Abstract
Hyperpolarized 13C MRSI can detect not only the uptake of the pre-polarized molecule but also its metabolic products in vivo, thus providing a powerful new method to study cellular metabolism. Imaging the dynamic perfusion and conversion of these metabolites provides additional tissue information but requires methods for efficient hyperpolarization usage and rapid acquisitions. In this work, we have developed a time-resolved 3D MRSI method for acquiring hyperpolarized 13C data by combining compressed sensing methods for acceleration and multiband excitation pulses to efficiently use the magnetization. This method achieved a 2 sec temporal resolution with full volumetric coverage of a mouse, and metabolites were observed for up to 60 sec following injection of hyperpolarized [1-13C]-pyruvate. The compressed sensing acquisition used random phase encode gradient blips to create a novel random undersampling pattern tailored to dynamic MRSI with sampling incoherency in four (time, frequency and two spatial) dimensions. The reconstruction was also tailored to dynamic MRSI by applying a temporal wavelet sparsifying transform in order to exploit the inherent temporal sparsity. Customized multiband excitation pulses were designed with a lower flip angle for the [1-13C]-pyruvate substrate given its higher concentration than its metabolic products ([1-13C]-lactate and [1-13C]-alanine), thus using less hyperpolarization per excitation. This approach has enabled the monitoring of perfusion and uptake of the pyruvate, and the conversion dynamics to lactate and alanine throughout a volume with high spatial and temporal resolution.
Keywords: compressed sensing, multiband pulses, dynamic MRSI, hyperpolarized C-13
Introduction
Recent studies have demonstrated the feasibility and potential clinical value of metabolic imaging using injected hyperpolarized [1-13C]-pyruvate for novel tissue characterization in vivo (1–11). With this method, the differential conversion of pyruvate to its metabolic products of lactate, alanine, and bicarbonate can be detected in vivo in sub-minute acquisition times. This is of particular value for cancer imaging in which this metabolic profile has been shown to distinguish normal and diseased tissues in preclinical animal models (3–9). This metabolic imaging method has also been used to monitor myocardium reperfusion in the heart following ischemia (11,12).
The high signal enhancement of hyperpolarized agents in vivo has been made possible by the development of methods utilizing Dynamic Nuclear Polarization (DNP) and rapid dissolution techniques that provide a SNR increase of over 40,000 for [1-13C]-pyruvate while producing an injectable solution with physiologic pH, osmolarity, and temperature (1,2). After injection of this solution, the distribution of pyruvate and its products provides metabolic information. The metabolite time courses contain additional dynamic information, such as the perfusion and uptake rate of the injected pyruvate, the duration of the metabolite signal, and the observation times of the metabolic products (13). For example, the lactate dynamics have been shown to be significantly different between tumors and normal tissues (14, 15). The use of hyperpolarized agents, however, requires rapid and efficient MR imaging techniques because the high polarization is irrecoverably lost fairly rapidly in vivo due to T1 relaxation to thermal equilibrium and T2 relaxation following any RF excitation.
Rapid MR spectroscopic imaging (MRSI) methods have been developed and employed to capture the spatial metabolite distributions and, in some cases, their dynamics. Acceleration with echo-planar spectroscopic imaging (EPSI) (16, 17) has been successfully applied for 3D MRSI acquisitions in less than 15 seconds (18), as well as 1D and 2D time-resolved MRSI (13, 19). EPSI has also been combined with compressed sensing methods for further acceleration to provide improved spatial resolution or coverage in 3D MRSI (20,21). Spiral imaging trajectories combined with a least-squares reconstruction have been used for both rapid single time-point and dynamic multislice MRSI (22–24). Parallel imaging can also accelerate the MRSI acquisition and has been applied to hyperpolarized 13C with a sensitivity encoding (SENSE) reconstruction (25). Rapid spectrally-selective imaging methods have been employed as well, including multi-echo chemical shift separation methods that allow for short readout durations (26, 27) and a single metabolite dynamic 3D imaging method using spectral-spatial excitation pulses (28).
In this paper, we present a novel technique that allows for time-resolved, volumetric spectroscopic imaging in vivo with hyperpolarized [1-13C]-pyruvate. The key features are a multiband excitation pulse that efficiently utilizes the hyperpolarized magnetization (19), a rapid acquisition with a randomized EPSI readout and a non-linear reconstruction with compressed sensing acceleration in four (time, frequency, and two spatial) dimensions. In this study, we demonstrate dynamic 13C 3D MRSI covering an entire mouse with 0.066 cc voxels, a 12×12×16 matrix and a time resolution of 2 s, providing the monitoring of spatially localized hyperpolarized metabolite signals for up to 60 s.
Methods
The pulse sequence we developed in this project for dynamic 13C 3D MRSI has two critical components that enable a rapid, efficient acquisition - a multiband excitation pulse and random gradient blips during the readout.
Multiband Excitation
As we have shown previously, a multiband excitation pulse efficiently uses the hyperpolarized magnetization, providing a balance of SNR between metabolites while preserving magnetization for the subsequent acquisitions (14, 19). Efficient magnetization usage is critical for hyperpolarized 13C imaging because the hyperpolarization is unrecoverable after T1 decay to thermal equilibrium or T2 decay following RF excitation. For example, a single 90° pulse will result in complete loss of magnetization after T2 decay, and in general, a θ excitation uses up 1 − cos(θ) of the magnetization.
