Abstract
Mitochondria are highly heterogeneous organelles that likely have unique isoelectric points (pI), which are related to their surface compositions and could be exploited in their purification and isolation. Previous methods to determine pI of mitochondria report an average pI. This article is the first report of the determination of the isoelectric points of individual mitochondria by capillary isoelectric focusing (cIEF). In this method, mitochondria labeled with the mitochondrial-specific probe 10-N-nonyl acridine orange (NAO) are injected into a fused-silica capillary in a solution of carrier ampholytes at physiological pH and osmolarity, where they are focused then chemically mobilized and detected by laser-induced fluorescence (LIF). Fluorescein-derived pI markers are used as internal standards to assign a pI value to each individually detected mitochondrial event, and a mitochondrial pI distribution is determined. This method provides reproducible distributions of individual mitochondrial pI, accurate determination of the pI of individual mitochondria by the use of internal standards, and resolution of 0.03 pH units between individual mitochondria. This method could also be applied to investigate or design separations of organelle subtypes (e.g. subsarcolemmal and interfibrillar skeletal muscle mitochondria) and to determine the pIs of other biological or non-biological particles.
Keywords: mitochondria, isoelectric point, capillary isoelectric focusing, fluorescence, q-q plot
INTRODUCTION
Mitochondria are heterogeneous organelles responsible for energy production and for cellular signaling pathways such as apoptosis.1 Mitochondrial dysfunction is associated with diseases such as cancer and Parkinson’s, and these organelles are thought to be involved in the aging process.2 It is still unclear how important functional differences may arise from mitochondrial heterogeneity.3 Due to this fact, methods that can distinguish between individual mitochondria are desirable. In particular, methods that underscore heterogeneity at the surface of mitochondria are desirable because it is the surface of these organelles that ultimately may regulate their participation in biological processes; for example, proteins such as the mitofusins are involved in regulating mitochondrial fusion and fission.4
Capillary electrophoresis (CE) is a technique useful for characterization of mitochondrial heterogeneity.5 This technique has been used to analyze individual organelles and other biological particles.6 In this method, organelles or particles are separated in an electric field based on differences in electrophoretic mobility. Individual mitochondria possess different electrophoretic mobilities due to differences in their morphology and outer membrane composition. However, some mitochondria may not exhibit differences in electrophoretic mobility but may still exhibit important functional differences stemming from variation in abundance of outer membrane proteins. Alternatively, isoelectric point (pI) is a property that may allow for characterization and separation of these mitochondria. There are no reports of determination of the pI of individual organelles.
Previous reports of mitochondrial pI vary depending on the source and method used to determine pI. Mitochondria from rat kidney and liver were found to have pIs of 3.9 and 4.4 by observing their migration time across a microscope slide in buffers of different pH.7 Cross-partitioning revealed that rat liver,8 bovine heart9 and rat brain10 mitochondria have pIs of 5.2 – 5.4, 5.6 and 4.5, respectively. The resolving power of this method is not well defined and the isoelectric point determination depends on the partitioning of the sample between phases of different electrostatic potential, requires multiple measurements at different pH, and consumes mL sample volumes. Microfluidic devices designed for free-flow isoelectric focusing have been used to determine that the pI of mitochondria from several cell lines lies between pI 4 and 5.11 The resolving power of these devices is typically on the order of 0.2 pH units or higher, and they use sample amounts larger than those used for cIEF.12 Clearly, while methods such as free-flow isoelectric focusing are ideal for sample enrichment on a preparative scale, they have inherent limitations in resolving power and detection sensitivity that make them unsuitable for investigation of small differences in mitochondrial pI and detection of individual mitochondria.
Capillary isoelectric focusing (cIEF) has been widely applied to the separation of proteins and is suitable for the analysis of biological particles such as microorganisms, viruses, and cells.13, 14 This technique has exceptional resolving power (e.g., ΔpI ~ 0.01 pH units) and requires only μL-scale volumes of sample. In the method described here, mitochondria are isolated from the cell and labeled with the cardiolipin-specific probe 10-N-nonyl acridine orange (NAO). The mitochondria are then suspended in a solution containing carrier ampholytes, pI marker internal standards,15 and sucrose to maintain physiological osmolarity. The carrier ampholytes are photobleached using a light-emitting diode to decrease their characteristic fluorescent background.16 A fused-silica capillary coated with the neutral polymer hydroxypropyl cellulose17 (HPC) to suppress electroosmotic flow is then filled with the mitochondrial suspension. The inlet and outlet are placed in anolyte and catholyte, respectively, and the mitochondria are then focused in the pH gradient formed by application of an electric field, and mobilized past an on-column detection point by replacing the catholyte with an acid.18 Mitochondria are detected as narrow spikes, and pI markers are detected as broad peaks. To demonstrate the sensitivity of this technique to changes in the composition of the mitochondrial surface, the mitochondria are treated with the protease trypsin. This treatment cleaves cytosolic domains of mitochondrial membrane proteins and other proteins associated with the outer surface of mitochondria, altering the pI. Changes in pI as low as 0.03 pH units are clearly detected by the proposed methodology.
