study population. Consistent with the findings that eosinophilic asthma represents a small subset of all asthmatic patients, our analysis of eosinophil numbers in this study population shows that the distribution of eosinophil counts is skewed towards zero. The presence of airway eosinophils or their degranulation products is characteristic of late onset asthma 33 and well known to correlate with clinical severity, 25 airway remodeling, and exacerbation‐prone disease. 34 Moreover, the presence of airway eosinophils is inversely related to PC20 methacholine, directly related to airway responsiveness 35 and is predictive of clinical response to high dose corticosteroid therapy. 36 Mechanistic studies have shown that tissue targeting, and activation of circulating eosinophils are regulated by GM‐CSF, IL‐5, and IL‐3 cytokines, 37 cytokines which also induce tissue survival by persistent signals mediated through a novel cross‐talk signaling pathway involving ICAM‐1, an adhesion molecule associated with tissue persistence. 38 Despite this understanding, tissue levels of BAL IL‐5 are not associated with sputum eosinophilia, 39 a finding consistent with our analysis that IL‐5 is not significantly different between the high and low eosinophil phenotypes ( Table 2 ). Instead in our study only the IL‐2 cytokine was different between high eosinophil and low eosinophil groups. This finding is of interest because inhibition of IL‐2 reduces eosinophils and improves airway function in patients with glucocorticoid‐resistant asthma. 40
Neutrophilic inflammation has been observed in severe asthma, occupational asthma, and childhood asthma. 41 In severe asthma, neutrophils have been identified in sputum, BAL and transbronchial biopsies of small airways. 42 The role of the neutrophil in asthma has not yet been well defined, although it has been suggested that this cell type may mediate chronic inflammation or remodeling. 41 Severe asthmatics have been characterized as having either neutrophil‐ or eosinophil‐predominant inflammation. 14 Consistently, our set analysis indicated that of the 18 subjects with either high neutrophils or high eosinophils, only 1 subject was a member of both classes ( Figure 3 ). This is similar to the findings of others, where neutrophils are found in greater numbers in patients with low eosinophils. 43 Our results indicating that the high neutrophil group are predominately distributed in the nonsevere asthmatic group are not consistent with a previous study where neutrophilia in induced sputum correlated with reduced responsiveness to inhaled glucocorticoids. 44 These differences may be due to differences in subject selection or how airway neutrophilia is assessed. Interestingly in our descriptive statistics, CXCL10/IP‐10 is the only cytokine that is significantly higher in the high neutrophil phenotype ( Table 2 ). We note CXCL10/IP‐10 is produced by IFN‐γ‐stimulated neutrophils, 45 and is a biomarker of rhinovirus‐induced asthma exacerbations.
A surprising finding from our studies is that specific BAL cytokines are related to indices of dynamic lung function, including bronchodilator response to β adrenergic agonists. This physiological response is of interest because bronchodilator response is both associated with improvement in FEV1 in response to chronic glucocorticoid treatment, 28 and a surrogate of severe asthma ( Figure 4 ).17 Our descriptive statistical analysis shows that IL‐1Ra, IL‐4, TNFa, and Eotaxin are significantly different in bronchodilators versus nonbronchodilator phenotype. Of these cytokines, Eotaxin is a CC chemokine inducibly secreted by a wide variety of airway cells, including Th2 lymphocytes. In response to IL‐4 and IL‐13, Eotaxin induces eosinophil accumulation in response to allergen challenge in vivo. 46 Because Eotaxin promotes eosinophil differentiation, migration and chemotaxis, it is interesting to us that the bronchodilator phenotype is largely distinct from the high eosinophil group. Eotaxin induces smooth muscle cell migration 47 and fibroblast proliferation, 48 cell types which may affect airway bronchodilator response and remodeling.