For in vivo imaging of [1-13C]-pyruvate, we want to observe the metabolic products of [1-13C]-lactate and [1-13C]-alanine. We have excluded 13C-bicarbonate in this pulse design because of its lower concentration, especially in cancer studies, but it could be included for other applications. The pyruvate concentration is generally significantly larger than lactate and alanine, and thus pyruvate is much easier to observe. For this reason, the pyruvate resonance can be excited with a smaller flip angle than its products and still have adequate SNR. Using a smaller flip angle also preserves more pyruvate magnetization, which is essential for dynamic imaging and allows for continued conversion to lactate and alanine over the course of the experiment. This is demonstrated by the simulation results in Figure 1a, where a constant 12° pulse and a multiband pulse with flip angles of 12° for lactate and 1.75° for pyruvate were applied every 250 ms. A conversion rate of kpyr→lac = .025s−1 and longitudinal relaxation rates of T1 = 50s were used. This simulation shows improved lactate signal with the multiband pulse (solid lines) as compared to a conventional constant flip angle pulse (dashed lines) while maintaining adequate pyruvate signal. We have also observed that the conversion from pyruvate to lactate and alanine is much larger than the reverse conversion, therefore a significant portion of the lactate and alanine magnetization can be used for each image while still faithfully observing the metabolic dynamics.
Figure 1.
Pulse sequence components. Multiband Excitation Pulse: (a) Simulated signal with a constant 12° pulse (dashed lines) as compared to the multiband pulse (solid lines). The multiband pulse both lengthens and increases the lactate signal because less pyruvate magnetization has been saturated, while still providing good signal for both metabolites. (b) Off-resonance profile for constant (dashed) and multiband (solid) pulse. The desired excitation bands for the multiband pulse are also shown with the solid gray lines. (c) RF and gradient waveforms. (d) Spectral-spatial profile. (e) Dynamic MRSI pulse sequence, which includes the multiband excitation pulse for efficient magnetization usage (19), two adiabatic pulses for a B1-insensitive spin-echo (18), and an EPSI readout gradient with random phase encoding blips for rapid data acquisition.
One of the RF pulses designed and used in our experiments along with its simulated profile are shown in Figure 1. This pulse has flip angles of 12° for lactate and alanine and 1.75° for pyruvate. We also used two other spectral-spatial pulses with different flip angles for pyruvate of 2.5° and 6° but the same 12° flip angle for lactate and alanine. Several different flip angles were tested empirically to find a good balance between metabolite SNRs as well as a sufficiently long window to observe the dynamics. The customized pulses were also designed to not excite pyruvate-hydrate because it is in rapid exchange with pyruvate, so pyruvate-hydrate excitation would result in additional pyruvate saturation. The pulse spectral selectivity provided bandwidths of ±2.0 ppm (±32 Hz at 3T) for pyruvate, ±1.5 ppm (±24 Hz at 3T) for lactate, and ±1.2 ppm (±19 Hz at 3T) for alanine.
Compressed Sensing
The key requirements for a compressed sensing (CS) acquisition and reconstruction are fourfold: firstly, the data must be sparse in some domain; secondly, there must be adequate SNR; thirdly, the undersampled acquisition must create incoherent aliasing interference in the sparse transform domain; and fourthly, a non-linear sparsity-enforcing reconstruction, e.g. minimum ℓ1-norm, must be used. We have previously applied CS methods to non-dynamic hyperpolarized 13C MRSI (20,21). In this work, we present new sampling strategies and reconstruction methods that are specialized for dynamic MRSI.
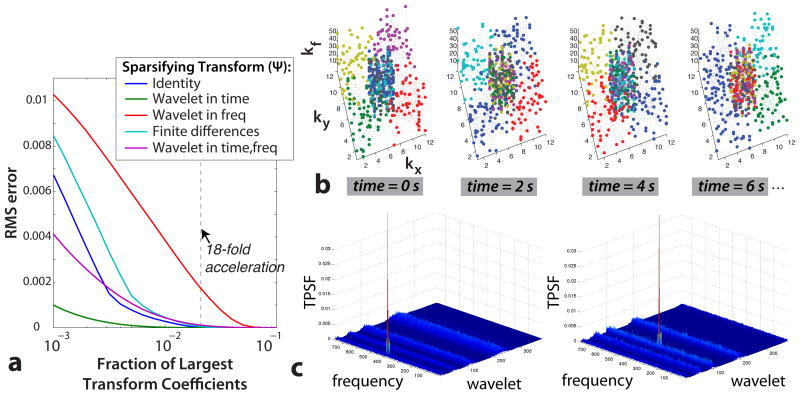
The first CS requirement is fulfilled by the nature of the hyperpolarized 13C signal. The spectrum following injection of hyperpolarized [1-13C]-pyruvate has four primary peaks in vivo -pyruvate, lactate, alanine, and pyruvate-hydrate - and this in itself is sparse. The sparsity is improved by not exciting pyruvate-hydrate with the multiband excitation pulse, which leaves only three peaks in the spectrum. The dynamics following injection are also redundant, and thus can be represented sparsely in a wavelet domain. Figure 2a shows that simulated dynamic hyperpolarized [1-13C]-pyruvate MRSI data can be accurately reconstructed with only a fraction of the largest coefficients in the temporal wavelet (“wavelet-in-time”) transform domain. The relatively low RMS error of the identity transform shows the inherent signal sparsity. The wavelet-in-time transform has the lowest RMS error, thus is the most sparse representation of the data.