EXPERIMENTAL SECTION
Reagents and Materials
House deionized water was further purified with a Synergy purification system (Millipore, Billerica, MA) and was used in preparation of all buffers and solutions. Sodium hydroxide (NaOH), potassium hydroxide (KOH), phosphoric acid (H3PO4), 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), hydroxypropyl cellulose (HPC, average Mw = 100,000), tris(hydroxymethyl)aminomethane (tris), methanol, DMSO, and gentamicin were from Sigma-Aldrich (St. Louis, MO). Ethylene glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid (EGTA) was from Amresco (Solon, OH). Acetic acid was from BDH Aristar (VWR, West Chester, PA). Sucrose was from Roche (Indianapolis, IN). D-mannitol was from Riedel de-Haën (Seelze, Germany). Fluorescein and 10-N-nonyl acridine orange (NAO) were from Molecular Probes (Invitrogen, Carlsbad, CA). Biolyte 3–10 and 4–6 carrier ampholytes (40% solutions) and phosphate buffered saline (PBS, 10×) were from Bio-Rad (Hercules, CA). Dulbecco’s modified Eagle medium (DMEM), fetal bovine serum (FBS), and 0.5% trypsin (10×, 5 g/L trypsin, 2 g/L EDTA·4Na, 8.5 g/L NaCl) were from Gibco (Invitrogen, Carlsbad, CA). Trypan blue stain (0.4%) was from Bio-Whittaker (Walkersville, MD). The Pierce BCA protein assay kit was used for protein quantitation (Thermo, Rockford, IL). Sequencing grade modified trypsin was from Promega (Madison, WI), and was reconstituted according to the manufacturer’s instructions. Fused-silica capillary tubing (50 μm I.D., 150 μm O.D., 12 μm polyimide coating) was from Polymicro (Phoenix, AZ). Fluorescent pI markers were a kind gift from Karel Slais, Academy of Sciences of the Czech Republic.
Buffers and solutions
Mitochondrial isolation buffer contained 210 mM mannitol, 70 mM sucrose, 10 mM HEPES, and 5.0 mM EDTA, adjusted to pH 7.4 with 1 M KOH. The HEPES buffer contained 10 mM HEPES, adjusted to pH 7.4 with KOH. Digestion buffer contained 10 mM tris-HCl, 250 mM sucrose, and 0.1 mM EGTA, adjusted to pH 7.4 with KOH. All buffers were filtered to 0.2 μm before use. A stock solution of 5.2 mM NAO was prepared in DMSO and diluted with mitochondrial isolation buffer before use. A stock solution of 2.25 mM fluorescein was prepared in methanol and diluted with HEPES buffer before use. Focusing solution contained 230 mM sucrose, 1% (w/v) Biolyte 3–10 carrier ampholytes, 1% (w/v) Biolyte 4–6 carrier ampholytes, and the four internal standard fluorescent pI markers at a concentration of 0.02 ng/mL each. The carrier ampholytes were first photobleached overnight with a 120 mW LED with λmax at 472 nm.16 Anolyte, catholyte, and chemical mobilizer solutions for cIEF were, respectively, 100 mM H3PO4, 20 mM NaOH, and 350 mM acetic acid.
Cell Culture
Adherent L6 rat myoblast cells (ATCC, Manassas, VA) were cultured in vented 75 cm2 flasks at 37° C and 5% CO2 in DMEM supplemented with 10% FBS and 0.01 mg/mL gentamicin. Cells were split every 2–4 days after reaching 90% confluence by rinsing with 1× PBS, releasing with 0.5% trypsin in PBS, and seeding back into the flask with fresh DMEM at a 1:40 splitting ratio.
Mitochondrial Preparation
Mitochondria were prepared using mechanical homogenization and differential centrifugation procedures.19 Cells were harvested with 0.5% trypsin in PBS and the cell suspension was added to an equal volume of DMEM. All subsequent procedures were performed on ice or at 4° C unless otherwise noted. Cells were washed three times with mitochondrial isolation buffer and resuspended at an approximate concentration of 5.6 × 106 cells/mL. The cells were disrupted using a glass Potter-Elvehjem homogenizer (Kontes, Vineland NJ) using a 0.06 mm clearance pestle. To measure cell density and homogenization efficiency, aliquots of cells were stained with trypan blue dye before and after homogenization and counted with a Fuchs-Rosenthal counting chamber (Hausser Scientific, Horsham, PA). Cell breakage above 90% was achieved by applying a sufficient number of strokes with the homogenizer and counting the remaining undisrupted cells. After homogenization, intact cells, nuclei, and cellular debris were eliminated from the preparation by centrifuging at 600 g for 10 minutes. Mitochondria in the supernatant were pelleted by centrifuging at 10,000 g for 10 minutes and resuspended in mitochondrial isolation buffer. Mitochondria were labeled by incubating with 5 μM NAO for 10 minutes in the dark. The mitochondria were then pelleted by centrifuging at 10,000 g for 10 minutes and resuspended in mitochondrial isolation buffer. This step was performed two additional times to wash the mitochondria. Mitochondria were then kept on ice until analysis by cIEF. Immediately prior to analysis, aliquots of this mitochondrial suspension were diluted 1:5 with mitochondrial isolation buffer.
