Abstract
A selective, oligonucleotide-based, label-free, turn-on fluorescence detection method for 3′ → 5′ exonuclease activity has been developed using crystal violet as a G-quadruplex-binding probe. The assay is highly simple and rapid, does not require the use of gel-based equipment or radioisotopic labeling, and is amenable to high-throughput and real-time detection. A proof-of-concept of this assay has been demonstrated for prokaryotic ExoIII and human TREX1.
Enzymes that contain 3′ → 5′ exonuclease activities play important roles in a variety of key cellular and physiological processes, such as DNA proofreading.1,2 3′ → 5′ exonuclease inhibitors have the potential to act synergistically with anticancer drugs, increasing their cytotoxic effects against cancer cells by inhibiting DNA repair.2,3 Commonly used 3′ → 5′ exonuclease activity assays are gel-based and/or require the use of radioisotopes such as 3H or γ-[32P]ATP-labeled DNA.4 However, these protocols tend to be unwieldy and time-consuming, and necessitate stringent safety measures to control radiographic exposure. Therefore, the development of an efficient detection method for 3′ → 5′ exonucleolytic activity amenable to high-throughput screening would greatly facilitate the identification of exonuclease modulators for potential therapeutic applications. We describe herein the first selective, label-free, high-throughput G-quadruplex-based turn-on fluorescence assay for 3′ → 5′ exonuclease activity.
The prokaryotic 3′ → 5′ exonuclease III was chosen to demonstrate the proof-of-concept of our approach. Exonuclease III (ExoIII) catalyzes the stepwise hydrolysis of mononucleotides from the 3′-terminus of double-stranded DNA.5 However, ExoIII is unable to catalyze the removal of bases from a single-stranded substrate. We thus designed an unlabelled oligonucleotide hairpin sequence G55 = [5′-AG3(T2AG3)3CAGA2G2AT2A(C3TA2)3C3T-3′] consisting of a 22-bp G-quadruplex-forming sequence at the 5′-terminus and its complementary cytosine-rich sequence at the 3′-terminus connected by a 11-bp flexible linker (Figure S1a). The oligonucleotide G55 was hybridized by annealing at 95 °C for 10 min and slowly cooling to room temperature, forming a stem-loop secondary DNA structure (Figure S1b). The circular dichroism (CD) spectrum of the annealed oligonucleotide confirmed the presence of duplex DNA (see below). We used the human telomeric G-quadruplex sequence [5′-AG3(T2AG3)3-3′] due to the strong fluorescent response of crystal violet (CV) to this G-quadruplex.6 CV is an inexpensive, commonly available triphenylmethane dye that has been demonstrated to display significant selectivity for the G-quadruplex secondary structure over single-stranded and double-stranded DNA.6 Celada and coworkers have measured the 3′ → 5′ exonucleolytic activity of TREX1 employing SYBR Green as a probe to monitor the double-stranded to single-stranded DNA transition of a short duplex.4a However, this “turn-off” fluorescence assay for exonuclease activity is readily subject to false positives due to fluorescence quenching by a variety of interfering mechanisms. Secondly, the presence of an intercalating dye in the reaction mixture may inhibit exonucleolytic activity by competing with the enzyme for the DNA substrate. Most importantly, this approach cannot effectively differentiate between the 3′ → 5′ and 5′ → 3′ exonucleases, or any other exo- or endonuclease acting on either single-stranded or double-stranded DNA
The principle of our 3′ → 5′ exonuclease detection assay is depicted schematically in Scheme 1. Exonuclease III digests DNA specifically from the 3′-terminus (blue line in Scheme 1) in the duplex stem region, but is arrested at the linker region (red line) due to its inability to accept single-stranded DNA as substrate. The 5′ guanine-rich sequence (black line), which is unaffected by the digestion, is released and folds into a G-quadruplex in the presence of potassium ions. CV binds strongly to the G-quadruplex and the emission of CV is high. In the absence of ExoIII, the G-quadruplex DNA secondary structure is not formed, and the emission of CV is low due to the weak interaction between CV and duplex DNA. Ren and co-workers have used a related assay to detect RNase activity.7
Scheme 1.