Figure 2.
Fulfillment of CS conditions. Sparsity of data in a transform domain: (a) Simulated data reconstructions using only a fraction of the largest transform coefficients. This data was sparsest in the wavelet-in-time transform, and the undersampling ratio used in vivo (dashed line) is several times the number of sparse coefficients. Sampling incoherence: (b) Random phase encoding pattern resulting from the gradient blips during the EPSI readout. The first four time points are shown, and are representative of all time points. The color corresponds to the temporal order of the encoding. (c) TPSF for two different coefficients with the wavelet-in-time sparsifying transform, and the x-y space has been tiled on the plots for different frequency and wavelet values. The aliasing interference is spread incoherently in all dimensions.
In order to fulfill the second CS requirement, there must be adequate SNR for the reconstruction to succeed. For the [1-13C]-pyruvate dose used, we empirically determined the flip angles and resolution for which it was obvious that the reconstruction succeeded, shown by the in vivo spectra and dynamic curves later in this paper. The adequate and expected SNR can also be more precisely derived based on CS theory and prior unaccelerated in vivo data. The CS reconstruction will fail when the noise dominates the signal in the sparse transform domain (29). Using the simulated dynamic MRSI data (described in the “Simulations” section) with the random sampling pattern and wavelet-in-time sparsifying transform, we found that the noise dominated the significant coefficients in the sparse transform domain for peak dynamic SNR values below 3. We can also estimate the expected SNR based on previous in vivo mice experiments using the same [1-13C]-pyruvate dose, but with an unaccelerated 3D MRSI (18) acquisition. The SNR across six of these in vivo studies, acquired 35 s after the start of injection with 0.135 cc voxel size and a progressive flip angle, was measured to be ≥ 60 for pyruvate, lactate, and alanine in the kidneys, liver, and throughout the gut. The expected SNR for the dynamic CS acquisition is reduced by the smaller voxel size of 0.066 cc, and is also decreased by the undersampled acquisition and the smaller excitation flip angles. For the 12° flip on lactate and alanine with 8 acquisitions per image, neglecting T1 decay, the expected dynamic SNR would be ≥ 15, enough for the reconstruction to succeed. For the smallest pyruvate flip angle used, 2.5°, the expected pyruvate SNR is ≥ 3.5, but this is based on in vivo data acquired at 35 s post-injection - well after the peak SNR of pyruvate. The peak pyruvate SNR will be at least 2–3 times greater (13), and thus also is enough for the reconstruction to succeed.
The third requirement, incoherent aliasing interference in the sparse transform domain, is fulfilled by using random phase encoding gradient blips during the echo-planar spectroscopic imaging readout (EPSI) (20, 21). For 3D imaging, these blips occurred in two sampling dimensions, kx and ky, during the EPSI readout in kz and kf, as shown in Figure 1e. The resulting k-space sampling pattern is shown in Figure 2b. It used a 12×12 kx-ky matrix and 8 TRs per time point. Each readout used gradient blips to travel up to 5 k-space phase encode steps. Variable density sampling was used, with 4 readouts randomly sampling the central 4×4 kx-ky region while the other 4 readouts sampled the remaining outer kx-ky regions. Since the majority of the signal energy is in the center of k-space, this significantly improved the reconstruction. The overall resulting CS acceleration factor was 18. The blip pattern was varied over time and the phase encode ordering was randomized across all acquisitions, represented by the encode colors in Figure 2b. These two variations added temporal randomness to the sampling, thus allowing CS acceleration from the dynamic compressibility of the signal. Figure 2c shows the transform point spread function (TPSF) for two different coefficients with a wavelet-in-time sparsifying transform. The TPSF, defined by Equation 2 in (29), shows the aliasing interference of the random sampling pattern in the wavelet-in-time domain. These two representative TPSFs show that the aliasing interference is incoherently spread in x, y, frequency, and the wavelet-in-time dimensions, thus fulfilling the CS sampling requirements.
The final requirement is fulfilled by applying a non-linear CS reconstruction based on the ℓ1-norm with an iterative conjugate gradient method (29). The sparsifying transform, Ψ, was a 1D length-4 Daubechies wavelet transform applied in the time dimension, exploiting the relative smoothness and redundancy in the temporal data. A total-variation (TV) penalty applied along the spectral and spatial dimensions, TVx⃗,f (m), was also included in the reconstruction to promote sparsity in the ℓ1-norm of the finite differences transform as done previously (20,21,29). The reconstruction solved the following problem:
| (1) |
where m is the reconstructed MRSI time series,  is the undersampled Fourier Transform corresponding to the sampling pattern, and y is the acquired data. The wavelet-in-time transformed image and the TV penalty were separately weighted prior to ℓ1-norm minimization, represented by λ and α. All reconstructions used the weights of 10−2 and 10−6, respectively, which were empirically determined through the phantom reconstructions. The missing points in kx-ky-kf-t space were filled in for each z location using the iterative conjugate gradient method, after which a phase correction was applied to compensate for the tilt of the EPSI readout. Every 20 conjugate gradient iterations, the data at the acquired points was filled in with the original data.
is the undersampled Fourier Transform corresponding to the sampling pattern, and y is the acquired data. The wavelet-in-time transformed image and the TV penalty were separately weighted prior to ℓ1-norm minimization, represented by λ and α. All reconstructions used the weights of 10−2 and 10−6, respectively, which were empirically determined through the phantom reconstructions. The missing points in kx-ky-kf-t space were filled in for each z location using the iterative conjugate gradient method, after which a phase correction was applied to compensate for the tilt of the EPSI readout. Every 20 conjugate gradient iterations, the data at the acquired points was filled in with the original data.