Trypsin Treatment
The procedure for trypsin treatment of mitochondria was reported by Forner et al.20 The above preparation procedure was followed until the step before NAO labeling. At this point, mitochondria were instead resuspended in digestion buffer and split into two aliquots. Total protein in the mitochondrial suspensions was determined by the BCA assay. Sequencing grade modified trypsin was reconstituted in the provided solution (1 M acetic acid), heated at 30° C for 30 minutes, and then added to one mitochondrial sample in a 50:1 protein: trypsin ratio. Mitochondrial suspensions were then gently mixed overnight on a rotational gel shaker at 4° C. After this overnight digestion, mitochondria were labeled by NAO and washed with mitochondrial isolation buffer according to the above procedure, then analyzed by cIEF according to the procedure below.
Instrument Description
A homebuilt capillary electrophoresis instrument which has been described previously21 has been adapted to cIEF for this work. A sample carousel was added to the outlet of the capillary for delivery of catholyte and chemical mobilizer. The 488 nm light from a 15 mW Ar+ laser (Melles-Griot, Carlsbad CA) was focused onto the detection window of the capillary. Light was collected at 90° to the laser by a 40×, 0.55 NA objective (New Focus, San Jose, CA). Scattered laser light was eliminated with a 488RU scattering filter (Semrock, Rochester, NY), and fluorescence was passed through a pinhole and a 535 ±17.5 nm band-pass filter (Omega Optical, Brattleboro, VT) onto a photomultiplier tube (R1477, Hamamatsu, Bridgewater, NJ) biased at 1000 V.
Capillary Preparation
Fused-silica capillaries were coated with HPC by a method adapted from Shen et al.17 Briefly, three capillaries were rinsed with methanol, 1.0 M NaOH, 0.1 M NaOH, and H2O for 10 minutes each by flushing under 10 psi nitrogen pressure. The capillaries were purged and flushed with a 5% (w/v) HPC solution for 1 hour. The solution was then purged from the capillaries. After purging, the capillaries were heated between two resistant plates with temperature controlled by a variable-voltage power source from 60 to 140 °C in ~15 minutes, and then holding at 140 °C for 30 minutes. The HPC flush, N2 purge, and heating steps were repeated once for a more robust coating. Capillaries were stored dry until use. Before use, the inlet and outlet were trimmed and the polyimide coating was burned off at the detection window to produce capillaries with total length 38.9 cm and effective length 35.7 cm.
The HPC-coated capillary was first installed in the instrument and aligned by flushing a 1 × 10−9 M solution of fluorescein in HEPES buffer by application of pressure to the inlet. The limit of detection (S/N =3), based on the peak height resulting from injecting 1 × 10−10 M fluorescein in HEPES buffer, was 29 ± 4 zeptomoles (average ± std. dev.; n = 3). The electroosmotic flow, calculated as the difference between fluorescein’s apparent electrophoretic mobility and its reported electrophoretic mobility (i.e., −3.0 ± 0.1 × 10−4 cm2·V−1·s−1 at pH 7.4),19 was 4.26 ± 0.07 × 10−5 cm2·V−1·s−1 (n = 3). This reduction from the value of 5.1 ± 0.1 × 10−4 cm2·V−1·s−1 (n = 15) for uncoated fused silica indicated an effective suppression of EOF by the capillary coating.
cIEF Procedure
To perform the cIEF procedure, the capillary was rinsed first with methanol for 15 minutes, water for 5 minutes, then a solution mixture of 230 mM sucrose, 1% Biolyte 3–10, and 1% Biolyte 4–6 for 10 minutes. A mitochondrial suspension to be analyzed by cIEF was first mixed gently with a pipettor, and then a 2 μL aliquot of this suspension was added to 50 μL of focusing solution and mixed. The sample was introduced to the capillary by application of 20 kPa to the inlet end. The PMT signal was monitored during injection to ensure complete filling of the capillary (a total volume of 760 nL). After injection, the outlet of the capillary was rinsed once in a 0.5 mL vial of catholyte, and then switched to the running vial of catholyte. The inlet was rinsed once in a vial of anolyte, and then switched to the running vial of anolyte. An electric field of 400 V/cm was applied, and the PMT and current signals were monitored during focusing. After the current decreased and reached a plateau indicating completion of focusing (for the experiment shown in Fig. 1, the current changed from −44 μA to −7 μA in 931 seconds), the outlet of the capillary was switched to a chemical mobilization solution of 350 mM acetic acid and the electric field was re-applied. Focused zones were then mobilized past the detector (cathodic mobilization). Between runs, the capillary was rinsed with methanol, water, and the sucrose-Biolyte solution. After the analysis, the capillary was flushed with methanol and water, then purged with air and stored dry.
