Abstract
Reductionist in vitro model systems which mimic specific extracellular matrix functions in a highly controlled manner, termed artificial extracellular matrices (aECM), have increasingly been used to elucidate the role of cell-ECM interactions in regulating cell fate. To better understand the interplay of biophysical and biochemical effectors in controlling three-dimensional cell migration, a poly(ethylene glycol)-based aECM platform was used in this study to explore the influence of matrix cross-linking density, represented here by stiffness, on cell migration in vitro and in vivo. In vitro, the migration behavior of single preosteoblastic cells within hydrogels of varying stiffness and susceptibilities to degradation by matrix metalloproteases was assessed by time-lapse microscopy. Migration behavior was seen to be strongly dependent on matrix stiffness, with two regimes identified: a nonproteolytic migration mode dominating at relatively low matrix stiffness and proteolytic migration at higher stiffness. Subsequent in vivo experiments revealed a similar stiffness dependence of matrix remodeling, albeit less sensitive to the matrix metalloprotease sensitivity. Therefore, our aECM model system is well suited to unveil the role of biophysical and biochemical determinants of physiologically relevant cell migration phenomena.
Introduction
Control of three-dimensional cell migration within and into biomaterial scaffolds plays a pivotal role in tissue engineering. Biomaterial implants, designed to attract endogenous stem/progenitor cells to the site of a tissue defect to induce regeneration, need to facilitate the repopulation and remodeling of an implant by host cells, a phenomenon that is controlled by large-scale cell migration (1). The lack of sufficient cell migration is arguably one of the most significant limitations in creating large tissue-engineered constructs; indeed impaired endothelial cell invasion can lead to lack of vascularization and, ultimately, to necrosis (2). Conversely, in biomaterials that are designed as carriers for cell delivery, encapsulated cells need to be able to leave the delivery system via extensive three-dimensional cell migration (3). As a result of these critical biological requirements, the engineering of biomaterials having optimized three-dimensional cell migration characteristics has gained increased attention in the biomaterials field.
The optimization of cell migration behavior within biomaterials necessitates a thorough understanding of three-dimensional cell migration mechanisms (4,5). Extensive research on cell migration through extracellular matrices, as it occurs in diverse tissues under physiological and pathological situations, suggests that cells can move in three dimensions using either proteolytic (mesenchymal) or nonproteolytic (amoeboid) strategies. In contrast to migration in two dimensions, cells in three dimensions must overcome the biophysical resistance of their surrounding milieu. In proteolytic migration, cells secrete active proteases which break down macromolecules of the extracellular matrix (ECM) and thus create macroscopic cavities which allow their movement. Importantly, matrix degradation is localized to the vicinity of the cell, which is possible because some proteases, such as membrane-type (MT) matrix metalloproteases (MMPs), are linked to the cell membrane.
Alternatively, a number of inflammatory cell types such as lymphocytes and dendritic cells, or tumor cells, are known to utilize strategies that allow them to overcome the biophysical matrix resistance by essentially squeezing through the ECM, or deforming it, independently of proteolysis (amoeboid migration). It has also been shown that the same cell type can take advantage of both mechanisms, depending on the specific ECM context (6).
Various approaches to engineering synthetic biomaterials which can support both of these types of cellular migration behaviors have been reported (1). The main focus has been on rendering materials conducive to migration through interconnected, preexisting pores (7). More recently, biomaterials have been developed as artificial extracellular matrices (aECM) that are sensitive to the action of cell-secreted proteases. For example, peptidic substrates for MMPs or plasmin placed in the backbone of cross-linked hydrophilic polymer chains have served as cleavage sites for the degradation of the resulting copolymer hydrogels. This simple design strategy, used in conjunction with the incorporation of integrin-binding peptide or protein components which enable cell adhesion, yields synthetic ECM analogs in which cells are able to migrate by proteolytic mechanisms.
Although aECMs are increasingly used in tissue engineering and three-dimensional cell culture to complement naturally derived matrices such as collagen or Matrigel (BD Biosciences, Franklin Lakes, NJ) (8,9), little is known about the interplay of the various biochemical and biophysical aECM characteristics in controlling three-dimensional cell migration behavior (4). For example, an open key question is whether certain aECM aspects favor proteolytically or nonproteolytically mediated cell migration modes. In this work, we have addressed this question using our previously developed aECM model system formed from poly(ethylene glycol)(PEG)-based macromers via the transglutaminase (TG) factor XIII (10,11).