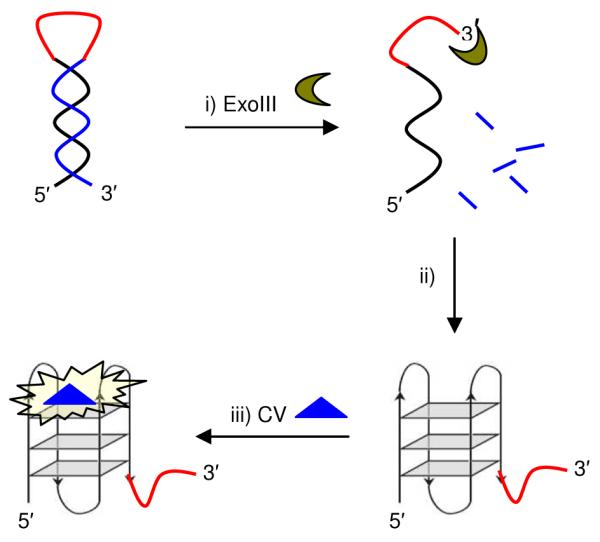
Principle of 3′ → 5′ exonuclease activity assay. i) ExoIII digests DNA from the 3′-terminus, but is halted at the loop region; ii) The released guanine-rich sequence folds into a G-quadruplex; iii) G-quadruplex-selective probe crystal violet (CV) exhibits a “turn-on” emission response.
In order to validate our approach, we incubated oligonucleotide G55 (15 μM) with various concentrations of ExoIII (0–2000 U/mL). Encouragingly, we observed that the fluorescence intensity of CV increased as the concentration of ExoIII was increased (Figure 1). The emission response of CV was significantly diminished when ExoIII was heat-inactivated before incubation with G55 (Figure S2). Furthermore, no significant increase in the background fluorescence signal of CV was observed upon addition of ExoIII enzyme in the absence of oligonucleotide (Figure S3). This suggests that the increase in fluorescence intensity of CV upon incubation of G55 with the enzyme is most likely due to the 3′ → 5′ exonuclease activity of ExoIII. A control experiment was performed with hairpin oligonucleotide G55m, which is identical to G55 but with four guanine residues replaced by other nucleotides [5′-A2G2T2AGCGT2AG2AT2ACGGCAGA2G2ATA2C2GTA2TC2TA2CGCTA2C2T2-3′]. Digestion of G55m by ExoIII releases a single-stranded sequence which cannot fold into a G-quadruplex. The emission response of CV is low due to the weak binding of CV to single-stranded DNA (Figure S4).
Figure 1.
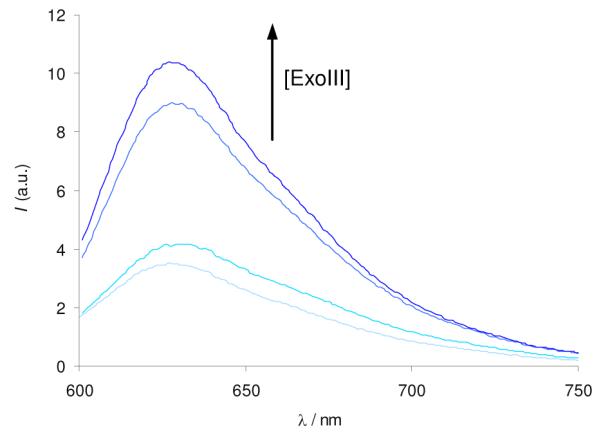
Fluorescence response of CV (1 μM) in the presence of oligonucleotide G55 (0.25 μM). G55 (15 μM) was incubated with ExoIII (0, 20, 200, 2000 U/mL) at 37 °C for 30 min.