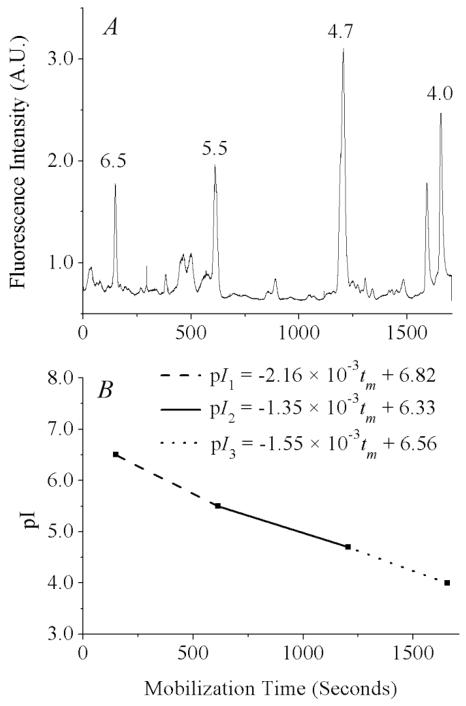
Figure 1.
pH gradient determination using internal standards. (A) Signal from fluorescent pI markers after median filtering to remove narrow spikes. Separation conditions: 1% Biolyte 3–10, 1% Biolyte 4–6, 230 mM sucrose, 0.02 ng/mL of each fluorescent pI marker. The anolyte, catholyte, and chemical mobilizer were 100 mM H3PO4, 20 mM NaOH, and 350 mM acetic acid. The electric field applied during focusing and mobilization was 400 V/cm. Detection was at 535 ± 17.5 nm, data acquisition was at 100 Hz, 1000 V was applied to the PMT. (B) pH gradient, as determined by mobilization times of the four pI markers.
Data Analysis
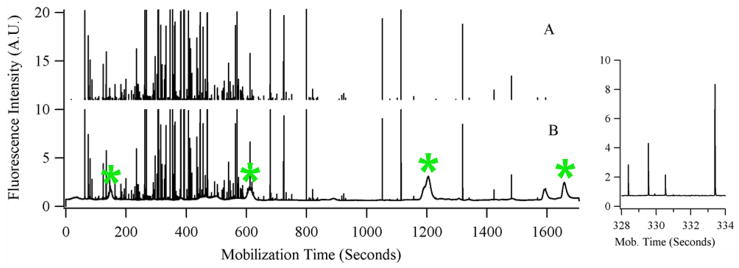
Data from the PMT was acquired at 100 Hz, digitized by an I/O data acquisition card (PCIMIO-16E-50) operated by Labview 5.1 software (National Instruments, Austin, TX), and stored as a binary file. Data was analyzed in Igor Pro software (Wavemetrics, Lake Oswego, OR) by a procedure written in-house which has been described previously, PeakPicks.22 Briefly, electropherograms were median filtered to separate narrow spikes (peaks with baseline widths smaller than 50 data points, or 0.50 sec) from broad peaks (pI markers and fluorescent species in the carrier ampholytes), as shown in Fig. 2. For example, the spikes selected in one experiment had baseline widths of 0.05 ± 0.01 seconds (avg. ± std. dev, n = 405) and the pI marker peaks had baseline widths of 25 ± 9 seconds (avg. ± std. dev, n = 4). The mobilization times of the pI markers were used to define the pH gradient, and the peak widths were used to determine the resolving power of the method. Spikes with signal intensity above a threshold of five standard deviations over the background signal were assigned to mitochondrial events. In addition to individual, intact mitochondria, some of these events may correspond to mitochondrial fragments, aggregates of multiple mitochondria, or a false positives (different organelles or particles exhibiting green fluorescence or a random deviation of the background above the threshold). The threshold limits the number of false positives detected to less than 5% of the total number of detected mitochondrial events (determined by analysis of both an unlabeled and NAO-labeled mitochondrial preparation, see Supplementary Information, page S-2, for details). Each mitochondrial event was then assigned an individual pI based on its mobilization time.
Figure 2.
Signal processing to select mitochondrial events. (B) Signal during mobilization step of focused mitochondria and pI marker internal standards. Narrow spikes represent individual mitochondrial events; broad peaks represent pI markers or fluorescent species from the carrier ampholytes. The pI markers are indicated with an asterisk (*). (A) Mitochondrial events selected after application of median filter and clipping of the lower signal. There are 360 mitochondrial events detected. The inset at the right is an expanded view of a crowded section of the electropherogram showing the resolution between individual spikes.
Peak Overlap
An issue in analysis of particles by capillary electrophoresis is whether peak crowding is a problem; i.e. whether observed peaks represent individual or multiple co-migrating components. Statistical overlap theory has been applied to this problem to determine a threshold for peak saturation, below which the number of observed peaks is a good estimate for the number of actual peaks, and so the effect of peak overlap on the observed distribution is minimal.23 We have applied this statistical test to each mitochondrial cIEF experiment and have found that peak overlap is not a significant problem (see Supplementary Information, page S-3, for details).