In contrast to existing three-dimensional cell culture matrices fabricated from naturally derived ECM components, these molecularly engineered TG-PEG gels are devoid of a microstructure and are therefore essentially nonporous. Indeed, these synthetic gels are made of a molecular meshwork consisting of flexible cross-linked polymers with a mesh size of approximately tens of nanometers (10). Similar to existing aECM models (e.g., (12–19)), the biochemical and biophysical properties of our TG-PEG hydrogel networks can be extensively specified, for example by incorporating protease-sensitive peptide domains and cell-adhesive ligands, or by tuning matrix stiffness via precursor polymer architecture or concentration. By exploring the modularity of this system, we systematically probed how three-dimensional single cell migration depends on gel stiffness, adjusted independently from matrix degradability.
Surprisingly, we find that in matrices with low stiffness, single cells can overcome the resistance of the matrix by engaging in a degradation-independent three-dimensional migration mode, suggesting the use of preexisting or de novo-formed macroscopic gel defects. This finding may also highlight that apart from porosity or protease sensitivity, the engineering of gel defects in seemingly homogeneous polymer gels could be a powerful means to increase cell permissiveness in relatively dense gels.
Materials and Methods
Materials and reagents
PEG-peptide conjugates
Production and characterization of different eight-arm PEG precursors containing pending factor XIIIa substrate peptides having either a glutamine acceptor substrate (n-PEG-Gln) or a lysine donor substrate containing a MMP-sensitive linker (n-PEG-MMPsensitive-Lys) or a MMP-insensitive linker (n-PEG-MMPinsensitive-Lys) was performed as described elsewhere (10,11). More information can be found in the Supporting Material.
PEG hydrogel preparation
PEG hydrogels were formed as described elsewhere (10,11). More information can be found in the Supporting Material.
PEG hydrogel characterization
Rheometry on swollen gels
Storage and loss moduli (G′ and G″) of swollen gels (n = 4 per condition) were obtained by small strain oscillatory shear rheometry (20).
Equilibrium swelling measurements
Swollen hydrogels were weighted just before rheometry, and the swelling ratio Q determined as the swollen gel mass divided by the gel's dry mass (calculated from the reaction conditions).
Detection of sol fractions by HPLC
The potential presence of defects in the network architecture due to uncross-linked PEG precursors was investigated by reverse phase high-performance liquid chromatography (RP-HPLC). First, a dilution series of unreacted PEG precursors was run on an RP-HPLC instrument using a Waters (Milford, MA) C18 symmetry column to establish a standard curve of ultraviolet intensities (recorded at 220 nm) as a function of PEG concentration. Then, gels (50 μL volume formed at 1.5% w/v) were prepared (n = 4) and each was incubated in 200 μL water for 24 h. The supernatant was then collected, run on the HPLC instrument and ultraviolet intensities were compared to the standard curve to determine the corresponding amount of unreacted polymer remaining in solution.
Cell culture
Mouse preosteoblastic cells MC3T3-E1 were purchased from American Type Culture Collection (ATCC, Manassas, VA) and grown under MC3T3-E1 culture medium (α-minimal essential medium, with 10% fetal bovine serum, 100 U/mL penicillin G, and 100 mg/mL streptomycin (GIBCO BRL, Life Technologies, Grand Island, NY) under standard cell culture conditions (37°C in humidified atmosphere and 5% CO2).
Cell encapsulation
Cells suspended in cell culture medium were added right after the FXIIIa enzyme to yield single dispersed cells at a final seeding density of 6 × 104 cells/mL of hydrogel. Subsequently the forming matrices were slowly rotated (10 min at RT) until the onset of gelation to prevent sedimentation of cells, then incubated at 37°C and 5% CO2 for an additional 30 min and finally immersed in cell culture medium.
Cell migration assay
Cell migration experiments were conducted at 37°C and 5% CO2 and high relative humidity. Hydrogel disks containing dispersed cells were equilibrated for 2 h in cell culture medium and then glued to the bottom of 24-well cell culture dishes by applying 10 μL of 5% hydrogel to the edge of the disks. After gelation was allowed to take place for 30 min, the samples were equilibrated for 1 h in 1 mL of pure cell culture medium or medium that contained 50 μM of the broad-range MMP inhibitor GM6001 (Chemicon, Billerica, MA). Three random (x-y-z) positions, carefully selected to be completely inside the matrix, were selected using an inverse wide-field microscope (DM IRBE; Leica, Wetzlar, Germany) equipped with a motorized stage and focus, and a black and white camera (ORKA ER; Hamamatsu, Hamamatsu City, Japan). Cell spreading and migration was followed for up to 36 h by software-controlled image acquisition (using the software Openlab; http://www.open-lab.com/index_en.html) every 15 min.
