Abstract
In Thailand, the term Hill Tribe is used to describe populations whose members traditionally practice slash and burn agriculture and reside in the mountains. These tribes are thought to have migrated throughout Asia for up to 5,000 years, including migrations through Southern China and/or Southeast Asia. There have been continuous migrations southward from China into Thailand for approximately the past thousand years and the present geographic range of any given tribe straddles multiple political borders. As none of these populations have autochthonous scripts, written histories have until recently, been externally produced. Northern Asian, Tibetan, and Siberian origins of Hill Tribes have been proposed. All purport endogamy and have non-mutually intelligible languages. In order to test hypotheses regarding the geographic origins of these populations, relatedness and migrations among them and neighboring populations, and whether their genetic relationships correspond with their linguistic relationships, we analyzed 2445 genome-wide SNP markers in 118 individuals from five Thai Hill Tribe populations (Akha, Hmong, Karen, Lahu, and Lisu), 90 individuals from majority Thai populations, and 826 individuals from Asian and Oceanean HGDP and HapMap populations using a Bayesian clustering method. Considering these results within the context of results of recent large-scale studies of Asian geographic genetic variation allows us to infer a shared Southeast Asian origin of these five Hill Tribe populations as well ancestry components that distinguish among them seen in successive levels of clustering. In addition, the inferred level of shared ancestry among the Hill Tribes corresponds well to relationships among their languages.
Keywords: Thailand, admixture, genomics, substructure
Mitochondrial DNA (mtDNA) or non-recombining Y-chromosomal (NRY) genotypic data have been the focus of numerous anthropological genetic studies. As these are haploid and nonrecombining, the unambiguous haplotype phase provides a direct way to trace the movement of lineages, past human behaviors, and language transmission (Garrigan and Hammer, 2006; Ségurel et al., 2008; Underhill and Kivisild, 2007; Xue et al., 2008). However, vast amounts of autosomal data are currently produced through medical research (Cichon et al., 2009; Ding and Jin, 2009). These data, when analyzed with statistical clustering methods provide a valuable resource to augment mtDNA and NRY studies (Reich et al., 2009; Novembre et al., 2008; McVean, 2009). The sheer volume of autosomal genetic data may partially make up for the drawback of unknown phase, and genetic drift is less likely to completely obliterate a signal in the manner of lineage sorting with mtDNA or NRY. In addition, recombination can be an advantage in that it allows for multiple ancestral components to be inferred, even within one individual. The expense and logistical difficulty of sample collection resulting in small sample sizes can be mitigated by using autosomal markers for which there are immense amounts of public data available from other relevant populations, resulting in statistically powerful studies. We used this strategy to test hypotheses regarding the geographic origins of five Hill Tribe populations sampled in Thailand; Hmong, Karen, Lahu, Lisu, and Akha, by inferring their genetic ancestry from genome-wide single nucleotide polymorphism (SNP) data in combination with corresponding publically available data.
In Thailand, the term Hill Tribe is used to describe populations traditionally residing in the hills and mountains whose members practice slash and burn agriculture. Chinese written records of military conflict indicate that these tribes have migrated throughout Asia for up to 5,000 years and the geographic range of any given tribe straddles multiple political borders (Duncan, 2004; Young, 1962). There have been continuous migrations southward from China into Thailand and its neighboring Southeast Asian countries for the past thousand years of both majority and minority populations (Race, 1974; Michaud, 1997; American Institutes for Research in the Behavioral Sciences. Cultural Information Analysis Center, 1970).
Languages of the Karen, Lisu, Lahu, and Akha are all in the Tibeto-Burman group of the Sino-Tibetan language family. However, geographic origins of Karen are disputed, as is the proper family to which their language belongs (American Institutes for Research in the Behavioral Sciences. Cultural Information Analysis Center, 1970). Not all dialects are mutually intelligible and it has been suggested that there are multiple geographic origins for Karen. Some linguists describe the Hmong language as part of the Miao-Yao group of the Austro-Thai language family or a branch of Sino-Tibetan, while others place the approximately twenty-nine dialects in the Hmongic group of the more isolated Hmong-Mien language family (Matisoff, 1991). In China, the Hmong are generally called Miao, and this designation includes other possibly closely-related groups such as Yao and non-Han mountain-dwelling people.
Theories on the geographic origins of various Hill Tribe populations have been based on interpretations of their physical appearance, oral histories, origin myths, language affiliation, and other cultural practices. The Sino-Tibeto-speaking populations are believed by most linguists to have originated in the Tibetan Plateau, which assumes their languages were transmitted vertically. Other proposed geographic origins, which rely on the accuracy of oral traditions as well as the accuracy of their translations and interpretations, include Mongolia and the far north of Asia or Siberia (Rajah, 2008). There also exist unsubstantiated theories such as Hmong having European ancestry (Quincy, 1995) or the Karen being a perennial Lost Tribe of Israel (Stern, 1968). All of the Hill Tribe populations are purported to be endogamous, however, there is an unknown degree of migration between them and between neighboring Thai or Chinese populations.
A limited number of Y-chromosomal and mtDNA genetic studies have been published with data on Hill Tribe populations and much of what has been published is based on small sample sizes and a small number of markers, in some cases only one marker (Fucharoen et al., 2001; Iwai et al., 2001; Lueangrangsiagun et al., 2005). The only published study to our knowledge including all of the Hill Tribe populations surveyed here, compared results of clustering analyses based on Y-chromosomal and mtDNA data to linguistic relationships between the same populations (Besaggio et al., 2007). The Hill Tribe population samples in the Bessagio et al study were collected in villages located either in the same Thai provinces in which sample collection took place for this study (Mae Hong Son and Chiang Mai) or one neighboring Thai province (Chiang Rai). By combining their data with public data from a number of other studies, for a total of 3044 individuals from 53 population samples with Y-chromosome data and 3644 individuals from 99 populations with mtDNA data, they were able to examine genetic variation of the Hill Tribes within the context of greater Asian genetic variation. Their results showed that, based on mtDNA data, Hmong clustered with Southeastern Chinese minority populations, Lahu, Lisu, Akha. One Karen population clustered with populations from Central Southern China who speak Sino-Tibetan languages, while another Karen population clustered with populations from India who speak Indo-European languages. These results are mostly concordant with historical accounts of the migrations of these populations. Results from 6 Y-chromosomal STRs were much less informative, which is not surprising given the small number of markers.