RESULTS AND DISCUSSION
Photobleaching of Carrier Ampholytes
Commercial preparations of carrier ampholytes are known to contain fluorescent species.24 This increases the background in cIEF with laser induced fluorescence detection, so it is necessary to photobleach the carrier ampholytes to reduce this background. Ramsay et al. demonstrated an elegant and simple method for reducing the background from the carrier ampholytes by photobleaching them with light emitting diodes with λmax matching that of the laser used for excitation.16 We employed this method using a blue LED with λmax at 472 nm to photobleach a 1:1 mixture of 40% Biolyte 3–10 and 40% Biolyte 4–6 overnight. After photobleaching, the reduction in background fluorescence from the carrier ampholytes was measured by focusing and mobilizing a solution with 2% Biolyte 3–10, 2% Biolyte 4–6, and 220 mM sucrose. The average signal from the untreated carrier ampholytes was 3.6 ± 1.8 V, as compared to 1.1 ± 0.4 V for the photobleached carrier ampholytes (see Supplementary Information, page S-4, for details). This reduction in background signal results in an increase in sensitivity necessary to detect individual mitochondria.
pH Gradient and pI Determination
After the signal from the PMT is median-filtered to separate narrow spikes from broader peaks, the mobilization times (tm) of the four pI markers are used to determine equations relating tm to pI. Since the shape of the pH gradient is dependent on the EOF, which is in turn dependent on the integrity of the capillary coating (we observed an increase in the EOF from 4.26 ± 0.07 × 10−5 cm2·V−1·s−1 directly after coating to 5.78 ± 0.04 × 10−5 cm2·V−1·s−1 after five days and 14 cIEF runs), these internal standards are necessary to compare pI data from run to run. For example, the equations for the pH gradient for the experiment shown in Fig. 1 are given below.
| (Eq. 1-A) |
| (Eq. 1-B) |
| (Eq. 1-C) |
Each mitochondrial event is assigned a pI using the appropriate equation for its mobilization time. Events which are mobilized past the detection point before the pI 5.5 marker are assigned a pI using the first equation, events which are detected between the pI 5.5 and 4.7 markers are assigned a pI using the second equation, and events detected after the 4.7 marker are assigned a pI using the third equation. We used this method, rather than using a single fit to describe the pH gradient, for two reasons. First, the pH gradients produced in cIEF with commercial preparations of carrier ampholytes are not linear.25 The causes of nonlinearity include compression of the pH gradient in the basic region by residual EOF, the presence of fewer basic than acidic carrier ampholytes in commercial preparations, and local effects on the gradient by the analytes themselves.26 Second, the gradient should not be linear across the entire pH range because we used a mixture of narrow (4–6) and wide-range (3–10) carrier ampholytes. Therefore, the most reliable determination of pI in a certain region should be made only with a linear fit between the two pI markers flanking that region.
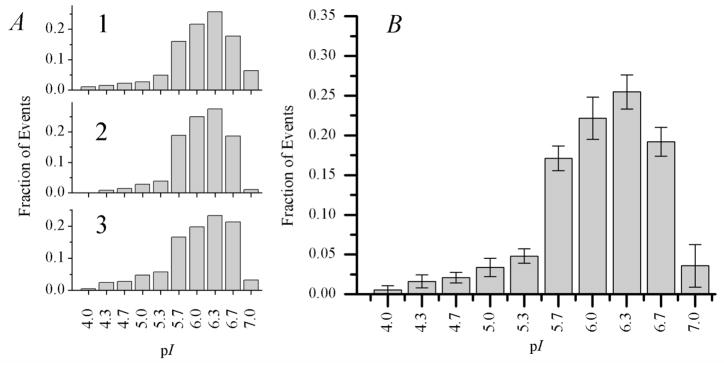
For three replicate analyses of isolated mitochondria, the mean pI was determined to be 5.9 ± 0.6 and the median pI was 6.0 (see Table 1). These values are slightly higher than previous reports of mitochondrial pI mentioned in the introduction, which range from 3.9 to 5.6. These differences could be accounted for by: (i) the different mitochondrial sources, (ii) variations in sample preparation and purification, and (iii) technique biases. Mitochondria in previous determinations were obtained from rat liver, kidney, and brain, bovine heart, and several types of cell cultures, including HT-29 cells, HeLa cells, and NR6wt murine fibroblasts.7–11 Different disruption procedures could cause variations in their outer surface characteristics such as those resulting from different levels of cytoskeletal components that were not removed during the isolation procedure.27 Lastly, some of the previous methods give an indirect measurement of pI (i.e. cross partitioning or observation of electrophoretic mobility at different pH), and did not include pI standards.
Table 1.
Mitochondrial pI statistics from three consecutive cIEF runs.
| Run number | pooled | |||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| number of mitochondrial events | 451 | 360 | 405 | 1216 |
| median pI | 6.0 | 6.0 | 6.0 | 6.0 |
| mean pI | 5.9 | 5.9 | 5.9 | 5.9 |
| standard deviation | 0.6 | 0.5 | 0.6 | 0.6 |
| minimum pI | 3.9 | 4.1 | 3.9 | 3.9 |
| maximum pI | 6.9 | 6.8 | 6.8 | 6.9 |
Resolving power and accuracy
It is informative to determine the resolving power of the method (the minimum difference in pI analytes must possess to be separated). The peak standard deviations of two adjacent pI markers, measured in time units, can be used to determine the resolving power in pI units using the following equation.
| (Eq. 2) |
For the experiment shown in Fig. 1, an average resolving power of 0.03 pH units is calculated. This is comparable to previous reports of resolving power in cIEF. However, the true resolving power of the method for mitochondria should be better than 0.03. Individual mitochondrial events are detected not as broad peaks consisting of a distribution of an analyte focused at its pI, but as narrow spikes with width defined by their migration time through the laser beam.