Enhanced sensitivity to methacholine‐induced bronchoconstriction is a highly sensitive and reproducible marker of airways hyper‐reactivity, and can be used as an objective outcome measure of therapeutic response. 29 In our study, IL‐1Ra and Eotaxin were identified in this study as being significantly different in methacholine hyper‐responders versus non‐hyper‐responders ( Table 2 ). We note that IL‐1Ra is a glucocorticoid‐inducible peptide that antagonizes IL‐1‐α and ‐β; the latter cytokines enhance airway smooth muscle contractile responses to cholinergic agents. 49
Relationship of intermediate phenotypes to asthma severity
The inter‐relationship of the intermediate asthma phenotypes bears further discussion. The Venn diagram analysis ( Figure 4 ) suggests that these subpopulations have distinct relationships with asthma severity classifications. For example, 73% of the high neutrophils and 66% of hyper‐responders are nonsevere asthmatics, whereas, by contrast, 65% of the bronchodilators are severe asthmatics ( Figure 3 ). Although the four phenotypes determined empirically by this study are largely distinct ( Figure 4 ), an exception is that 15.8% of the bronchodilators also fall within the hyper‐responder class. This functional overlap is reinforced by overlap in shared discriminant cytokines, with IL‐1Ra, IL‐5, and Eotaxin being shared between the two classes ( Table 2 ). Moreover, IL‐1Ra, IL‐4, and Eotaxin have significant χ 2 scores in both the LR models for bronchodilators and hyper‐responders. The overlap in associated cytokines may imply that common pathophysiolgical processes underly these intermediate phenotypes. It will be of interest in future studies to examine the clinical and therapeutic response of this bronchodilator‐hyper‐responder group.
Statistical learning approaches for predicting asthma phenotypes
A systematic comparison of distinct statistical learning approaches for relating BAL cytokine concentrations to intermediate asthma phenotypes is warranted because currently it is not possible presently to select a priori the machine learning tool that performs best for any given data set. This is because all statistical learning approaches are sensitive to the underlying data structure. For example, LR is a parametric approach that identifies main effects of candidate cytokines and assumes a global linear relationship between the independent (cytokine) and the dependent (asthma phenotype) variables. Although LR performs well if the relationship between the dependent and independent variables is well described, the performance of LR is reduced with high dimensional data or when there are multiple interactions between independent variables. By contrast, MARS is a nonparametric, piecewise linear approach that can establish relationships over smaller intervals of independent variables, as well as detect interactions between independent variables. 50
Our findings in this study show that LR and MARS result in quite similar accuracy for relating proteomic profiles to clinical phenotypes in asthma, with both clearly outperforming CART and RF classifiers. Interestingly to us, for many of the best performing models developed in LR and MARS, the cytokine features important for the model are different. For example, with the high eosinophil class, Eotaxin has the highest χ 2 score statistic in the LR model, whereas IL‐8 is the cytokine with the greatest variable importance in MARS (Supporting Table SII). We interpret these data to mean that Eotaxin values are globally linearly related to eosinophil numbers, whereas the relationship with IL‐8 and eosinophils is strong for small regions of the feature‐response relationship.
Model validation and future studies
Our findings are significant because they support a conclusion that proteomic profiling will be fruitful to identify distinct subtypes of asthma and identify two statistical learning methods for doing so. However, the models are not highly sensitive; this may be due to the low number of subjects in some of the classes in this study. These specific models and major predictive features will need to be replicated on a larger number of subjects.
Several additional points are worthy of mentioning. First, because our study is cross‐sectional in design, we do not formally know whether the cellular phenotypes are “stable,” meaning whether a subject with high eosinophils will demonstrate BAL high eosinophils over extended time, or whether these phenotypes are distinct “meta‐stable” states of asthma, where a subject may have a different BAL phenotype as the disease evolves. Further analysis using repeated measures of BAL sampling and physiological assessment in longitudinal clinical studies will indicate how stable each phenotype is. Second, a multidimensional profiling‐machine learning approach can be extended and used to identify other intermediate phenotypes. Further work will be required to determine what these phenotypes are, if they are stable, how they relate to asthma severity and if they can be predicted with markers of airway inflammation. It is entirely reasonable to expect that some phenotypes may not be associated with cytokines, a finding that, in itself, is informative, suggesting other underlying pathophysiological mechanisms. For example, in data not shown, we have been unable to produce acceptable models of subjects with low FEV1, suggesting that FEV1 reduction may not be the direct consequence of on‐going mucosal inflammation, or may be associated with as of yet unmeasured proteins or metabolites. For this reason, it will be of interest to conduct unbiased discovery proteomics on airway biofluids. In addition, the application of systematic approaches combining genetic and metabolomic measurements with protein profiles may add discriminating information to developing more accurate predictors.