Two recent large studies of East Asian populations that had been genotyped for hundreds of thousands of SNPs, examining Han Chinese (the majority population in China and the ethnic affiliation of 20% of the world’s population) in particular, that also included public data from HapMap and Human Genome Diversity Project (HGDP) (Cann et al., 2002), reported evidence for two main ancestral populations for present-day Han; and reported that of the two Han HapMap samples, Han Chinese from Beijing (CHB) is closer than Han Chinese from Denver, CO (CHD) to Japanese from Tokyo (JPT) (Xu et al., 2009; Chen et al., 2009) but these and other studies found that there is a closer relationship, overall, between Han and Northeast Asian populations than to Southeast Asian populations (Tian et al., 2008; Teo et al., 2009). Based on inclusion of multiple regional samples collected in China, the two main Han hypothesized parental populations were designated by Xu et al (2009) and Chen et al (2009) as Northern and Southern. This could be interpreted to represent a two-pronged settlement pattern of Asia which has been previously inferred from mtDNA (Kivisild et al., 2002; Yao et al., 2002, 2000) and Y-chromosomal studies (Su et al., 1999; Shi et al., 2005; Tajima et al., 2002). However, a similarly recent and large Human Genome Organization (HUGO) Pan-Asian study (The HUGO Pan-Asian SNP Consortium et al., 2009) concluded that there was one migration into Asia through a southern route, subsequently migrating north and that the present North-South genetic cline represents this migration in which populations have become differentiated through isolation by distance. Regardless of whether this situation was created by a single or two-pronged migration, the Yangtze River has served as a geographic barrier to maintain these differences, and clustering algorithms infer two source populations for Han which correspond to geography.
We used the East Asian North-South gradient of genetic variation established by these three studies to examine genetic variation of Thai Hill Tribe samples using enough data to be able to do this with a higher degree of detail than in any previous reports of which we are aware. We combined genome-wide single nucleotide polymorphism (SNP) data from Thai Hill Tribe and ethnic Thai samples with those of large public data sets such that some population samples and data were overlapping between our study, theHUGO Pan-Asian SNP Consortium (2009), Xu et al (2009) and Chen et al (2009) studies. By comparing the results of Bayesian clustering analysis of our SNP data set with results of these three other studies, we were able to test hypotheses regarding the geographic origins of Thai Hill Tribe population samples as well as the extent of concordance between their linguistic and genetic affinities.
MATERIALS AND METHODS
Populations and sampling
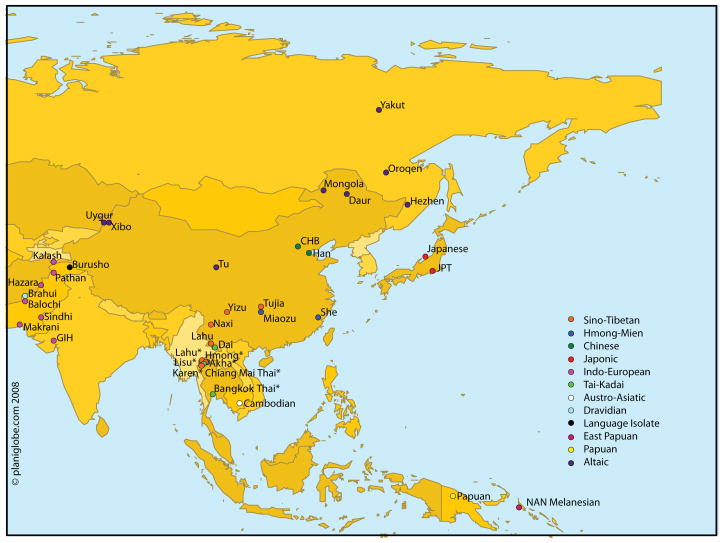
A total of 1023 unrelated subjects were initially selected for inclusion in this study: 118 unrelated individuals from five Hill Tribe population samples collected in Northern Thailand (Hmong, N=25, Akha, N=23, Lahu, N=24, Lisu, N=24, Karen, N=22), 86 unrelated individuals who reported having four Thai grandparents (Chiang Mai, Thailand, N=52 and Bangkok, Thailand, N=34), unrelated individuals from Bangkok, Thailand who reported having four Chinese grandparents (N=4), 348 unrelated individuals from four HapMap samples (International HapMap Consortium, 2005) (Han Chinese in Beijing, China (CHB), N=86, Japanese in Tokyo, Japan (JPT), N=89, Gujarati Indians in Houston, TX (GIH), N=88, and Chinese in Metropolitan Denver, CO (CHD), N=85) and twenty-eight Human Genome Diversity Project (HGDP) population samples (Cann et al., 2002) totaling 467 individuals. All populations and sample sizes are listed in Table 1. A map of the collection locations of our samples, HGDP samples, and the assumed geographic origin of HapMap samples (some of which were collected in the United States) is shown in Figure 1.
Table 1.