Statistical analysis of migration parameters
Projections of real three-dimensional tracks were followed manually by using a “Manual Tracking” plugin in ImageJ software. The resulting x and y coordinates were used in a correlated random walk model, as described by Raeber et al. (17) and as detailed in the Supporting Material.
Staining and confocal microscopy
MC3T3-E1 cells were stained for f-actin and nuclei. Samples were fixed and permeabilized in 4% paraformaldehyde containing 0.2% Triton X-100 in phosphate-buffered saline (PBS) for 20 min at 4°C. Samples were incubated for 10 min in 0.1 M glycine followed by a wash step in PBS. For f-actin staining, the gels were incubated with 0.4 U/mL rhodamine-labeled phalloidin (R-415; Molecular Probes, Eugene, OR) in PBS with 1% bovine serum albumin for 1 h at 4°C. After washing the samples three times for 5 min in PBS, cell nuclei were costained with 1 ng/μL DAPI (4′,6-diamidino-2-phenylindole) (D-1306; Molecular Probes) in PBS for 10 min at 4°C. Z-series of ∼30 equidistant x-y scans at 0.272-μm intervals (63×) were acquired and processed in Imaris software (Bitplane, Zürich, Switzerland).
For time-lapse confocal imaging, cells were stained with PKH-26 (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. Fluorescently labeled cells were encapsulated in hydrogels containing covalently linked TG-Lys-FITC (21–24). The resulting hydrogel disks were equilibrated for 2 h in cell culture medium and then glued to the bottom of 24-well cell culture dishes by applying 10 μL of 5% hydrogel to the edge of the disks. Z-series of ∼100 equidistant x-y scans at 1.5-μm intervals (20×) were acquired for 8 h at 15-min intervals in resonant scanning mode with a Leica TCS SP5 confocal microscope and processed with Imaris software.
Animal experiments
Animal experiments were authorized by the Veterinary Authority of the Canton of Zurich (Zurich, Switzerland). Adult female Sprague-Dawley albino rats (300–350 g) were used for bone regeneration experiments as previously described (25). More information can be found in the Supporting Material.
Results
Characterization of aECM properties
Cell behavior in three-dimensional microenvironments is largely influenced by cross-link density, degradability, porosity/topology, and cell-ECM adhesion, parameters which can, at least to some extent, be fine-tuned in synthetic matrices. We produced TG-PEG hydrogel matrices (10) where swelling and viscoelastic properties were modulated by different precursor concentrations (1.5, 2, and 2.5% v/w), and where sensitivity to degradation by MMPs was modulated by the linker peptide (sensitive: Ac-FKGG↓GPQG IWGQ-ERCG-NH2, ↓ indicates the cleavage site, or insensitive: Ac-FKGG-GPQGIAGF-ERCG-NH2) (Fig. 1). The viscoelastic properties of swollen gels were determined as a function of frequency by small strain oscillatory shear rheometry (20) (Fig. S1 in the Supporting Material).
Figure 1.
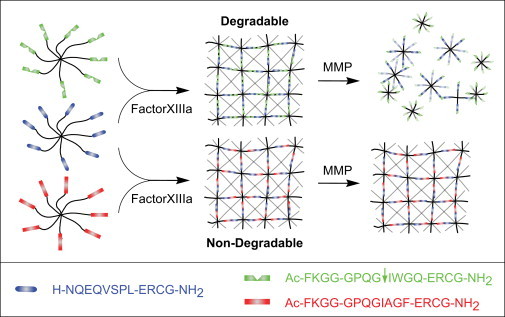
Scheme of the modular design of PEG-based aECMs. Stoichiometrically balanced ([Lys]/[Gln] = 1) 8-arm PEG macromers in a buffer solution are enzymatically cross-linked via their pending glutamine acceptor [Gln] and lysine-donor [Lys] FXIIIa substrate sequences to form a hydrogel. By variation of the linker sequence and the initial precursor concentration, aECMs with different stiffness and MMP sensitivities in presence of constant RGD concentrations can be generated.
The storage moduli (G′) at a frequency of 1 Hz ranged from 94 ± 25 Pa at 1.5% to 482 ± 77 Pa at 2.5% for MMP-sensitive gels, and from 62 ± 32 Pa (1.5%) to 347 ± 19 Pa (2.5%) for MMP-insensitive gels (for simplicity we refer to the 1.5% gels as soft, the 2.5% as stiff, and those formed at 2% as intermediate) (Fig. S2 A). TG-PEG hydrogels with a dry mass content during gel formation of 1.5–2.5% thus exhibited relatively similar elastic properties as biologically derived hydrogel networks such as collagen or fibrin at concentrations of 2 mg/mL (17). The phase angle δ (corresponding to the ratio of storage and loss modulus) of 12 ± 7° of soft gels is in the range of collagen matrices, suggesting the presence of network defects in the form of noncovalently cross-linked and therefore elastically inactive network components.