The accuracy of the pI determination is limited by the accuracy to which the true pIs of the standards are known. Since pI is temperature-dependent and the pIs of the standards are reported to one decimal place, we report mitochondrial pIs with an accuracy of ±0.1 pH units. A full propagation of error on the formula used to calculate individual pI from the internal standards supports this conclusion (see Supplementary Information, pages S-5 through S-7, for details).
Reproducibility of the Mitochondrial pI Distribution
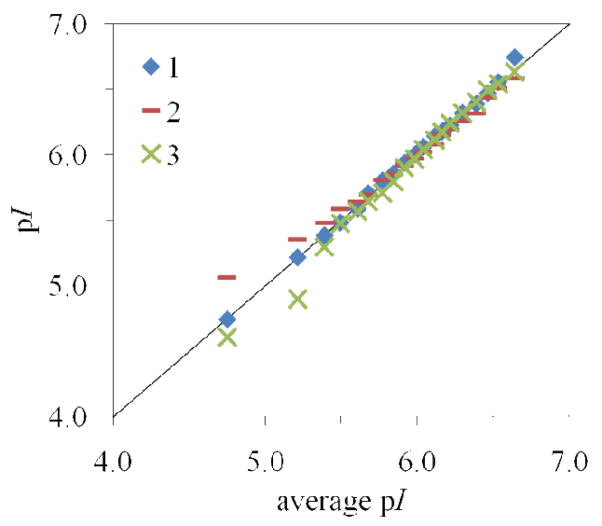
Reproducibility of this technique was assessed by performing three consecutive analyses of the same mitochondrial sample. While the data can be represented statistically as in Table 1 or graphically as in Fig. 3, the quantile-quantile plot (q-q plot) can be used to demonstrate the reproducibility of the distributions more clearly. This plot is produced by graphing the 5th through 95th percentiles of each individual run against the average percentiles of the three runs, and has been used previously to compare mitochondrial migration time distributions in capillary electrophoresis.28 The mitochondrial pI distributions are considered to be qualitatively similar since the majority of the data falls very close to the line. Visual inspection of the q-q plot in Fig. 4 indicates that the distributions are more reproducible above pI 5.5. Additionally, the sum of squares of the residuals (ssres) can be used as a metric for reproducibility:
| (Eq. 3) |
Figure 3.
Mitochondrial pI distributions from three consecutive cIEF runs. (A) Individual distributions from each run. The total number of mitochondrial events detected is 451, 360, and 405 for 1, 2, and 3. Each plot is normalized to the total number of mitochondrial events from that run. (B) Mitochondrial pI distribution, pooled from the three consecutive cIEF runs. Error bars for each bin represent plus or minus one standard deviation between the three runs. The total number of mitochondrial events detected is 1216.
Figure 4.
Q-Q plot to assess reproducibility of three consecutive cIEF runs. The percentiles from each run (y-axis) are plotted against the percentiles of the pooled data (x-axis). This plot is a qualitative indication that the distributions are very reproducible above pI 5.5. The sums of squares of the residuals are 0.015, 0.150, and 0.145 respectively for runs 1, 2, and 3.
For the three consecutive runs, the ssres are 0.015, 0.150, and 0.145 respectively for runs 1, 2, and 3. We will therefore consider distributions to be different if their q-q plot produces a sum of squares of residuals larger than 0.150.
Variation of mitochondrial pI
Biological variation and variations in sample preparation are expected to explain the observed mitochondrial pI distributions. First, natural biological variation between cells and individual mitochondria could be responsible for differences in pI – mitochondria from cells at different stages in the cell cycle, undergoing fusion and fission, or at different energetic status may exhibit different surface characteristics.29 Secondly, differences in sample preparation could cause variation in pI. Variations in the homogenization of the cells and in sample preparation could affect mitochondrial surface characteristics and have previously shown effects on mitochondrial electrophoretic mobility.27 In fact, there is a difference in the pI distributions of freshly-isolated mitochondria described in Table 1 and untreated mitochondria stored overnight described in Table 2. We attribute this difference to variations in sample preparation and the different storage conditions. The experiment described in Table 1 was performed with freshly-isolated mitochondria, and the experiment described in Table 2 used mitochondria that were stored overnight at 4° C in a different buffer.
Table 2.
Effect of overnight trypsin treatment.*
| untreated | trypsin-treated | |
|---|---|---|
| number of mitochondrial events | 461 | 432 |
| median pI | 5.8 | 5.7 |
| mean pI ** | 5.7 | 5.6 |
| standard deviation | 0.7 | 0.6 |
| minimum pI | 3.9 | 3.9 |
| maximum pI | 6.8 | 6.8 |
Data is pooled from two cIEF runs of each sample.
The difference in the means is statistically significant, p = 0.0181.