The digestion of oligonucleotide G55 by ExoIII could be monitored by CD spectroscopy. The CD spectrum of untreated G55 exhibits an intense positive peak at 270 nm and a strong negative peak at 240 nm, which is characteristic for duplex DNA (Figure 2). Upon incubation with ExoIII, the spectrum changes to reveal a positive band at 290–300 nm, a weak negative peak at 240 nm, and a characteristic positive band at 210 nm (Figure 2). This spectrum suggests the formation of a mixture of parallel and anti-parallel G-quadruplex structures,8 which have been reported to be the dominant quadruplex topologies of the human telomeric sequence under K+ ions, and is consistent with the CD spectrum of the human telomeric 21-bp G3(T2AG3)36 or 22-bp AG3(T2AG3)38 G-quadruplexes. These results confirm that ExoIII efficiently degrades the initial duplex DNA structure formed by G55, resulting in a G-quadruplex secondary structure formed presumably from the release of the guanine-rich strand. Real-time analysis of the CD spectra of G55 reveals a time-dependent decrease of the duplex peak at 270 nm and a concomitant increase of the G-quadruplex signal at 295 nm (Figure S5a). With G55m however, the CD signal at 295 nm continually decreased, suggesting that the digestion of the duplex structure of G55m by ExoIII was not accompanied by G-quadruplex formation (Figure S5b). To provide further confirmation of the DNA hydrolysis reaction, we monitored the time-course of the digestion reaction using a gel electrophoresis (Figure S6). We noticed the time-dependent appearance of a faster-running band, which matched the band containing the expected DNA hydrolysis product, [5′-A(GGGTTA)3GGGCAGAAGGATAA-3′]. These results are consistent with the previous data and support the hypothesis that the observed fluorescence increase of CV for oligonucleotide G55 is most likely due to G-quadruplex formation after digestion of the hairpin structure by ExoIII.
Figure 2.
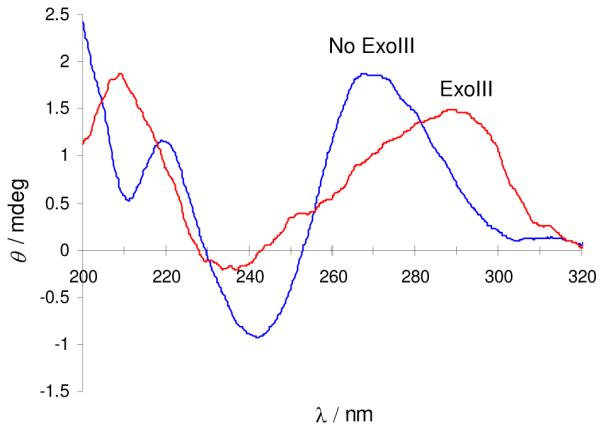
Circular dichroism spectra of G55 (5 μM) in Tris buffer (10 mM Tris-HCl, 10 mM KCl, pH 7.5). G55 was incubated in the presence (red) or absence (blue) of ExoIII. The red trace has been scaled by a factor of 2 for clarity.
We next examined whether our assay could selectively differentiate between different types of nucleases. The design of the stem-loop motif of G55 ensures that a 3′ → 5′ exonuclease will invariably excise nucleotides from the cytosine-rich strand at the 3′-termini. Furthermore, only a 3′→ 5′ exonuclease accepting duplex DNA as a substrate can promote the release of the G-quadruplex sequence. Therefore, this system is selective for ExoIII over T7 exonuclease, exonuclease I (ExoI) and DNase I. T7 exonuclease acts on duplex DNA in the 5′→ 3′ direction.9 This releases the cytosine-rich human telomeric strand, which is not expected to form the i-motif DNA secondary structure under the experimental conditions (pH 7.5).10 The interaction between CV and single-stranded DNA is weak, resulting in a low emission response for T7 exonuclease (Figure 3). ExoI is a 3′ → 5′ exonuclease that only catalyzes the removal of nucleotides from single-stranded DNA.11 Hence, ExoI is unable to act on the G55 stem-loop, and the emission response of CV is similar to that of untreated G55 (Figure 3). This result demonstrates that our assay is able to differentiate between different 3′ → 5′ exonucleases depending on their preferred DNA substrates. Finally, DNase I non-discriminately cleaves all types of DNA to release nucleotide products.12 G55 becomes fully digested and the emission response of CV is low (Figure 3). Taken together, these results suggested that our assay is capable of distinguishing between different types of nucleases. Importantly, however, only ExoIII results in a “turn-on” fluorescent response (Figure 3).