Other Parameters
The flyback EPSI readout gradient, with 16 kz and 59 kf samples, acquired data only on the plateaus of the gradient (16). This gradient was 101.48 ms with a spectral resolution of 9.83 Hz and a 581 Hz spectral bandwidth to include all metabolites of interest. A full echo acquisition was used for improved SNR and to allow for magnitude spectral reconstructions. The acquisitions used a TE = 160ms, TR = 250ms, 3.5×3.5×5.4 mm resolution (0.066 cc), a time resolution of 2 s.
Simulations
A single slice of simulated spectra was created using actual data from a 13C phantom with the metabolites of interest, which were completely denoised by thresholding while Lorentzian lineshapes were maintained (21). The peak amplitudes were modulated in time using one-directional two-site exchange model:
| (2) |
| (3) |
The pyruvate signal, P(t), and subsequent metabolic product signal, M(t), were seeded by single and double-peak input functions, I(t). The relaxivity (RP, RM) and exchange (kPM) rates were chosen such that the dynamic shapes were similar to previous in vivo non-localized, 1D, and 2D dynamic acquisitions (19).
Both the previously described “One-at-a-time” ℓ1-norm reconstruction (21), which applies only for individual time points, and the proposed “Wavelet-in-time” ℓ1-norm reconstruction, which was applied to the collection of all time points, were used with reconstruction parameters that were empirically optimized on this simulated data. Complex-valued, white gaussian noise was optionally added to the simulated k-space data. The SNR values for the fully sampled simulation data were normalized by the square root of the undersampling factor. This is because the undersampling reduces the inherent acquisition SNR by approximately the square root of the undersampling factor, which for our trajectory is 18-fold and results in a 4.24-fold decrease in SNR.
Animal Experiments
All animal studies were carried out under a protocol approved by the Institutional Animal Care and Use Committee. Experiments were performed on a 3T MRI system (GE Health-care, Waukesha, WI, USA) with 40 mT/m, 150 mT/m/ms gradients and a broadband RF amplifier. A custom built, dual-tuned mouse birdcage coil was used for RF transmission and signal reception (30). The 13C RF transmitter gain was determined based on previous phantom experiments, and was estimated to be within ±10% of the appropriate value, while the 13C center frequency was calibrated using syringes filled with a 1.77 M concentration of [1-13C]-lactate and 8 M 13C-urea inserted next to the animal in the coil. A preparation consisting of [1-13C] pyruvic acid and 15 mM trityl radical (GE Healthcare, Oslo, Norway) was polarized in a HyperSense DNP system (Oxford Instruments, Abingdon, UK) at 3.35 T and a temperature of 1.4° K. The hyperpolarized pyruvate was dissolved to 80 mM, and 350 μL of this solution was injected into the animal over 10 seconds. An aliquot was taken and injected into a polarimeter to measure the percent polarization. The pH was monitored using the aliquot and other excess pyruvate solution. Both a Tet-o-MYC/LAP-tTA double transgenic mouse model of liver cancer (31, 32) and transgenic adenocarcinoma of mouse prostate (TRAMP) mouse model (33) were used for the experiments.
Results
Simulations
As shown in Figure 3, the reconstruction algorithms were tested at various sampling fractions and for various peak SNR levels. The new wavelet-in-time method consistently outperformed the one-at-a-time method, and it had a low RMS error of less than 0.001 at the 18-fold acceleration factor used in these experiments (Figure 3a). The RMS error becomes much larger at small sampling fractions as the CS reconstruction breaks down, and the one-at-a-time method is worse across all sampling fractions. At 18-fold undersampling, the one-at-a-time method appears to be breaking down, which is due to not enough data sparsity in the transform domain (frequency wavelet).
Figure 3.
Sampling fraction and noise properties of the CS reconstruction. (a) The wavelet-in-time reconstruction had a very low RMS error for sampling fractions down to 18-fold acceleration. The wavelet-in-time always had a lower RMS error than the one-at-a-time method. (b) In the presence of noise, the wavelet-in-time outperforms the one-at-a-time reconstruction consistently. Both CS methods also denoise, resulting in lower RMS errors than the original noisy data. (c) Peak area ratios between the original noiseless data (Sorig) and a reconstruction of noisy data (Srecon) with SNR values up to 66, plotted for both the different peaks as well over their time series. The fits show bias in both methods towards smaller peak areas than the original data. The error bars show one standard deviation from the fit. The data points in (a) and (b) are averages from 3 simulations. The 18-fold accelerated sampling in Figure 2b was used in (b) and (c).
The wavelet-in-time method also outperformed the one-at-a-time method in the presence of noise (Figure 3b,c). The wavelet-in-time method has a lower RMS error than the one-at-a-time method down to low peak SNRs ≤10. The spectra in Figure 4 show how the wavelet-in-time outperforms the one-at-a-time method at a peak SNR of 10. (The RMS error for peak SNRs ≤10 is dominated by the CS denoising and not the peak reconstruction accuracy.) In our conjugate gradient iterations, the original data was periodically reinserted into the reconstructed data. Simulation tests showed larger errors when this reinsertion was not performed (dashed and solid lines in Figure 3b).