Effect of Trypsin Treatment
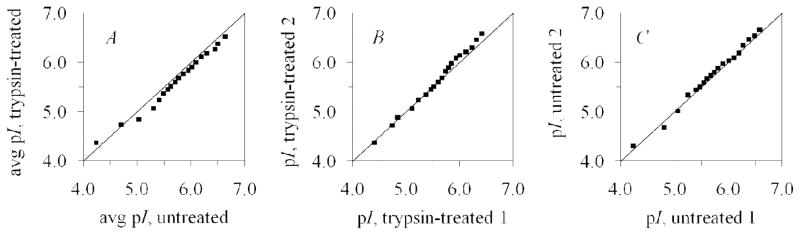
Trypsin is a protease that digests proteins by hydrolytic cleavage of the peptide backbone at the C-terminus of lysine and arginine residues.30 Treatment of a mitochondrial preparation with trypsin should cleave proteins associated with the mitochondrial outer membrane, the cytosolic domains of mitochondrial outer membrane proteins, and cytoskeletal proteins attached to mitochondria.20, 31 We analyzed untreated and trypsin-treated mitochondria by cIEF (two replicate analyses of each sample) and found the mitochondrial pI distribution to be shifted. Examining the q-q plots in Fig. 5 reveals that there is a greater difference between the pI distributions of untreated and trypsin-treated mitochondria than there is between replicate analyses of either sample. When comparing the pooled data from the two cIEF runs of trypsin-treated mitochondria to the pooled data from the untreated mitochondria (Table 2), the ssres is 0.335 (Fig. 5A). This value is larger than the ssres of 0.143 for the comparison of two replicate runs of trypsin-treated mitochondria (Fig. 5B) and 0.081 for the comparison of two replicate runs of untreated mitochondria (Fig. 5C). The ssres of 0.143 and 0.081 are also comparable to those calculated for replicate runs of the same sample in the above section. The median pI of the trypsin-treated mitochondria (5.7) was 0.1 pH units lower than that of the untreated mitochondria. This pI difference could not have been detected with other techniques with resolving power higher than 0.1. The mean pI was also lower (see Table 2). The means were statistically different, p = 0.0181. More information about the pI distribution can be gained from examining the q-q plot; the plot in Fig. 5A indicates that the trypsin-treated pI distribution is shifted to lower pI but approximately the same shape as the untreated distribution. From this we can conclude that trypsin treatment decreases the net charge on the surface of mitochondria, by cleavage of mitochondrial and mitochondrial-associated proteins and cytoskeletal components.
Figure 5.
Q-Q plots showing the effect of trypsin treatment on the pI of mitochondria. The sums of squares of the residuals are 0.335, 0.143, and 0.081 respectively for A, B, and C, indicating that trypsin treatment changes the mitochondrial pI distribution. The majority of points in A fall below the line, indicating that the trypsin-treated pI distribution is shifted to lower pI.
CONCLUSIONS
We have presented a method for the determination of the pI of individual mitochondria by cIEF with laser-induced fluorescence detection. The use of fluorescent internal standards provides an accurate determination of mitochondrial pI. The ability to determine the pIs of individual mitochondria is an important tool for characterizing mitochondrial heterogeneity and detecting changes to the composition of the surface of mitochondria. The fact that mitochondria exhibit a pI distribution reveals that they are a heterogeneous population with different surface characteristics; e.g. mitochondria with a lower pI may express a higher abundance of a protein with a low pI. This method is sensitive to changes in the mitochondrial pI distribution upon treatment with trypsin, which modifies the proteins associated with the surface of the mitochondria. This method could also be used to determine the pI of other biological or synthetic particles, provided an appropriate fluorescent labeling strategy exists. One limitation of the method is the definition of the pH gradient above the pI 6.5 marker – it has been established that commercial preparations of carrier ampholytes are limited in the basic regions, and EOF distorts the pH gradient more at higher pH. This issue could be improved by using different ranges of carrier ampholytes or an improved capillary coating to better suppress EOF.32 Additionally, there is a need for fluorescent pI standards with higher pI for more accurate definition of the pH gradient in the basic regions. The speed and throughput of cIEF is also not ideal. Multicapillary33 and microfluidic34 IEF devices are promising in adapting this technique to a preparative format to collect isolated fractions separated by pI. Further developments of isoelectric focusing techniques may also have an impact on the characterization and purification of synthetic nanoparticles that exhibit differences in ζ-potential.35
Supplementary Material
Acknowledgments
We thank Karel Slais (Institute of Analytical Chemistry of the ASCR, Brno, Czech Republic) for the kind gift of the pI markers. This work was supported by the National Institutes of Health (R01-AG20866). G.G.W. was supported by NIH Training Grant T32-AG029796.