Figure 3.
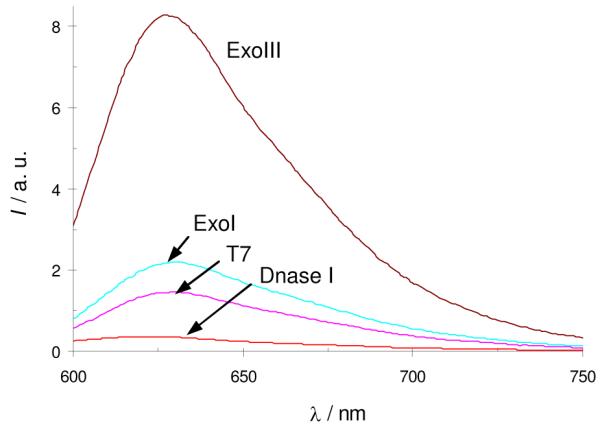
Selectivity for ExoIII over other nucleases. Fluorescence response of CV (1 μM) with oligonucleotide G55 (0.25 μM). G55 (15 μM) was treated with ExoIII, ExoI, T7 or DNase I (all 200 U/mL) at 37 °C for 30 min.
We next explored whether our 3′ → 5′ exonuclease detection assay could be adapted to a high-throughput format. A high-throughput, microplate-based method carries the following advantages: i) lower amounts of reagents can be used, reducing the cost of the assay; ii) the emission measurement can be performed on standard fluorescence microplate readers which are generally more accessible than fluorescence spectrophotometers; iii) a large number of samples can be processed simultaneously on a microplate. In order to demonstrate the feasibility of the high-throughput method, we performed an exonuclease III activity assay in a 96-well microplate employing nine different enzyme concentrations in duplicate spanning three orders of magnitude. Under these conditions, the first significant increase in CV emission intensity was observed at 5 U mL−1 of ExoIII, and the maximum intensity was reached at 50 U mL−1 (Figure 4). This result demonstrates that our assay for 3′ → 5′ exonuclease activity is easily amenable to a high-throughput format. We envisage that this high-throughput assay may be readily adapted for the rapid screening of exonuclease activity modulators. The emission intensity of CV decreased slowly after reaching a maximum value (Figure S7), a phenomenon that was also observed in the RNase assay by Ren and co-workers.7 The real-time CD analysis suggests that the G-quadruplex is not a substrate for the exonuclease, as evidenced by the CD signal at 295 nm reaching a stable value (Figure S5a). Another possibility that could contribute to this decrease is the photobleaching of the organic dye. We anticipate that further optimization of the assay protocol could mitigate this effect. Given that the rate of fluorescence increase can be used to evaluate exonuclease activity, the subsequent decrease is considered to be non-critical for the practical application of this assay.
Figure 4.