SNP data cleaning results for marker overlap and quality and for sample genotyping rate
| SNP data cleaning | ||
| autosomal, non x-linked SNPs with HapMap/Illumina linkage 12 overlap | 5737 | |
| after removal of SNPs with < 0.90 call rates | 4177 | |
| after removal of SNPs with different missing rates between platforms (p ≤ 0.001) | 4115 | |
| after combining with HGDP data | 2445 | |
| Population | N after removal of samples for low genotyping | Genotyping rate for 2445 SNPs |
| Thai samples | ||
| Hmong | 25 | 0.993 |
| Karen | 22 | 0.999 |
| Lahu | 24 | 0.994 |
| Lisu | 24 | 0.989 |
| Akha | 23 | 0.989 |
| Bangkok Thai | 34 | 0.998 |
| Bangkok Chinese | 2 | |
| Chiang Mai Thai | 52 | 0.999 |
| HapMap samples | ||
| GIH | 88 | 0.998 |
| CHD | 85 | 0.996 |
| CHB | 84 | 0.997 |
| JPT | 86 | 0.996 |
| HGDP samples | ||
| Brahui | 25 | 0.999 |
| Balochi | 24 | 0.999 |
| Hazara | 22 | 0.999 |
| Makrani | 25 | 0.999 |
| Sindhi | 24 | 0.999 |
| Pathan | 22 | 0.999 |
| Kalash | 23 | 0.999 |
| Burusho | 25 | 0.999 |
| NAN Melanesian | 10 | 0.999 |
| Papuan | 17 | 0.999 |
| Cambodian | 10 | 0.999 |
| Japanese | 28 | 0.999 |
| Han_N | 44 | 0.999 |
| Yakut | 25 | 0.999 |
| Tujia | 10 | 0.999 |
| Yizu | 10 | 0.999 |
| Miaozu (Hmong) | 10 | 0.999 |
| Oroqen | 9 | 0.999 |
| Daur | 9 | 0.999 |
| Mongola | 10 | 0.999 |
| Hezhen | 8 | 0.999 |
| Xibo | 9 | 0.999 |
| Uygur | 10 | 0.999 |
| Dai | 10 | 0.998 |
| HGDPLahu | 8 | 0.999 |
| She | 10 | 0.999 |
| Naxi | 8 | 0.999 |
| Tu | 10 | 0.999 |
Figure 1.
The Hill Tribe samples and the Chiang Mai Thai in this study are a subset of samples collected as part of an ongoing gene mapping and population genetics study in Thailand in which multiple villages in two Northern Thai provinces were sampled for each Hill Tribe and each participant reported four grandparents from a single population. Samples of self-identified Bangkok Thai (N = 34) and Chinese (N = 4) were obtained from a blood drive in Bangkok, Thailand. DNA was extracted directly from blood using PaxGene materials and the manufacturer’s specified protocol (Qiagen, Valencia CA, USA) (Hmong) or standard phenol/chloroform methods. All Hill Tribe samples and Chiang Mai Thai were collected using Oragene saliva collection kits and DNAs were extracted according to the manufacturer’s protocol (DNA Genotek Inc., Ontario, Canada). Detailed pedigree information was available for the Hmong samples and their relationships were checked with 2445 unlinked SNPs using PLINK v1.07 software (Purcell et al., 2007). Any close relatives in additional samples collected in Thailand were previously identified and excluded using genotypes from 32 unlinked microsatellite markers (Listman, 2009). All subjects provided informed consent as approved by the appropriate institutional review boards.
Markers and genotyping
All 118 Hill Tribe and 90 Thai samples were genotyped using the Illumina Linkage Panel 12 (San Diego, CA, USA), which includes 6090 SNP markers, with an average intermarker distance of 0.65 cM. Genotypes were called and reports were produced using Illumina BeadStudio software (San Diego, CA, USA). Autosomal genotypes for HapMap samples were downloaded from the HapMap website and incorporated into a merged file using IGG3 software (Li et al., 2009), which uses HapMap and chip manufacturer annotation files to correct for strand differences. This produced a merged file with data from 5735 SNPs from the Illumina Linkage Panel 12 chip that also had genotypes reported for some of the HapMap populations included in this study. Overlapping data for Asian and Oceanean Human Genome Diversity Panel (HGDP) population samples that had been genotyped using Illumina 650Y arrays were downloaded from the Stanford Human Genome Center http://hagsc.org/hgdp/files.html (Li et al, 2008) and merged with the above data set, resulting in a data set of 2445 markers. Plink was used to check for strand differences.
Statistical analyses
Data cleaning
PLINK v1.07 software (Purcell et al., 2007) was used to remove from the data set SNPs (< 0.90) and individuals (< 0.95) with low genotyping rates. A Fisher’s exact test was used in PLINK to identify SNPs that had significant differences in missing rates between Hill Tribe and HapMap samples to avoid false clustering due to missing genotypes specific to genotyping platforms or batches. A conservative p-value (considering adjustment for multiple testing would require p ≤ 0.000012) of p ≤ 0.001 was chosen as a cutoff.
Hardy Weinberg Equilibrium (HWE)
Markers were tested for significant deviation from Hardy-Weinberg equilibrium expectations using the Wigginton et al (2005) exact test in PLINK, for each population separately. A Holm-Bonferroni correction for multiple testing was then applied to evaluate the exact test p-values.
Population structure
Since our marker set is a subset of the same markers used to analyze population substructure and evaluate ancestry components in theHUGO Pan-Asian SNP Consortium (2009), Xu et al (2009) and Chen et al (2009) studies and we included JPT, CHB, and CHD, three HapMap populations which were included in those studies, as well, we used these three populations as a reference to infer geographic ancestry components for the Hill Tribe population samples in our study (similar to methods used in our previous study of Jewish populations, (Listman et al. 2010)). Relative to their other population samples, these studies identified JPT as Northern Asian and the HapMap CHB sample had a larger Northern Asian component than that of CHD. We analyzed our data set to determine where the Hill Tribe populations clustered against this established backdrop of a north-south gradient of genetic variation.