Footnotes
SUPPORTING INFORMATION AVAILABLE: Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Green DR, Reed JC. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 3.Kuznetsov AV, Margreiter R. Int J Mol Sci. 2009;10:1911–1929. doi: 10.3390/ijms10041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen HC, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffy CF, Fuller KM, Malvey MW, O’Kennedy R, Arriaga EA. Anal Chem. 2002;74:171–176. doi: 10.1021/ac010939i. [DOI] [PubMed] [Google Scholar]
- 6.Kostal V, Arriaga EA. Electrophoresis. 2008;29:2578–2586. doi: 10.1002/elps.200700917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plummer DT. Biochem J. 1965;96:729–732. doi: 10.1042/bj0960729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ericson I. Biochimica Et Biophysica Acta. 1974;356:100–107. doi: 10.1016/0005-2736(74)90297-1. [DOI] [PubMed] [Google Scholar]
- 9.Lundberg P, Ericson I. Biochemical and Biophysical Research Communications. 1975;65:530–536. doi: 10.1016/s0006-291x(75)80179-3. [DOI] [PubMed] [Google Scholar]
- 10.Sanchezprieto J, Lopezperez MJ. Biochimica Et Biophysica Acta. 1984;778:81–86. [Google Scholar]
- 11.Lu H, Gaudet S, Schmidt MA, Jensen KF. Anal Chem. 2004;76:5705–5712. doi: 10.1021/ac049794g. [DOI] [PubMed] [Google Scholar]
- 12.Kohlheyer D, Eijkel JCT, Schlautmann S, van den Berg A, Schasfoort RBM. Anal Chem. 2007;79:8190–8198. doi: 10.1021/ac071419b. [DOI] [PubMed] [Google Scholar]
- 13.Horka M, Ruzicka F, Horky J, Hola V, Slais K. Anal Chem. 2006;78:8438–8444. doi: 10.1021/ac061200h. [DOI] [PubMed] [Google Scholar]
- 14.Horka M, Kubicek O, Kubesovi A, Kubickova Z, Rosenbergova K, Slais K. Electrophoresis. 2010;31:331–338. doi: 10.1002/elps.200900310. [DOI] [PubMed] [Google Scholar]
- 15.Slais K, Horka M, Novackova J, Friedl Z. Electrophoresis. 2002;23:1682–1688. doi: 10.1002/1522-2683(200206)23:11<1682::AID-ELPS1682>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 16.Ramsay LM, Dickerson JA, Dada O, Dovichi NJ. Anal Chem. 2009;81:1741–1746. doi: 10.1021/ac8025948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen YF, Smith RD. J Microcolumn Sep. 2000;12:135–141. [Google Scholar]
- 18.Hjerten S, Zhu MD. J Chromatogr. 1985;346:265–270. [Google Scholar]
- 19.Kostal V, Fonslow BR, Arriaga EA, Bowser MT. Anal Chem. 2009;81:9267–9273. doi: 10.1021/ac901508x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forner F, Arriaga EA, Mann M. J Proteome Res. 2006;5:3277–3287. doi: 10.1021/pr060361z. [DOI] [PubMed] [Google Scholar]
- 21.Poe BG, Duffy CF, Greminger MA, Nelson BJ, Arriaga EA. Anal Bioanal Chem. 2010;397:3397–3407. doi: 10.1007/s00216-010-3751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duffy CF, Gafoor S, Richards DP, Admadzadeh H, O’Kennedy R, Arriaga EA. Anal Chem. 2001;73:1855–1861. doi: 10.1021/ac0010330. [DOI] [PubMed] [Google Scholar]
- 23.Davis JM, Arriaga EA. J Chromatogr A. 2009;1216:6335–6342. doi: 10.1016/j.chroma.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Righetti PG, Righetti AB, Galante E. Anal Biochem. 1975;63:423–432. doi: 10.1016/0003-2697(75)90365-6. [DOI] [PubMed] [Google Scholar]
- 25.Shimura K, Zhi W, Matsumoto H, Kasai K. Anal Chem. 2000;72:4747–4757. doi: 10.1021/ac000387o. [DOI] [PubMed] [Google Scholar]
- 26.Righetti PG, Simo C, Sebastiano R, Citterio A. Electrophoresis. 2007;28:3799–3810. doi: 10.1002/elps.200700232. [DOI] [PubMed] [Google Scholar]
- 27.Fuller KM, Arriaga EA. J Chromatogr B. 2004;806:151–159. doi: 10.1016/j.jchromb.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 28.Whiting CE, Arriaga EA. J Chromatogr A. 2007;1157:446–453. doi: 10.1016/j.chroma.2007.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benard G, Rossignol R. Antioxid Redox Signal. 2008;10:1313–1342. doi: 10.1089/ars.2007.2000. [DOI] [PubMed] [Google Scholar]
- 30.Olsen JV, Ong SE, Mann M. Mol Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Anesti V, Scorrano L. Biochim Biophys Acta-Bioenerg. 2006;1757:692–699. doi: 10.1016/j.bbabio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Gao L, Liu SR. Anal Chem. 2004;76:7179–7186. doi: 10.1021/ac049353x. [DOI] [PubMed] [Google Scholar]
- 33.Dada OO, Ramsay LM, Dickerson JA, Cermak N, Jiang R, Zhu CR, Dovichi NJ. Anal Bioanal Chem. 2010;397:3305–3310. doi: 10.1007/s00216-010-3595-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen J, Wilker EW, Yaffe MB, Jensen KF. Anal Chem. 2010;82:1253–1260. doi: 10.1021/ac902157e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyell U. Electrophoresis. 2010;31:814–831. doi: 10.1002/elps.200900555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.