Figure 4.
Simulated data reconstruction results: representative spectra with a peak SNR = 10. (a) Wavelet-in-time MRSI data from the 6th time point, showing the spectral quality. The highlighted voxels are plotted in Figure 5. (b) Representative voxels at different locations and time points, scaled to the Sorig SNR values shown. The wavelet-in-time reconstruction outperformed the one-at-a-time method, better reproducing the peaks.
The reconstruction noise response is also illustrated by the peak area ratios of the reconstructions to the original, noiseless phantom (Figure 3c). The fits demonstrate a systematic underestimation that is worse for lower SNRs, as was also shown in Figure 2 in (21). As mentioned in this previous work, the ℓ1-norm reconstruction is equivalent to iterative soft thresholding, which has an underestimation bias for detected peaks (34). This bias of Srecon can be described by a constant, β, and thus a fitting function of (Sorig − β)/Sorig was used in the plot. The underestimation is predictable and depends only on the reconstruction parameters, thus one could estimate β in simulation and then use this estimate to correct the bias at the expense of increasing the variance. In Figure 3c, β = 4.36 for the one-at-a-time while the wavelet-in-time had a smaller bias of β = 1.27. The wavelet-in-time also had less variance in Srecon/Sorig, as shown by the error bars.
Figures 4 and 5 show representative voxels and dynamic curves of simulated and reconstructed data using zero-filling and both ℓ1 reconstruction methods. The proposed wavelet-in-time method performed well in the presence of noise, accurately reproducing the different dynamic shapes and reconstructing low SNR peaks not retained by the one-at-a-time method, as shown in Figure 4b. The failure of the one-at-a-time method - predicted by Figure 3a - can be seen by the substantial temporal instability in the dynamic curves in Figure 5, while the wavelet-in-time method curves better match the simulated data. A double-peak input (I(t) in Eq. 2) was used in the upper-right curve in Figure 5 to simulate two passes of pyruvate in the vasculature, and it was accurately reconstructed by the wavelet-in-time method. These figures also show the underestimation bias of the peak amplitudes, which was worse for the one-at-a-time method.
Figure 5.
Simulated data reconstruction results: peak amplitude dynamic curves with peak SNRs of 10 and infinity. The curves were generated using Equations 2 and 3, and the top row is pyruvate, P(t), while the bottom row is the metabolic product, M(t), such as lactate or alanine. In both cases, the wavelet-in-time curves match the simulated data better than the one-at-a-time method.
In Vivo Experiments
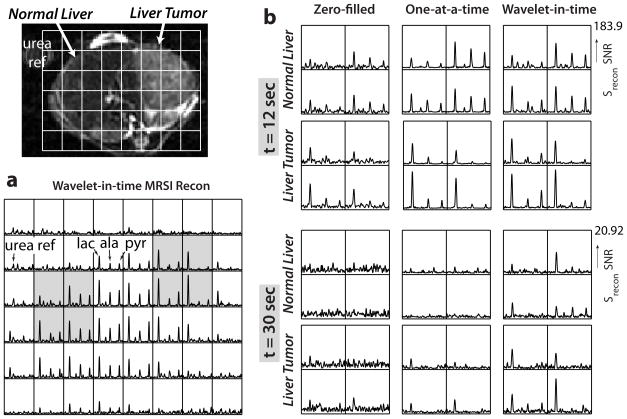
Spectra and dynamic curves from experiments in transgenic liver and prostate cancer mouse models are shown in Figures 6 and 7. They illustrate how the ℓ1 wavelet-in-time reconstruction deblurs and denoises when filling in the undersampled data. There was also a substantial improvement over the one-at-a-time reconstruction, which cannot support the 18-fold undersampling, particularly at later time points where there is less signal - shown in Figure 6b at t = 30 sec by the overall improved peak detection. The wavelet-in-time spectra at t = 12 sec shows some small noise peaks that do not correspond to the chemical shift of any metabolites but illustrate the difficulty in choosing the reconstruction parameters to provide a balance between denoising and reconstruction fidelity. The spectra show a clear difference between the the normal and cancerous liver at both time points.
Figure 6.
Transgenic liver cancer mouse experimental results. The 13C-urea reference syringe can be seen next to the animal. (a) Wavelet-in-time reconstruction at 12 sec after injection, showing the deblurring and representative spectral quality. (b) Reconstructions of the highlighted voxels with zero-filling and both ℓ1 methods. The improved peak detection of the wavelet-in-time method is most evident at 30 sec after injection when the SNR was lower. The liver tumor had higher lactate but lower alanine and pyruvate than the normal liver.