The program STRUCTURE 2.2.3 (Falush et al., 2003; Pritchard et al., 2000) uses Bayesian clustering of multilocus genotypes to assign individuals to populations, estimate admixture proportions for individuals, and infer the number of parental populations (K) for a sample. To account for variance of STRUCTURE results, each run was repeated 3 times with all 2445 markers. The parameters used were K=2 through K=13 and 10,000 burn-in and 10,000 Markov chain Monte Carlo (MCMC) iterations. The self-reported population of origin was not used as additional data by STRUCTURE and the presence of admixture was assumed. Runs were carried out using the Computational Biology Service Unit from Cornell University http://cbsuapps.tc.cornell.edu/structure.aspx.
The authors of STRUCTURE recommend using the maximal value for lnP(D) to determine the best value of K for the data. However, it has been observed that lnP(D) will plateau while continuing to increase slightly as assumed K increases past a biologically relevant K. Therefore, identifying the K for which lnP(D) is greatest may not be sufficient to identify the optimal (underlying) K. We employed the ΔK statistic of Evanno et al (2005) within the online version of Structure Harvester v0.56.1 software (Earl, 2009) that uses the output from STRUCTURE to identify the K for which lnP(D) is maximized while both |lnP(D)K+1 − (lnP(D)K − lnP(D)K−1)| and variance of lnP(D)are minimized. This identifies the highest value of K, prior to the plateau of lnP(D).
STRUCTURE runs were unsupervised, using the admixture model and correlated allele frequencies. Structure randomly assigns clusters in each run such that the correspondence between runs is non-obvious. CLUMPP software (Jakobsson and Rosenberg, 2007) determines which clusters from different runs correspond, then averages the assignment values between runs for each individual. To account for cluster label switching between runs, we used the full search option and non-weighted alignment procedure in CLUMPP v1.1.1 to identify corresponding clusters between runs for a set of three runs with a given K and to produce average membership coefficients for each individual for each cluster. These average assignment values were used with the program, DISTRUCT (Rosenberg, 2004), to produce graphs of STRUCTURE output.
RESULTS
Data cleaning
After removal of SNPs from the original 5735 that overlapped between our samples and HapMap samples with low call rates (< 0.90), many of which were SNPs that have not yet been reported for HapMap III samples, there were 4177 SNPs remaining. An additional 62 SNPs were removed due to significant differences in missing rates between HapMap database genotypes and the Hill Tribe samples, leaving 4115 SNPs. SNPs with overlapping data available for the HGDP samples reduced the number of SNPs to 2445. Two individuals from CHB, 3 individuals from JPT, 2 individuals from Bangkok with four Chinese grandparents, and 12 HGDP individuals were removed for low genotyping rates (< 0.95). Final sample sizes and genotyping rates for each population for the 2445 SNPS remaining after data cleaning are in Table 1.
Hardy Weinberg Equilibrium (HWE)
Markers were tested for significant deviation from Hardy-Weinberg equilibrium for each population using the Wigginton et al (2005) exact test. After correction for multiple testing using the Holm-Bonferonni method, no loci showed significant deviation from HWE expectations over all populations.
Population structure
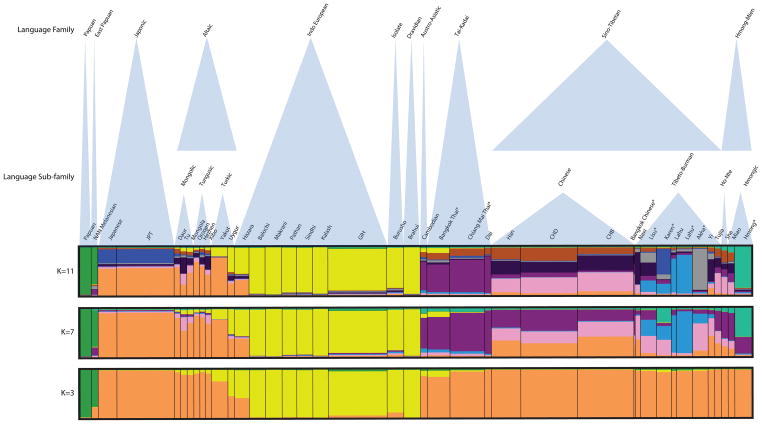
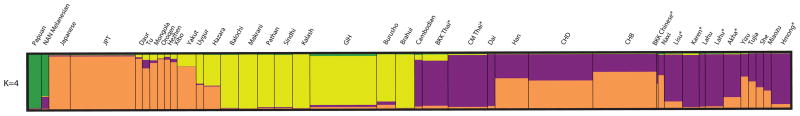
Using the Evanno et al (2005) method to identify statistically significant K, a hierarchical level of clustering was inferred with sharp increases of ΔK at K=3 and K=7 and the highest ΔK at K=11 (Fig 2), indicating that the most likely number of clusters given the data is eleven. The HUGO Pan-Asian SNP Consortium (2009), Xu et al (2009) and Chen et al (2009) studies inferred Northeast Asian ancestry for JPT and a combination of Northeast and Southeast Asian ancestry for CHB, and CHD with greater Southeast Asian ancestry in the CHD sample. Using these three populations as references, we were able to infer a majority Southeast Asian ancestral component (Table 1 and Fig 3, purple portion) for Akha, Karen, Lahu, Lisu, and Hmong population samples in our study when K=4 and all non-Oceanean, non-Indo-European speaking populations are assigned to two primary clusters. However, as the assumed K increased, genetic differentiation between the Hill Tribe samples was detectable in spite of their geographic overlap which has lasted for centuries if not millennia. The level of genetic differentiation as shown by clustering patterns correlated closely with the branching patterns of their language groups. (Fig 2)
Figure 2.