Finally, even when this study is validated on a large independent population, we recognize that a relatively invasive BAL sampling approach is not yet ready for application as a clinical test for molecular profiling. However, identification of specific groups of cytokines that would indicate specific responsive phenotype is important information that can be used to develop point‐of‐care diagnostic assay. In this regard, cytokine measurements have been successfully performed on exhaled bronchial breath condensates; 51 once stable markers and predictive statistical models have been identified using BAL samples, we suggest that these assays can be adapted to breath condensate analysis for larger scale clinical application.
In conclusion, this study provides proof‐of‐principle for predicting intermediate, quantitative asthma phenotypes based on multidimensional BAL cytokine profiling. Our analysis further indicates the application of LR and MARS as appropriate statistical learning methods for developing predictive models. We anticipate that this work will inform clinical studies, but our findings will need to be validated and better approaches for biomarker measurement will need to be developed before these findings can be applied to clinical management.
Supporting information
Figure S1. Inter‐relationship of asthmatic phenotypes with SARP classification. Venn diagram analysis of phenotypes with clinical asthmatic groups. Shown is the intersection for various groups. (A) high BAL eosinophils (eosinophils); (B), high BAL neutrophils (neutrophils); (C) bronchodilation in response to 4 puffs of albuterol (bronchodilators); (D), methacholine sensitivity (``hyper‐responder'') class. Note similar distribution as the larger SARP study.
Figure S2. Inter‐relationship of asthmatic phenotypes. Shown is a Venn diagram of the membership for all classes. Note similar class relationships as the larger SARP study.
Figure S3 CART decision tree for eosinophils
Figure S4. CART for neutrophils
Figure S5 CART for bronchodilators
Figure S6 CART for hyper‐responders
Table S1. Clinical/demographic features for the SARP subjects on whom cytokine arrays were performed.
Table SII. Logistic regression models for asthma phenotypes. Logistic regression with information criterion (AIC) was performed for high eosinophils, high neutrophil, bronchodilator and hyper‐responder class. Abbreviations: OR5odds ratio; CI5confidence interval; AUC5area under ROC curve.
Table SIII. MARS models for asthma phenotypes Abbreviations: BF5basis function; OR5odds ratio; CI5confidence interval; AUC5area under ROC curve. At right is shown the performance metrics for the overall model.
Table SIV. Features used for high Eosinophil classifiers. Shown is rank‐ordered feature for the two highest performance machine learning classifiers (LR and MARS) for predicting the high eosinophil phenotype. For the LR, the x2 score statistic is used, whereas rank‐ordered variable importance is shown for the MARS model. Abbreviations: 5feature does not appear in model.
Table SV. Rank ordered features for high neutrophil classifiers. Rank‐ordered feature list for LR and MARS models predicting the high neutrophil phenotype.
Table SVI. Rank ordered feature list for bronchodilators. Rank‐ordered feature list for LR and MARS models predicting bronchodilator phenotype.
Table SVII. Rank ordered feature list for hyper‐responders. Rank‐ordered feature list for LR and MARS models predicting hyper‐responder phenotype.
Please note: Wiley‐Blackwell Publishing is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
This material is available as part of the online article from http://www.ctsjournal.com.
Supporting info item
Acknowledgments
The authors would like to thank Heidi Weiss PhD, UTMB, for constructive comments on the manuscript. This work was supported, in part, by NIH grants 1U54RR02614 UTMB CTSA(ARB), AI062885 (ARB), NHLBI contract BAA‐HL‐02‐04 (ARB), HL69130 US SARP (WJC), Integrated Health Science Facility Core P30 ES06676 (to K. Elferink, UTMB) and HL69149 (MC).