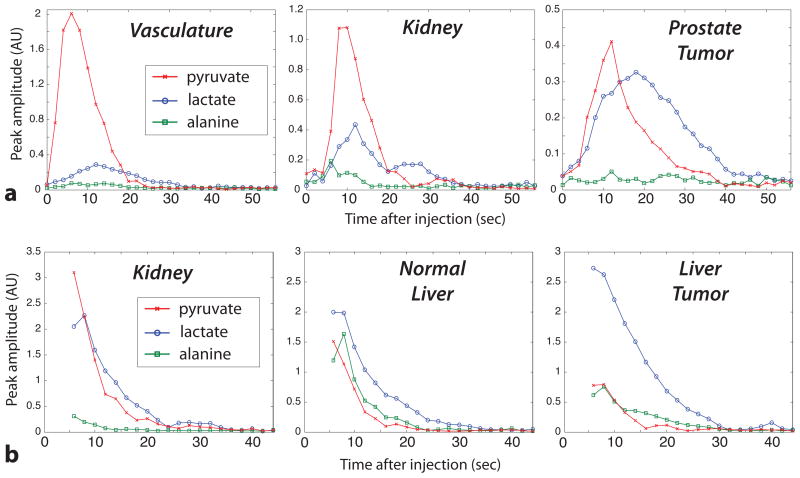
Figure 7.
Reconstructed dynamic curves. (a) Transgenic prostate cancer mouse (TRAMP) experiment. Imaging was started at the beginning of the pyruvate injection with a 12° flip for lactate and alanine and 2.5° for pyruvate. (b) Transgenic liver cancer mouse experiment (same as Fig. 6). Imaging was started 6 sec after the beginning of the pyruvate injection with a 12° flip for lactate and alanine and 6° for pyruvate. The vasculature and kidney voxels had very high pyruvate shortly after the injection, likely in the blood. The highest alanine was seen in the liver. Both tumor types had high lactate signals with a long duration.
With 3D mouse coverage, we were able to compare the metabolite dynamics in various regions, such as the liver, kidneys, vasculature, and tumors (Figure 7). The vasculature mirrored the pyruvate injection, and the pyruvate also passed relatively quickly through the kidneys. The prostate tumor demonstrated a relatively late and persistent lactate signal, which agreed with previous observations in 2D dynamic MRSI (14) and metabolite-specific imaging studies (15). The pyruvate arrival, which is influenced by the vascularity and perfusion, varied throughout the animal, and was later in the prostate tumor when compared to the vasculature and kidneys. In the transgenic liver tumor mouse, the tumor had higher lactate but lower alanine and pyruvate when compared to normal liver, as was seen previously (21), and the pyruvate curves provided perfusion information across the animal.
Color overlay movies of the dynamics in several mice at several slices are also available at http://www.mrsc.ucsf.edu/~plarson/dynamic_c13_mrsi/.
Discussion
The new wavelet-in-time reconstruction method presented uses metabolite spectral and spatial location information obtained from all time points to effectively constrain the reconstruction. Jointly employing all of the available information allowed for accurate reconstructions, even with relatively high noise levels. This method consistently outperformed the previous one-at-a-time method, which only uses information from individual time points, both in simulations and in vivo. Low SNR metabolite peaks were poorly reconstructed by the one-at-a-time method. Similarly, the one-at-a-time method also resulted in substantial temporal instability and fluctuations even with noiseless data, which did not occur in the wavelet-in-time reconstructions (Figure 5). These reconstruction failures with noiseless data are a result of not enough sparsity of the data in the frequency wavelet transform domain; the results in Figure 3a and also Figure 1b in (21) show how the one-at-a-time method breaks down at high undersampling factors. This was not the case for the wavelet-in-time method, which performed similarly in Figure 3a from no acceleration up to the 18-fold acceleration used.
Achieving a rapid temporal resolution of 2 sec allowed us to resolve physiologic processes such as bolus delivery and vascular flow, as seen in Figure 7. Initial implementations of our method were limited to a 6 sec temporal resolution, a period over which the hyperpolarized signal can vary substantially. With this longer temporal resolution, there were blurring and spreading artifacts due to flow, motion and T1 decay, especially around the time of the bolus injection. We found that improving the temporal resolution to 2 sec substantially reduced these artifacts and their accompanying signal losses (data not shown). In humans, who have slower heart rates than mice, these artifacts will likely be less significant, allowing this method to even more precisely resolve flowing metabolites.
In designing the undersampling pattern, we experimented with the variable density sampling, which in general improves the SNR and reconstruction quality because of the high concentration of energy at the center of k-space for typical in vivo spatial distributions. Our final design samples the center 4×4 with 4-fold undersampling while the rest of the 12×12 matrix has 32-fold undersampling. This resulted in a good balance of SNR and high frequency sampling coverage. With this sampling pattern, the data can easily be reconstructed with resolutions of any multiple of 2 sec by combining time points prior to the ℓ1 reconstruction. The sampling pattern was designed such that combined data will also have uniformly distributed random undersampling while still providing sampling incoherence. Using a 4 sec resolution reconstruction - an overall 9-fold acceleration - could be valuable to improve studies with low SNR and when the dynamics are sufficiently slowly varying.
Using the multiband pulses requires accurate frequency calibration and they are susceptible to B0 inhomogeneity. The pulses used had a limited spectral bandwidth achievable at each resonance. The achievable bandwidth is proportional to the pulse length, which should be limited to minimize profile blurring during the pulse. Artifacts can result when the inhomogeneity exceeds the spectral bandwidths and the metabolites do not receive the desired flip angles. The spectral bandwidths used in these mice experiments (≥ ±1.2 ppm) were sufficient to cover the B0 inhomogeneities. In larger animals, such as humans, and over larger volumes, these inhomogeneities will be more likely to cause artifacts. Performing more precise shimming across the volume of interest, which can be done using the proton signal, will reduce the chances of B0 inhomogeneity artifacts.