Figure 3.
DISCUSSION
We collected a large set of autosomal SNP data for 118 unrelated individuals from five Southeast Asian Hill Tribe populations, each of which espouses endogamy but also has a long history of migration throughout East Asia, during which has geographically overlapped with multiple ethnic groups. The geographic origins of these populations are unknown, in part because their histories have been entirely oral until relatively recently and when written, have not been done so by members of the populations, themselves. We took advantage of the availability of public SNP data sets for a large number of East Asian, Oceanean, and South Asian populations and the East Asian North-South gradient of autosomal genetic variation established by multiple published studies to examine genetic variation of our samples within a detailed context and infer their geographic origins.
We inferred genetic ancestry components of the Hill Tribe samples using Bayesian clustering techniques with a subset of genome-wide SNPs from the Illumina Linkage 12 chip and corresponding HapMap phase I, II, and III data, HGDP data from Asian and Oceanean population samples, and Illumina Linkage 12 data from ethnic Thai samples from Bangkok and Chiang Mai, Thailand. Then, by comparing our results with those of several other recent studies (Chen et al., 2009; HUGO Pan-Asian SNP Consortium, 2009; Xu et al., 2009), we inferred a shared Southeast Asian origin for Akha, Hmong, Karen, Lahu, and Lisu (Table 2). The clustering patterns, as assumed K increased, corresponded closely with branching of the language phylogeny for these populations. Culturally-influenced migration and isolation between populations is reflected in the ancestry component patterns of individuals.
Table 2.
Mean % assignment per population sample for STRUCTURE clusters when K=4.
| Mean % assignment for STRUCTURE clusters when K=4 | |||||
|---|---|---|---|---|---|
| population | N | South East Asian | South Asian | North East Asian | Oceanean |
| Lahu | 8 | 0.961 | 0.005 | 0.021 | 0.013 |
| Lahu* | 24 | 0.957 | 0.007 | 0.026 | 0.011 |
| Karen* | 22 | 0.955 | 0.027 | 0.013 | 0.004 |
| Dai | 10 | 0.953 | 0.004 | 0.041 | 0.002 |
| Chiang Mai Thai | 52 | 0.951 | 0.016 | 0.026 | 0.006 |
| Hmong* | 25 | 0.928 | 0.003 | 0.065 | 0.004 |
| Lisu* | 24 | 0.862 | 0.014 | 0.117 | 0.008 |
| Cambodian | 10 | 0.857 | 0.090 | 0.035 | 0.018 |
| Bangkok Thai | 34 | 0.827 | 0.114 | 0.049 | 0.010 |
| Akha* | 23 | 0.785 | 0.013 | 0.194 | 0.008 |
| Miaozu | 10 | 0.682 | 0.003 | 0.313 | 0.002 |
| She | 10 | 0.621 | 0.002 | 0.374 | 0.003 |
| Bangkok Chinese | 2 | 0.552 | 0.003 | 0.444 | 0.001 |
| Tujia | 10 | 0.511 | 0.003 | 0.482 | 0.005 |
| CHD | 85 | 0.484 | 0.005 | 0.507 | 0.004 |
| Han | 44 | 0.443 | 0.005 | 0.550 | 0.003 |
| Yizu | 10 | 0.412 | 0.010 | 0.571 | 0.007 |
| Naxi | 8 | 0.372 | 0.009 | 0.599 | 0.019 |
| CHB | 84 | 0.328 | 0.005 | 0.663 | 0.004 |
| NAN Melanesian | 10 | 0.220 | 0.012 | 0.011 | 0.757 |
| Tu | 10 | 0.156 | 0.094 | 0.747 | 0.004 |
| Xibo | 9 | 0.061 | 0.077 | 0.859 | 0.003 |
| Burusho | 25 | 0.061 | 0.865 | 0.068 | 0.006 |
| Hezhen | 8 | 0.051 | 0.039 | 0.905 | 0.004 |
| Mongola | 10 | 0.049 | 0.102 | 0.846 | 0.004 |
| GIH | 88 | 0.038 | 0.898 | 0.030 | 0.034 |
| Daur | 9 | 0.029 | 0.044 | 0.923 | 0.004 |
| Uygur | 10 | 0.024 | 0.533 | 0.441 | 0.002 |
| Japanese | 28 | 0.021 | 0.005 | 0.970 | 0.005 |
| JPT | 86 | 0.020 | 0.006 | 0.969 | 0.006 |
| Sindhi | 24 | 0.018 | 0.957 | 0.012 | 0.013 |
| Pathan | 22 | 0.017 | 0.962 | 0.013 | 0.008 |
| Hazara | 22 | 0.015 | 0.565 | 0.412 | 0.008 |
| Oroqen | 9 | 0.014 | 0.068 | 0.914 | 0.004 |
| Papuan | 17 | 0.014 | 0.002 | 0.003 | 0.982 |
| Balochi | 24 | 0.010 | 0.973 | 0.008 | 0.009 |
| Kalash | 23 | 0.006 | 0.985 | 0.007 | 0.002 |
| Yakut | 25 | 0.005 | 0.219 | 0.774 | 0.002 |
| Brahui | 25 | 0.005 | 0.980 | 0.005 | 0.009 |
| Makrani | 25 | 0.004 | 0.989 | 0.004 | 0.004 |
Samples collected in Thailand for current study are denoted with *.