The SARP is a multicenter asthma research group funded by the NHLBI consisting of the following contributors (Principal Investigators are marked with an asterisk): Brigham and Women‘s Hospital—Elliot Israel*, Bruce D. Levy, Gautham Marigowda; Cleveland Clinic—Serpil C. Erzurum*, Raed A. Dweik, Suzy A.A. Comhair, Emmea Cleggett‐Mattox, Deepa George, Marcelle Baaklini, Daniel Laskowski; Emory University—Anne M. Fitzpatrick, Eric Hunter, Denise Whitlock; Imperial College School of Medicine—Kian F. Chung*, Mark Hew, Patricia Macedo, Sally Meah, Florence Chow; University of Pittsburgh—Sally E. Wenzel*, Erin Aiken; University of Texas‐Medical Branch—William J. Calhoun*, Bill T. Ameredes, Dori Smith; University of Virginia—Benjamin Gaston*, W. Gerald Teague*, Mike Davis; University of Wisconsin—William W. Busse*, Nizar Jarjour, Ronald Sorkness, Sean Fain, Erin Billmeyer, Cheri Swenson, Gina Crisafi, Laura Frisque, Dan Kolk; Wake Forest University—Eugene R. Bleecker*, Deborah Meyers, Wendy Moore, Stephen Peters, Annette Hastie, Gregory Hawkins, Jeffrey Krings, Regina Smith; Washington University in St Louis—Mario Castro*, Leonard Bacharier, Iftikhar Hussain, Jaime Tarsi; Data Coordinating Center—Douglas Curran‐Everett*, Ruthie Knowles, Lori Silveira; NHLBI—Patricia Noel*, Robert Smith.
References
- 1. Busse WW, Lemanske RF. Asthma. N Engl J Med. 2001; 344: 350–362. [DOI] [PubMed] [Google Scholar]
- 2. Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, Chu HW. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med. 1999; 160(3): 1001–1008. [DOI] [PubMed] [Google Scholar]
- 3. Fahy JV, Corry DB, Boushey HA. Airway inflammation and remodeling in asthma. Curr Opin Pulm Med. 2000; 6(1): 15–20. [DOI] [PubMed] [Google Scholar]
- 4. Tan WC. Viruses in asthma exacerbations. Curr Opin Pulm Med. 2005; 11(1): 21–26. [DOI] [PubMed] [Google Scholar]
- 5. Aysola RS, Hoffman EA, Gierada D, Wenzel S, Cook‐Granroth J, Tarsi J, Zheng J, Schechtman KB, Ramkumar TP, Cochran R, Xueping E, Christie C, Newell J, Fain S, Altes TA, Castro M. Airway remodeling measured by multidetector CT is increased in severe asthma and correlates with pathology. Chest. 2008; 134(6): 1183–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Szefler SJ, Martin RJ, King TS, Boushey HA, Cherniack RM, Chinchilli VM, Craig TJ, Dolovich M, Drazen JM, Fagan JK, Fahy JV, Fish JE, Ford JG, Israel E, Kiley J, Kraft M, Lazarus SC, Lemanske RF Jr, Mauger E, Peters SP, Sorkness CA. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol. 2002; 109(3): 410–418. [DOI] [PubMed] [Google Scholar]
- 7. Rifai N, Gerszten RE. Biomarker discovery and validation. Clin Chem. 2006; 52(9): 1635–1637. [DOI] [PubMed] [Google Scholar]
- 8. Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotech. 2006; 24(8): 971–983. [DOI] [PubMed] [Google Scholar]
- 9. Chanez P, Wenzel SE, Anderson GP, Anto JM, Bel EH, Boulet LP, Brightling CE, Busse WW, Castro M, Dahlen B, Dahlen SE, Fabbri LM, Holgate ST, Humbert M, Gaga M, Joos GF, Levy B, Rabe KF, Sterk PJ, Wilson SJ, Vachier I. Severe asthma in adults: what are the important questions J Allergy Clin Immunol. 2007; 119(6): 1337–1348. [DOI] [PubMed] [Google Scholar]
- 10. Wenzel SE, Busse WW. Severe asthma: lessons from the Severe Asthma Research Program. J Allergy Clin Immunol. 2007; 119(1): 14–21. [DOI] [PubMed] [Google Scholar]
- 11. Godard P, Chanez P, Siraudin L, Nicoloyannis N, Duru G. Costs of asthma are correlated with severity. Eur Respir J. 2002; 19: 61–67. [DOI] [PubMed] [Google Scholar]
- 12. Serra‐Battles J, Plaza V, Morejon E, Comella A, Brugues J. Costs of asthma according to the degree of severity. Eur Respir J. 1998; 12: 1322–1326. [DOI] [PubMed] [Google Scholar]
- 13. Wenzel S. Pathology of difficult asthma. Paediatr Respir Rev. 2003; 4(4): 306–311. [PubMed] [Google Scholar]