Conclusion
We have developed and demonstrated a time-resolved 3D MRSI acquisition scheme for hyperpolarized 13C studies that uses customized multiband excitation pulses along with a compressed sensing acquisition and reconstruction. For these studies, there is a very short imaging window due T1 decay and metabolic conversion, thus the acceleration from CS is critical for optimally sampling the temporal metabolite dynamics. Furthermore, each RF excitation accelerates the signal decay by using some of the available hyperpolarized magnetization. This makes the multiband excitation pulses, which efficiently utilize and preserve the magnetization, invaluable for the time-resolved acquisition.
The new CS sampling patterns and reconstruction methods were designed specifically for dynamic hyperpolarized 13C studies and validated in simulations. These new methods enabled more than twice as much acceleration (18-fold vs. 7.53-fold) as previous single time-point 3D MRSI schemes with CS (21). Due to the 18-fold CS acceleration, this sequence acquires a full 3D volume in the same time as our previous time-resolved 2D MRSI sequence (19). It achieves full mouse coverage every 2 sec, and allows for up to 60 sec of metabolite observation in vivo following injection. The time-resolved acquisition provides perfusion, uptake and conversion dynamics for enhanced metabolic imaging and tissue characterization.
Acknowledgments
The authors would like to acknowledge Dr. Robert Bok, Kristen Scott, Dr. Cornelius Von Morze, and Mark VanCriekinge for assistance performing the experiments, and Dr. James Tropp for the 1H/13C mouse coil. This work was supported by an American Cancer Society Postdoctoral Fellowship (grant #PF-09-036-01-CCE), NIH grants (RO1-EB007588, R21-EB005363 & RO1CA111291), the Sir Peter & Lady Michael Foundation, and UC Discovery Grant ITLbio04-10148 with GE Healthcare.
References
- 1.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golman K, Ardenkjaer-Larsen JH, Petersson JS, Mansson S, Leunbach I. Molecular imaging with endogenous substances. Proc Natl Acad Sci U S A. 2003;100:10435–10439. doi: 10.1073/pnas.1733836100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golman K, in ‘t Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci U S A. 2006;103:11270–11275. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen AP, Albers MJ, Cunningham CH, Kohler SJ, Yen YF, Hurd RE, Tropp J, Bok R, Pauly JM, Nelson SJ, Kurhanewicz J, Vigneron DB. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T–initial experience. Magn Reson Med. 2007;58:1099–1106. doi: 10.1002/mrm.21256. [DOI] [PubMed] [Google Scholar]
- 5.Kohler SJ, Yen Y, Wolber J, Chen AP, Albers MJ, Bok R, Zhang V, Tropp J, Nelson S, Vigneron DB, Kurhanewicz J, Hurd RE. In vivo 13carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn Reson Med. 2007;58:65–69. doi: 10.1002/mrm.21253. [DOI] [PubMed] [Google Scholar]
- 6.Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13:1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 7.Nelson SJ, Chen AP, Bok R, Albers MJ, Zierhut ML, Kurhanewicz J, Vigneron DB, Kohler S, Yen YF, Tropp J, Gram A, Wolber J, Dirven H, Hurd RE. Hyperpolarized C-13 MRSI data of dog prostate at 3T. Proceedings of the 15th Annual Meeting of ISMRM; Berlin. 2007. p. 536. [Google Scholar]
- 8.Park I, Larson PEZ, Zierhut ML, Hu S, Bok R, Ozawa T, Kurhanewicz J, Vigneron DB, VandenBerg SR, James CD, Nelson SJ. Hyperpolarized 13C MR metabolic imaging: application to brain tumors. Neuro Oncol. 2010;12:133–44. doi: 10.1093/neuonc/nop043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen YF, Nelson SJ, Vigneron DB, Kurhanewicz J. Hyperpolarized C-13 lactate, pyruvate, and alanine: Noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008;68:8607–8615. doi: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder MA, Cochlin LE, Heather LC, Clarke K, Radda GK, Tyler DJ, Shulman RG. In vivo assessment of pyruvate dehydrogenase flux in the heart using hyperpolarized carbon-13 magnetic resonance. Proc Natl Acad Sci U S A. 2008;105:12051–12056. doi: 10.1073/pnas.0805953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merritt ME, Harrison C, Storey C, Jeffrey FM, Sherry AD, Malloy CR. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proc Natl Acad Sci U S A. 2007;104:19773–19777. doi: 10.1073/pnas.0706235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merritt ME, Harrison C, Storey C, Sherry AD, Malloy CR. Inhibition of carbohydrate oxidation during the first minute of reperfusion after brief ischemia: NMR detection of hyperpolarized 13CO2 and . Magn Reson Med. 2008;60:1029–36. doi: 10.1002/mrm.21760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zierhut ML, Yen YF, Chen AP, Bok R, Albers MJ, Zhang V, Tropp J, Park I, Vigneron DB, Kurhanewicz J, Hurd RE, Nelson SJ. Kinetic modeling of hyperpolarized (13)C(1)-pyruvate metabolism in normal rats and TRAMP mice. J Magn Reson. 2010;202:85–92. doi: 10.1016/j.jmr.