The recent studies that included numerous Asian population samples as well as the HapMap samples JPT, CHB, and CHD, inferred Northeast Asian and Southeast Asian ancestral gene pools after analyzing their data with the same Bayesian clustering tool, STRUCTURE or with principal components analysis (PCA). Although these studies began with between 400,000 and 600,000 possible genotypes for each subject, in all cases the data were reduced to 100,000 to 150,000 for PCA and to between 15,000 and 20,000 markers for STRUCTURE both to reduce linkage disequilibrium between markers and to reduce computational difficulty. The greater similarity between CHB than CHD to JPT seen in their results and which they attributed partly to Northern/Southern differentiation was replicated here and this consistency indicates that our current marker set of 2445 SNPs should be adequate to reliably infer population structure and Northeastern versus Southeastern Asian ancestry components.
Sampling of Hill Tribe populations took place in Northern Thailand, while the geographic range of each tribe straddles multiple political borders. However, each Hill Tribe was sampled from multiple villages and when possible from multiple provinces within Thailand (a practice not always considered in anthropological genetics, population genetics, or medical genetics studies) to avoid drawing inferences based on what could, in effect, be a single extended family residing in one village (Curran and Buckleton, 2007; Overall and Nichols, 2001). In addition, samples were tested for the presence of first or second degree relatives by evaluating genotype data with statistical methods, followed by the exclusion of all but one of the relatives from further analyses. The genetic similarity between the HGDP Lahu and our Lahu sample indicate that substructure among the Hill Tribe samples is not likely due to unknown close relatives within each sample.
Because the Hill Tribe populations have histories which include migration through Southern China and some of their oral histories were interpreted in the past by missionaries to indicate may have come from Northern China (Lee, 2008) it was logical to compare them with CHB and CHD and to include JPT because there are numerous studies including these HapMap population samples as well as HGDP East Asian populations that are useful reference points. In addition, because the samples were collected in Thailand, we included samples from ethnic Thai populations both from Northern and Southern Thailand. Written history indicates that Karen migrated through Northern India and may have multiple geographic origins (American Institutes for Research in the Behavioral Sciences. Cultural Information Analysis Center, 1970) and one Y-chromosomal study clustered some Karen samples with Indo-European speakers (Besaggio et al., 2007). Thus, we included data from GIH and Southern Asian HGDP samples (collected in India and Pakistan) in our analysis. However, it does not appear that there is a significant Southern Asian component in any of the Hill Tribe population samples.
STRUCTURE can provide information on migration between populations and admixture levels for individuals, shedding light on the reality of human behavior among purportedly endogamous cultures. Results must be interpreted while keeping in mind two issues that affect outcomes of this clustering algorithm. Differences in population sample sizes can affect the results because additional subdivision may be found in a larger population sample while not in smaller samples within the same analysis as a result of additional information provided by more individuals (McVean, 2009; Rosenberg et al., 2005). The demographic histories of Hill Tribe populations may include multiple migrations, bottlenecks, small population size, relatively recent founding, or polygyny, in addition to espoused endogamy. Such populations may form distinct clusters as a result of a small effective population size and genetic drift-driven redistribution of population allele frequencies, but which are not necessarily parental populations contributing to other samples (Listman, 2009).
The level of genetic contribution from each of the Northeast Asian and Southeast Asian ancestral populations for a given East Asian population provides information about the geographic origins of its gene pool but not necessarily its language. However, known histories of the tribal populations in this study and their language relationships are strongly, but not perfectly, reflected in the admixture and clustering patterns seen for a given tribal population. STRUCTURE results (Fig. 2, K=7 and K=11) show a closer genetic relationship between Lisu and Akha than Lahu and Akha, although Lahu and Akha are closer linguistically. These results may be due, in part, to the ancestral Han Chinese (as represented by CHB/CHD and HGDP Han) admixture in Lisu and Akha shown in STRUCTURE. Blurry genetic relationships among the three populations on the Lolo-Burmese-speaking branch of the Sino-Tibetan, Tibeto-Burman language family (Akha, Lahu, and Lisu) are not surprising given the known practices of intermarriage and financial interactions between them as well as the likelihood of Akha and Lisu men speaking Lahu. Lisu men who speak Lahu may marry Lahu women and Lisu intermarriage with Chinese is common in Southern China (Young, 1962; Gordon, 2005). Such couples probably reside with Lisu because traditional marriage practices are more strictly enforced in patrilocal than matrilocal Hill Tribe populations (Besaggio et al., 2007). Therefore genetic evidence of Lahu and Chinese migration into the Lisu population should be expected. Lisu and Akha were not sampled for the HGDP nor are they included in any of the samples for which there is published data for the HUGO Pan-Asian study, as of yet (The HUGO Pan-Asian SNP Consortium et al., 2009) so there were no publically available samples with which to compare ours.
The Hmong population has a recent history of repeated fractioning and migration throughout Southeast Asia as well as loss of numbers due to military conflict, a known history that may contribute to our results (Quincy, 1995). Based on our results, Hmong are the most genetically distinct of the Hill Tribes as well as the most linguistically distinct. The primary ancestry component in our Hmong sample is also represented in the HGDP Miao (an alternative name for Hmong in China) and HGDP She samples, both of which speak Hmong-Mien languages. It would appear that linguistic barriers and possibly other cultural barriers have effectively influenced mating behavior, maintaining genetic differentiation between Hmong presently in Thailand and their neighbors throughout their migration process.