- 14. Jatakanon AN, Uasuf CA, Maziak WA, Lim SA, Chung KF, Barnes PJ. Neutrophilic inflammation in severe persistent asthma. Am J Respir Crit Care Med. 1999; 160(5): 1532–1539. [DOI] [PubMed] [Google Scholar]
- 15. Wenzel SE, Schwartz LB, Langmack EL, Halliday JL, Trudeau JB, Gibbs RL, Chu HW. Evidence that severe asthma can be divided pathologically into two inflammatory subtypes with distinct physiologic and clinical characteristics. Am J Respir Crit Care Med 1999; 160(3): 1001–1008. [DOI] [PubMed] [Google Scholar]
- 16. Venarske D, Busse W, Griffin M, Gebretsadik T, Shintani AK, Minton PA, Peebles RS, Hamilton R, Weisshaar E, Vrtis R, Higgins SB, Hartert TV. The relationship of rhinovirus‐associated asthma hospitalizations with inhaled corticosteroids and smoking. J Infect Dis. 2006; 193(11): 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brasier AR, Victor S, Boetticher G, Ju H, Lee C, Bleecker ER, Castro M, Busse W, Calhoun WJ. Molecular phenotyping of severe asthma using pattern recognition of bronchoalveolar lavage‐derived cytokines. J Allergy Clin Immunol 2008; 121: 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. American Thoracic Society . Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med. 2000; 162: 2341–2351. [DOI] [PubMed] [Google Scholar]
- 19. Hankinson J, Odencrantz J, Fedan K. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999; 159(1): 179–187. [DOI] [PubMed] [Google Scholar]
- 20. Moore WC, Bleecker ER, Curran‐Everett D, Erzurum SC, Ameredes BT, Bacharier L, Calhoun WJ, Castro M, Chung KF, Clark MP, Dweik RA, Fitzpatrick AM, Gaston B, Hew M, Hussain I, Jarjour NN, Israel E, Levy BD, Murphy JR, Peters SP, Teague WG, Meyers DA, Busse WW, Wenzel SE. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007; 119(2): 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bronchoalveolar Lavage Cooperative Steering Committee . BAL constituents in healthy individuals, idiopathic pulmonary fibrosis, and selected comparison groups. Am Rev Respir Dis. 1990; 141: S169–S202. [DOI] [PubMed] [Google Scholar]
- 22. Weng X, Liu Y, Ma J, Wang W, Yang G, Caballero B. Use of body mass index to identify obesity‐related metabolic disorders in the Chinese population. Eur J Clin Nutr. 2006; 60(8): 931–937. [DOI] [PubMed] [Google Scholar]
- 23. Stone M. An asymptotic equivalence of choice of model by cross‐validation and Akaikes’ criterion. J Royal Stat Soc, Ser B (Meth) 1977; 39(1): 44–47. [Google Scholar]
- 24. Friedman JH. Multivariate adaptive regression splines. Annals Stat. 1991; 19(1): 1–67. [DOI] [PubMed] [Google Scholar]
- 25. Bousquet J, Chanez P, Lacoste JY, Barnéon G, Ghavanian N, Enander I, Venge P, Ahlstedt S, Simony‐Lafontaine J, Godard P. Eosinophilic inflammation in asthma. N England J Med. 1990; 323(15): 1033–1039. [DOI] [PubMed] [Google Scholar]
- 26. Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efhimiadis A, Pizzichini E, Hargreave FE, O‘Byrne PM. Mepolizumab for prednisone‐dependent asthma with sputum eosinophilia. N Engl J Med. 2009; 360(10): 985–993. [DOI] [PubMed] [Google Scholar]
- 27. Fahy JV. Eosinophilic and neutrophilic inflammation in asthma: insights from clinical studies. Proc Am Thorac Soc. 2009; 6(3): 256–259. [DOI] [PubMed] [Google Scholar]
- 28. Kerstjens HA, Overbeek SE, Schouten JP, Brand PL, Postma DS. Airways hyperresponsiveness, bronchodilator response, allergy and smoking predict improvement in FEV1 during long‐term inhaled corticosteroid treatment. Dutch CNSLD Study Group. Eur Respir J. 1993; 6(6): 868–876. [PubMed] [Google Scholar]
- 29. Guidelines for Methacholine and Exercise Challenge Testing—1999 . This official statement of the American thoracic society was adopted by the ATS board of directors, July 1999. Am J Respir Crit Care Med. 2000; 161(1): 309–329. [DOI] [PubMed] [Google Scholar]