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larson PEZ, Bok R, Kerr AB, Lustig M, Hu S, Chen AP, Nelson SJ, Pauly JM, Kurhanewicz J, Vigneron DB. Investigation of tumor hyperpolarized [1-13C]-pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magn Reson Med. 2010;63:582–591. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupo JM, Chen AP, Zierhut ML, Bok RA, Cunningham CH, Kurhanewicz J, Vigneron DB, Nelson SJ. Analysis of hyperpolarized dynamic 13C lactate imaging in a transgenic mouse model of prostate cancer. Magn Reson Imaging. 2010;28:153–62. doi: 10.1016/j.mri.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham CH, Vigneron DB, Chen AP, Xu D, Nelson SJ, Hurd RE, Kelley D, Pauly JM. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005;54:1286–1289. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 17.Yen YF, Kohler SJ, Chen AP, Tropp J, Bok R, Wolber J, Albers MJ, Gram KA, Zierhut ML, Park I, Zhang V, Hu S, Nelson SJ, Vigneron DB, Kurhanewicz J, Dirven HA, Hurd RE. Imaging considerations for in vivo (13)C metabolic mapping using hyperpolarized (13)C-pyruvate. Magn Reson Med. 2009;62:1–10. doi: 10.1002/mrm.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham CH, Chen AP, Albers MJ, Kurhanewicz J, Yen YF, Hurd RE, Pauly JM, Nelson SJ, Vigneron DB. Double spin-echo sequence for rapid spectroscopic imaging of hyperpolarized 13C. J Magn Reson. 2007;187:357–362. doi: 10.1016/j.jmr.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Larson PEZ, Kerr AB, Chen AP, Lustig M, Zierhut ML, Hu S, Cunningham CH, Pauly JM, Kurhanewicz J, Vigneron DB. Multiband excitation pulses for hyperpolarized 13C dynamic chemical shift imaging. J Magn Reson. 2008;194:121–127. doi: 10.1016/j.jmr.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu S, Lustig M, Chen AP, Crane J, Kerr A, Kelley DAC, Hurd RE, Kurhanewicz J, Nelson SJ, Pauly JM, Vigneron DB. Compressed sensing for resolution enhancement of hyperpolarized 13C flyback 3D-MRSI. J Magn Reson. 2008;192:258–264. doi: 10.1016/j.jmr.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu S, Lustig M, Balakrishnan A, Larson PEZ, Bok R, Kurhanewicz J, Nelson SJ, Goga A, Pauly JM, Vigneron DB. 3D compressed sensing for highly accelerated hyperpolarized 13C MRSI with in vivo applications to transgenic mice models of cancer. Magn Reson Med. 2010;63:312–321. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM. Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized 13C imaging. Magn Reson Med. 2006;56:932–7. doi: 10.1002/mrm.21025. [DOI] [PubMed] [Google Scholar]
- 23.Levin YS, Mayer D, Yen YF, Hurd RE, Spielman DM. Optimization of fast spiral chemical shift imaging using least squares reconstruction: application for hyperpolarized (13)C metabolic imaging. Magn Reson Med. 2007;58:245–52. doi: 10.1002/mrm.21327. [DOI] [PubMed] [Google Scholar]
- 24.Mayer D, Yen YF, Tropp J, Pfefferbaum A, Hurd RE, Spielman DM. Application of subsecond spiral chemical shift imaging to real-time multislice metabolic imaging of the rat in vivo after injection of hyperpolarized 13C1-pyruvate. Magn Reson Med. 2009;62:557–64. doi: 10.1002/mrm.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arunachalam A, Whitt D, Fish K, Giaquinto R, Piel J, Watkins R, Hancu I. Accelerated spectroscopic imaging of hyperpolarized C-13 pyruvate using SENSE parallel imaging. NMR Biomed. 2009;22:867–873. doi: 10.1002/nbm.1401. [DOI] [PubMed] [Google Scholar]
- 26.Reeder SB, Brittain JH, Grist TM, Yen YF. Least-squares chemical shift separation for (13)C metabolic imaging. J Magn Reson Imaging. 2007;26:1145–52. doi: 10.1002/jmri.21089. [DOI] [PubMed] [Google Scholar]
- 27.Leupold J, Månsson S, Petersson JS, Hennig J, Wieben O. Fast multiecho balanced SSFP metabolite mapping of (1)H and hyperpolarized (13)C compounds. MAGMA. 2009;22:251–6. doi: 10.1007/s10334-009-0169-z. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham CH, Chen AP, Lustig M, Hargreaves BA, Lupo J, Xu D, Kurhanewicz J, Hurd RE, Pauly JM, Nelson SJ, Vigneron DB. Pulse sequence for dynamic volumetric imaging of hyperpolarized metabolic products. J Magn Reson. 2008;193:139–146. doi: 10.1016/j.jmr.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 30.Derby K, Tropp J, Hawryszko C. Design and evaluation of a novel dual-tuned resonator for spectroscopic imaging. J Magn Reson. 1990;86:256–262. [Google Scholar]
- 31.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–7. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 32.Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med. 2007;13:820–7. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg N, DeMayo F, Finegold M, Medina D, Tilley W, Aspinall J, Cunha G, Donjacour A, Matusik R, Rosen J. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stern AS, Donoho DL, Hoch JC. NMR data processing using iterative thresholding and minimum l(1)-norm reconstruction. J Magn Reson. 2007;188:295–300. doi: 10.1016/j.jmr.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]