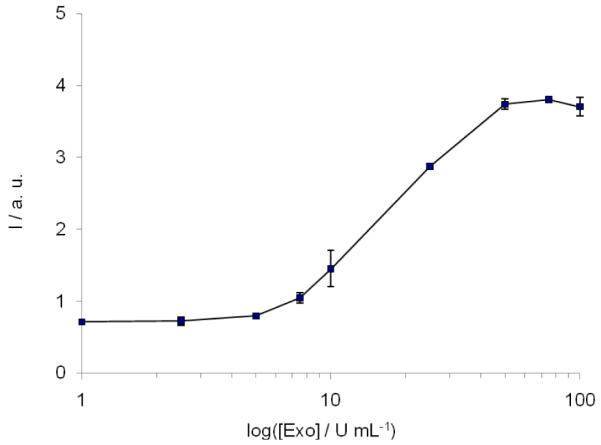
High-throughput assay for ExoIII activity. Fluorescence of CV (2 μM) with oligonucleotide G55 (0.5 μM) vs. ExoIII concentration. G55 (1 μM) was incubated with ExoIII (0–200 U/mL) at 37 °C for 30 min. Error bars represent the standard error of duplicate results.
We next applied our G-quadruplex-based fluorescence assay to detect the exonucleolytic activity of human TREX1, the major 3′ → 5′ exonuclease in mammalian cells.13 Interestingly, recent research has shown that TREX1 may not be involved in DNA repair but could instead be vital for controlling autoimmunity.14 The inactivation of TREX1 and the related TREX2 has been associated with impaired apoptosis and autoimmune-like inflammatory diseases.15 Additionally, we wished to investigate whether the exonucleolytic activity of TREX1 could be monitored in real-time. Since CV binds strongly to the G-quadruplex digestion product and weakly to the duplex hairpin substrate, we hypothesized that the addition of CV as a probe to monitor the time course of the reaction should not significantly affect TREX1 activity. We observed that the emission intensity of CV increased with time in the presence of TREX1 (Figure 5). The results indicate that the exonucleolytic activity of TREX1 can be monitored in real-time using this assay, contrasting favourably with gel-based radiographic techniques that quantitate only the endpoint of the digestion reaction. The sensitivity of this assay for nanomolar concentrations of TREX1 enzyme is comparable to that of gel-based and radiographic methods previously reported.4a,b
Figure 5.
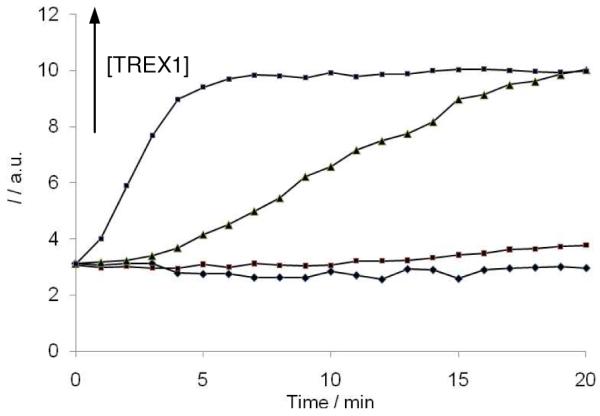
Time-course of TREX1 activity. Emission intensity of CV (2 μM) with G55 (0.5 μM) vs. time in the presence of TREX1 (0, 0.60, 1.80, 6.02 nM) at 25 °C.
In conclusion, we have described the first selective, label-free, high-throughput G-quadruplex-based turn-on fluorescence assay for 3′ → 5′ exonuclease activity. Our method is highly simple and rapid, uses unmodified oligonucleotides, and avoids the requirement for gel-based equipment and/or radioactive labels. Based on the critical design of the oligonucleotide and the selectivity of CV for the G-quadruplex, this assay can effectively discriminate between various nucleases. This assay is readily amenable to a high-throughput format and the exonucleolytic activity of the enzyme can be monitored in real-time. We have demonstrated a proof-of-concept of this assay for the detection of the 3′ → 5′ exonucleolytic activities of the prokaryotic ExoIII and the human TREX1 proteins.
Supplementary Material
Acknowledgement
This work was supported by the Hong Kong Baptist University (FRG1/09-10/064 and FRG2/09-10/070) and the National Institutes of Health U.S.A (CA-063477 and AI038204).