- 30. Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006; 27: 861–874. [Google Scholar]
- 31. Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic curve. Radiology. 1982; 143: 29–36. [DOI] [PubMed] [Google Scholar]
- 32. Jayaram L, Pizzichini MM, Cook RJ, Boulet JP, Lemiére C, Pizzichini E, Cartier A, Hussack P, Goldsmith CH, Laviolette M, Parameswaran K, Hargreave FE. Determining asthma treatment by monitoring sputum cell counts: effect on exacerbations. Eur Respir J. 2006; 27(3): 483–494. [DOI] [PubMed] [Google Scholar]
- 33. Miranda C, Busacker A, Balzar S, Trudeau J, Wenzel SE. Distinguishing severe asthma phenotypes: role of age at onset and eosinophilic inflammation. J Allergy Clin Immunol. 2004; 113(1): 101–108. [DOI] [PubMed] [Google Scholar]
- 34. Green RH, Brightling CE, McKenna S, Hargadon B, Parker D, Bradding P, Wardlaw AJ, Pavord ID. Asthma exacerbations and sputum eosinophil counts: a randomised controlled trial. Lancet. 2002; 360(9347): 1715–1721. [DOI] [PubMed] [Google Scholar]
- 35. Louis R, Lau LC, Bron AO, Roldaan AC, Radermecker M, Djukanovic R. The relationship between airways inflammation and asthma severity. Am J Respir Crit Care Med. 2000; 161(1): 9–16. [DOI] [PubMed] [Google Scholar]
- 36. Ten Brinke A, Zwinderman AH, Sterk PJ, Rabe KF, Bel EH. “Refractory” eosinophilic airway inflammation in severe asthma: effect of parenteral corticosteroids. Am J Respir Crit Care Med. 2004; 170(6): 601–605. [DOI] [PubMed] [Google Scholar]
- 37. Foster P, Mould A, Yang M, Mackenzie J, Mattes J, Hogan SP, Mahalingam S, Mckenzie AN, Rothenberg ME, Young IG, Matthaei KI, Webb DC. Elemental signals regulating eosinophil accumulation in the lung. Immunol Rev. 2001; 179: 173–181. [DOI] [PubMed] [Google Scholar]
- 38. Pazdrak K, Young TW, Stafford S, Olszewska‐Pazdrak B, Straub C, Starosta V, Brasier A, Kurosky A. Crosstalk betwen ICAM‐1 and granulocyte‐macrophage colony‐stimulating factor receptor signaling modulates eosinophil survival and activation. J Immunol. 2007; 180: 4182–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shannon J, Ernst P, Yamauchi Y, Olivenstein R, Lemiere C, Foley S, Cicora L, Ludwig M, Hamid Q, Martin JG. Differences in airway cytokine profile in severe asthma compared to moderate asthma. Chest. 2008; 133: 420–426. [DOI] [PubMed] [Google Scholar]
- 40. Busse WW, Israel E, Nelson HS, Baker JW, Charous BL, Young DY, Vexler V, Shames RS. Daclizumab improves asthma control in patients with moderate to severe persistent asthma: a randomized, controlled trial. Am J Respir Crit Care Med 2008; 178(10): 1002–1008. [DOI] [PubMed] [Google Scholar]
- 41. Wenzel S. The significance of the neutrophil in asthma. Clin Exp Allergy Rev. 2001; 1: 89–92. [Google Scholar]
- 42. Balzar S, CHU HW, Strand M, Wenzel S. Relationship of small airway chymase‐positive mast cells and lung function in severe asthma. Am J Respir Crit Care Med. 2005; 171(5): 431–439. [DOI] [PubMed] [Google Scholar]
- 43. Kiley J, Smith R, Noel P. Asthma phenotypes. Curr Opin Pulm Med. 2007; 13: 19–23. [DOI] [PubMed] [Google Scholar]
- 44. Green RH, Brightling CE, Woltmann G, Parker D, Wardlaw AJ, Pavord ID. Analysis of induced sputum in adults with asthma: identification of subgroup with isolated sputum neutrophilia and poor response to inhaled corticosteroids. Thorax. 2002; 57(10): 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cassatella MA, Gasperini S, Calzetti F, Bertagnin A, Luster A, McDonald P. Regulated production of the interferon–inducible protein‐10 (IP‐10) chemokine by human neutrophils. Eur J Immunol. 1996; 27: 111–115. [DOI] [PubMed] [Google Scholar]