The HUGO Pan-Asian study shows their Karen sample to cluster inconsistently, in the sense that it does not cluster genetically with other Sino-Tibetan-speaking populations rather with geographically close Austro-Asiatic speakers, raising the question of vertical vs. horizontal transmission of language. The Karen sample in our study did cluster with the three other Sino-Tibetan speaking Hill Tribe populations; however there is only one Austro-Asiatic speaking population (HGDP Cambodian) in our study. At the same time, our Karen sample contained significant ancestry from the same Northern Asian component (Fig 2, K=11, dark blue) which identifies the HapMap and HGDP Japanese samples we included. Previous Y-chromosomal studies showed the D-M174 haplogroup to be at high frequencies in some Tibetan populations as well as Andaman and Japanese, but nearly absent elsewhere and suggest that in Tibetan populations this represents the remnants of a population that made an early migration to Northern Asia (Stoneking and Delfin, 2010). It is possible that the inconsistencies in the genetic, linguistic, and geographic affinities of the Karen are explained by partial ancestry from the same remnant in addition to horizontal language transmission.
Acknowledgments
Greg Kay and Ann Marie Lacobelle provided excellent technical assistance. The authors would like to thank Dr. Shizong Han for his help with formatting data. This work was supported in part by the National Institutes of Health [grant numbers R01 DA12849, R01 DA12690, K24 DA15105, K24 AA13736, NIDA R01 DA018363 NIDA K24 DA017899] and the National Institutes of Health/National Institute on Drug Abuse/Fogarty International Center [Thai-US Drug Dependence Genetics Research Training Grant D43-TWO6166]. AM was supported in part by the Thailand Research Fund. JBL was supported in part by a National Institutes of Health/National Institute on Drug Abuse Ruth L. Kirschstein National Research Service Award for Individual Predoctoral Fellows [grant number FDA019761A], a National Science Foundation Doctoral Dissertation Research Improvement Grant [grant number 0622348], and a Wenner Gren Foundation for Anthropological Research Dissertation Fieldwork Grant. Part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University which is partially funded by Microsoft Corporation.
Grant support: JB Listman: NIH/NIDA Ruth L. Kirschstein NRSA Predoctoral Fellows [FDA019761A], NSF Doctoral Dissertation Research Improvement Grant [0622348], Wenner Gren Foundation for Anthropological Research Dissertation Fieldwork Grant. J Gelernter and R Malison: NIH [R01 DA12849, R01 DA12690, K24 DA15105, K24 AA13736, R01 DA018363, K24 DA017899], NIH/NIDA/Fogarty International Center [Thai-US Drug Dependence Genetics Research Training Grant D43-TWO6166]. A Mutirangura: Thailand Research Fund.
Footnotes
Web resources:
IGG3 http://bioinfo1.hku.hk:13080/iggweb/
PLINK http://pngu.mgh.harvard.edu/purcell/plink/
STRUCTURE http://pritch.bsd.uchicago.edu/structure.html
Server for STRUCTURE http://cbsuapps.tc.cornell.edu/structure.aspx
Structure Harvester http://taylor0.biology.ucla.edu/struct_harvest/
Planiglobe http://www.planiglobe.com/omc_set.html
LITERATURE CITED
- American Institutes for Research in the Behavioral Sciences. Cultural Information Analysis Center. Minority groups in Thailand. Washington: Headquarters, Dept. of the Army; 1970. [Google Scholar]
- Besaggio D, Fuselli S, Srikummool M, Kampuansai J, Castrì L, Tyler-Smith C, Seielstad M, Kangwanpong D, Bertorelle G. Genetic variation in Northern Thailand Hill Tribes: origins and relationships with social structure and linguistic differences. BMC Evol Biol. 2007;7(Suppl 2):S12. doi: 10.1186/1471-2148-7-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand M, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng H, Bei J, Sun L, Jia W, Li T, Zhang F, Seielstad M, Zeng Y, Zhang X, et al. Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am J Hum Genet. 2009;85:775–785. doi: 10.1016/j.ajhg.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, Kelsoe J, Lehner T, Levinson DF, Moran A, Sklar P, et al. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166:540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran JM, Buckleton J. The appropriate use of subpopulation corrections for differences in endogamous communities. Forensic Sci Int. 2007;168:106–111. doi: 10.1016/j.forsciint.2006.06.073. [DOI] [PubMed] [Google Scholar]
- Ding C, Jin S. High-throughput methods for SNP genotyping. Methods Mol Biol. 2009;578:245–254. doi: 10.1007/978-1-60327-411-1_16. [DOI] [PubMed] [Google Scholar]
- Duncan CR, editor. Civilizing the margins: Southeast Asian government policies for the development of minorities. Ithaca: Cornell University Press; 2004. [Google Scholar]
- Earl DA. Structure Harvester. 2009 Available from: http://users.soe.ucsc.edu/~dearl/software/struct_harvest/
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucharoen G, Fucharoen S, Horai S. Mitochondrial DNA polymorphisms in Thailand. J Hum Genet. 2001;46:115–125. doi: 10.1007/s100380170098. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Hammer MF. Reconstructing human origins in the genomic era. Nat Rev Genet. 2006;7:669–680. doi: 10.1038/nrg1941. [DOI] [PubMed] [Google Scholar]
- Gordon R. Ethnologue: languages of the world. 15. Dallas, TX: SIL International; 2005. [Google Scholar]
- The HUGO Pan-Asian SNP Consortium. Science. Vol. 326. 2009. Mapping human genetic diversity in Asia; pp. 1541–1545. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K, Hirono A, Matsuoka H, Kawamoto F, Horie T, Lin K, Tantular IS, Dachlan YP, Notopuro H, Hidayah NI, et al. Distribution of glucose-6-phosphate dehydrogenase mutations in Southeast Asia. Hum Genet. 2001;108:445–449. doi: 10.1007/s004390100527. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Tolk H, Parik J, Wang Y, Papiha SS, Bandelt H, Villems R. The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol. 2002;19:1737–1751. doi: 10.1093/oxfordjournals.molbev.a003996. [DOI] [PubMed] [Google Scholar]