Footnotes
Supporting Information Available: Experimental procedures and supplementary figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).(a) Byrnes JJ, Downey KM, Black VL, So AG. Biochemistry. 1976;15:2817. doi: 10.1021/bi00658a018. [DOI] [PubMed] [Google Scholar]; (b) Simon M, Giot L, Faye G. EMBO J. 1991;10:2165. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Paull TT, Gellert M. Mol. Cell. 1998;1:969. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- (2).Shevelev IV, Hübscher U. Nat. Rev. Mol. Cell Biol. 2002;3:1. doi: 10.1038/nrm804. and the references therein.
- (3).(a) Belmont P, Jourdan M, Demeunynck M, Constant J-F, Garcia J, Lhomme J. J. Med. Chem. 1999;42:5153. doi: 10.1021/jm9901428. [DOI] [PubMed] [Google Scholar]; (b) Belmont P, Demeunynck M, Constant J-F, Lhomme J. Bioorg. Med. Chem. Lett. 2000;10:293. doi: 10.1016/s0960-894x(99)00681-2. [DOI] [PubMed] [Google Scholar]
- (4).(a) Brucet M, Querol-Audí J, Bertlik K, Lloberas J, Fita I, Celada A. Protein Sci. 2009;17:2059. doi: 10.1110/ps.036426.108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nimonkar AV, Ozsoy AV, Genschel J, Modrich P, Kowalczykowski SC. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16906. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ishino Y, Iwasaki H, Kato I, Shinagawa H. J. Biol. Chem. 1994;269:14655. [PubMed] [Google Scholar]; (d) Elisseeva E, Mandal SS, Reha-Krant LJ. J. Biol. Chem. 1989;274:25151. doi: 10.1074/jbc.274.35.25151. [DOI] [PubMed] [Google Scholar]; (e) Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW. J. Biol. Chem. 2008;46:31649. doi: 10.1074/jbc.M806155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rogers GS, Weiss B. In: Methods Enzymol. Grossman L, Moldave K, editors. Vol. 65. Academic Press; New York: 1980. pp. 201–211. [Google Scholar]
- (6).Kong D-M, Ma Y-E, Wu J, Shen H-X. Chem. – Eur. J. 2009;15:901. doi: 10.1002/chem.200801441. [DOI] [PubMed] [Google Scholar]
- (7).Hu D, Pu F, Huang Z, Ren J, Qu X. Chem. – Eur. J. 2010;16:2605. doi: 10.1002/chem.200902166. [DOI] [PubMed] [Google Scholar]
- (8).Paramasivan S, Rujan I, Bolton PH. Methods. 2007;43:324. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- (9).Kerr C, Sadowski PD. J. Biol. Chem. 1972;247:305. [PubMed] [Google Scholar]
- (10).Phan AT, Mergny J-L. Nucleic Acids Res. 2002;30:4618. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lehman IR, Nussbaum AL. J. Biol. Chem. 1964;239:2628. [PubMed] [Google Scholar]
- (12).Vanecko S, Laskowski M., Sr. J. Biol. Chem. 1961;236:3312. [PubMed] [Google Scholar]
- (13).(a) Lindahl T, Gally JA, Edelman GM. J. Biol. Chem. 1969;244:5014. [PubMed] [Google Scholar]; (b) Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. EMBO J. 1999;18:3868. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mazur DJ, Perrino FW. J. Biol. Chem. 1999;274:19655. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- (14).(a) Kavangh D, Spitzer D, Kothari PH, Shaikh A, Liszewski MK, Richards A, Atkinson JP. Cell Cycle. 2008;7:1718. doi: 10.4161/cc.7.12.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang C-J, Lam W, Bussom S, Chang H-M, Cheng Y-C. DNA Repair. 2009;8:1179. doi: 10.1016/j.dnarep.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).(a) Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Mol. Cell. Bio. 2004;15:6719. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Crow YJ, et al. Nat. Genet. 2006;38:917. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]; (c) Stetson DB, Ko JS, Heidmann T, Medzhitov R. Cell. 2008;134:587. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Richards A, et al. Nat. Genet. 2007;39:1068. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