- 46. Jose PJ, Griffiths‐Johnson DA, Collins PD, Walsh DT, Moqbel R, Totty NF, Truong O, Hsuan JJ, Williams TJ. Eotaxin: a potent eosinophil chemoattractant cytokine detected in a guinea pig model of allergic airways inflammation. J Exp Med. 2008; 179: 881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kodali RB, Kim WJH, Galaria II, Miller C, Schecter AD, Lira SA, Taubman MB. CCL11 (Eotaxin) induces CCR3‐dependent smooth muscle cell migration. Arterioscler Thromb Vasc Biol. 2004; 24(7): 1211–1216. [DOI] [PubMed] [Google Scholar]
- 48. Puxeddu I, Bader R, Piliponsky AM, Reich R, Levi‐Schaffer F, Berkman N. The CC chemokine eotaxin/CCL11 has a selective profibrogenic effect on human lung fibroblasts. J Allergy Clin Immunol. 2006; 117(1): 103–110. [DOI] [PubMed] [Google Scholar]
- 49. Hakonarson H, Maskeri N, Carter C, Chuang S, Grunstein MM. Autocrine interaction between IL‐5 and IL‐1+¦ mediates altered responsiveness of atopic asthmatic sensitized airway smooth muscle. J Clin Invest. 1999; 104(5): 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cook NR, Zee RYL, Ridker PM. Tree and spline based association of gene‐gene interaction models for ischemic stroke. Stat Med. 2005; 23: 1439–1453. [DOI] [PubMed] [Google Scholar]
- 51. Matsunaga K, Yanagisawa S, Ichikawa T, Ueshima K, Akamatsu K, Hirano T, Nakanishi M, Yamagata T, Minakata Y, Ichinose M. Airway cytokine expression measured by means of protein array in exhaled breath condensate: correlation with physiologic properties in asthmatic patients. J Allergy Clinl Immunol. 2006; 118: 84–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Inter‐relationship of asthmatic phenotypes with SARP classification. Venn diagram analysis of phenotypes with clinical asthmatic groups. Shown is the intersection for various groups. (A) high BAL eosinophils (eosinophils); (B), high BAL neutrophils (neutrophils); (C) bronchodilation in response to 4 puffs of albuterol (bronchodilators); (D), methacholine sensitivity (``hyper‐responder'') class. Note similar distribution as the larger SARP study.
Figure S2. Inter‐relationship of asthmatic phenotypes. Shown is a Venn diagram of the membership for all classes. Note similar class relationships as the larger SARP study.
Figure S3 CART decision tree for eosinophils
Figure S4. CART for neutrophils
Figure S5 CART for bronchodilators
Figure S6 CART for hyper‐responders
Table S1. Clinical/demographic features for the SARP subjects on whom cytokine arrays were performed.
Table SII. Logistic regression models for asthma phenotypes. Logistic regression with information criterion (AIC) was performed for high eosinophils, high neutrophil, bronchodilator and hyper‐responder class. Abbreviations: OR5odds ratio; CI5confidence interval; AUC5area under ROC curve.
Table SIII. MARS models for asthma phenotypes Abbreviations: BF5basis function; OR5odds ratio; CI5confidence interval; AUC5area under ROC curve. At right is shown the performance metrics for the overall model.
Table SIV. Features used for high Eosinophil classifiers. Shown is rank‐ordered feature for the two highest performance machine learning classifiers (LR and MARS) for predicting the high eosinophil phenotype. For the LR, the x2 score statistic is used, whereas rank‐ordered variable importance is shown for the MARS model. Abbreviations: 5feature does not appear in model.
Table SV. Rank ordered features for high neutrophil classifiers. Rank‐ordered feature list for LR and MARS models predicting the high neutrophil phenotype.
Table SVI. Rank ordered feature list for bronchodilators. Rank‐ordered feature list for LR and MARS models predicting bronchodilator phenotype.
Table SVII. Rank ordered feature list for hyper‐responders. Rank‐ordered feature list for LR and MARS models predicting hyper‐responder phenotype.
Please note: Wiley‐Blackwell Publishing is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
This material is available as part of the online article from http://www.ctsjournal.com.
Supporting info item