- Lee GY. Diaspora and the predicament of origins: interrogating Hmong postcolonial history and identity. Hmong Stu J. 2008;8:1–25. [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Li M, Jiang L, Kao PY, Sham P, Song Y. IGG3: a tool to rapidly integrate large genotype datasets for whole-genome imputation and individual-level meta-analysis. Bioinformatics. 2009;25:1449–1450. doi: 10.1093/bioinformatics/btp183. [DOI] [PubMed] [Google Scholar]
- Listman JB. Thesis. New York University; 2009. Biases in study design affecting the inference of evolutionary events and population structure in closely-related human populations. [Google Scholar]
- Listman JB, Malison RT, Sughondhabirom A, Yang B, Raaum RL, Thavichachart N, Sanichwankul K, Kranzler HR, Tangwonchai S, Mutirangura A, et al. Demographic changes and marker properties affect detection of human population differentiation. BMC Genet. 2007;8:21. doi: 10.1186/1471-2156-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listman JB, Hasin D, Kranzler HR, Malison RT, Mutirangura A, Sughondhabirom A, Aharonovich E, Spivak B, Gelernter J. Identification of population substructure among Jews using STR markers and dependence on reference populations included. BMC Genet. 2010;11:48. doi: 10.1186/1471-2156-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueangrangsiagun T, Bhoopat T, Steger HF. Data on eight STR loci in Shan, Akha, Lisu, Lahu, and Hmong populations of Northern Thailand. J Forensic Sci. 2005;50:482–484. [PubMed] [Google Scholar]
- Matisoff JA. Sino-Tibetan linguistics: present state and future prospects. Anthropology. 1991;20:469–504. [Google Scholar]
- McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5:e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud J. From Southwest China into Upper Indochina: an overview of Hmong (Miao) migrations. Asia Pacific Viewpoint. 1997;38:119–130. [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall AD, Nichols RA. A method for distinguishing consanguinity and population substructure using multilocus genotype data. Mol Biol Evol. 2001;18:2048–2056. doi: 10.1093/oxfordjournals.molbev.a003746. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quincy K. Hmong, history of a people. Cheney: Eastern Washington University Press; 1995. [Google Scholar]
- Race J. The war in Northern Thailand. Modern Asian Studies. 1974;8:85–112. [Google Scholar]
- Rajah A. Remaining Karen: a study of cultural reproduction and the maintenance of identity. Australian National University E Press; 2008. Available from: http://epress.anu.edu.au/karen_citation.html. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségurel L, Martínez-Cruz B, Quintana-Murci L, Balaresque P, Georges M, Hegay T, Aldashev A, Nasyrova F, Jobling MA, Heyer E, et al. Sex-specific genetic structure and social organization in Central Asia: insights from a multi-locus study. PLoS Genet. 2008:4. doi: 10.1371/journal.pgen.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Dong Y, Wen B, Xiao C, Underhill P, Shen P, Chakraborty R, Jin L, Su B. Y-chromosome evidence of southern origin of the East Asian–specific haplogroup O3-M122. Am J Hum Genet. 2005;77:408–419. doi: 10.1086/444436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern T. Ariya and the Golden Book: A millenarian Buddhist sect among the Karen. J Asian Studies. 1968;27:297–328. [Google Scholar]
- Stoneking M, Delfin F. The human genetic history of East Asia: weaving a complex tapestry. Curr Biol. 2010;20:R188–R193. doi: 10.1016/j.cub.2009.11.052. [DOI] [PubMed] [Google Scholar]
- Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J. Y-chromosome evidence for a northward migration of modern humans into Eastern Asia during the Last Ice Age. Am J Hum Genet. 1999;65:1718–1724. doi: 10.1086/302680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima A, Pan I, Fucharoen G, Fucharoen S, Matsuo M, Tokunaga K, Juji T, Hayami M, Omoto K, Horai S. Three major lineages of Asian Y chromosomes: implications for the peopling of East and Southeast Asia. Hum Genet. 2002;110:80–88. doi: 10.1007/s00439-001-0651-9. [DOI] [PubMed] [Google Scholar]
- Teo Y, Sim X, Ong RTH, Tan AKS, Chen J, Tantoso E, Small KS, Ku C, Lee EJD, Seielstad M, et al. Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations. Genome Res. 2009;19:2154–2162. doi: 10.1101/gr.095000.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Kosoy R, Lee A, Ransom M, Belmont JW, Gregersen PK, Seldin MF. Analysis of East Asia genetic substructure using genome-wide SNP arrays. PLoS ONE. 2008;3:e3862. doi: 10.1371/journal.pone.0003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Kivisild T. Use of y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Yin X, Li S, Jin W, Lou H, Yang L, Gong X, Wang H, Shen Y, Pan X, et al. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet. 2009;85:762–774. doi: 10.1016/j.ajhg.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue F, Wang Y, Xu S, Zhang F, Wen B, Wu X, Lu M, Deka R, Qian J, Jin L. A spatial analysis of genetic structure of human populations in China reveals distinct difference between maternal and paternal lineages. Eur J Hum Genet. 2008;16:705–717. doi: 10.1038/sj.ejhg.5201998. [DOI] [PubMed] [Google Scholar]
- Yao Y, Kong Q, Bandelt H, Kivisild T, Zhang Y. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet. 2002;70:635–651. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YG, Watkins WS, Zhang YP. Evolutionary history of the mtDNA 9-bp deletion in Chinese populations and its relevance to the peopling of East and Southeast Asia. Hum Genet. 2000;107:504–512. doi: 10.1007/s004390000403. [DOI] [PubMed] [Google Scholar]
- Young G. The Hill Tribes of Northern Thailand: a socio-ethnological report. 2. Bangkok: Siam Society; 1962. [Google Scholar]