To further characterize gel properties, we determined the swelling ratio Q. As expected, Q decreased with increasing precursor concentration (Fig. S2 B). Matrices containing MMP-sensitive peptide building blocks showed a lower swelling ratio than those with MMP-insensitive peptides. Employing the Flory-Rehner model (26), Q values allowed an estimation of the mesh size of both gel types of ∼40 nm, which is orders-of-magnitude below cellular features (not shown). It should be noted, however, that the mesh size is a crude, averaged approximation of the gel network microstructure, which does not provide any information on the defect size distribution. We would expect that three-dimensional cell migration would instead depend on the maximal defect size. Nevertheless, we hypothesized that three-dimensional cell migration would only be possible by local proteolytic matrix degradation, as reported for a similar chemically cross-linked aECM system (17,27).
Finally, to approximate the degree of defects in the gels, we evaluated the sol fraction of the networks by RP-HPLC (Fig. S3). The unreacted control PEG precursor was seen to have a retention time of ∼11.7 min, and the smallest detectable amount using our method was established to be 7.5 μg per 100 μL injection. Based on our detection limit of 7.5 μg, we could evaluate whether up to 2.5% of PEG remained unreacted in solution. Inasmuch as we could not detect any signal from our supernatant sample injections, we conclude that no soluble PEG precursors (above our detection limit of 2.5%) were present after gelation, which is not surprising because the probability that none of the octafunctional macromer had reacted is very low.
Increasing cross-linking density impedes cell spreading
To test the influence of matrix cross-link density on three-dimensional cell spreading, we entrapped single murine MC3T3-E1 preosteoblastic cells in degradable gels with variable stiffness. Representative confocal images of cells that were cultured for 24 h revealed different morphologies in response to variable network stiffness (Fig. S4). Cells in soft gels quickly adopted a spindle-shaped morphology. With increasing stiffness the morphology became less elongated and reticulate filopodia were formed. In the stiff gels, the cells generally remained round with frayed filopodia.
Increasing cross-linking density impedes cell migration
Time-lapse imaging revealed that matrix stiffness not only influenced initial cell spreading but also three-dimensional cell migration (Fig. 2). The onset of migration was observed as early as 4 h after encapsulation in soft gels, whereas at higher stiffness the cells started to migrate after ∼6 h. Notably, the overall mobility of cells entrapped in the stiffest gels was dramatically reduced compared to the intermediate and soft gels (Fig. 2 A and Movies S1, S3, and S5). Based on time-lapse imaging, we analyzed cell displacement data and created polar plots to confirm the directional independence of cell movement (Fig. 2 B) (17).
Figure 2.
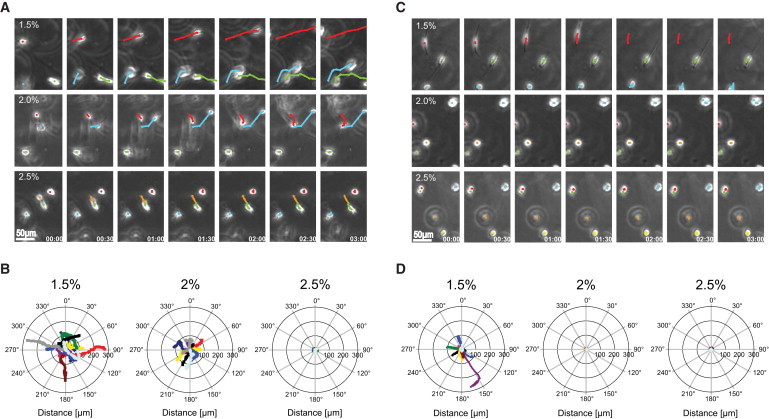
Time-lapse images and representative track plots of MC3T3-E1 cells in three-dimensional culture. (A and C Typical time-lapse images of cells in three-dimensional culture (time interval t = 30 min). The track of three individual cells is indicated by the sequentially determined center of the cell. (A) Efficient migration in soft MMP-sensitive hydrogels was observed that is reduced with increasing stiffness. (C) In soft MMP-insensitive hydrogels, cells also migrated, whereas in intermediate or stiff gels, cells remained round. (B and D) The projection of 10 migrating cells was acquired and overlaid with a common starting point. The tracks representing a 14.5 h period (58 × 15 min) do not show a preferred orientation.
MMP-dependent cell migration behavior
To verify that three-dimensional cell migration in TG-PEG hydrogels is not only dependent on matrix cross-link density but also on susceptibility to proteolytic degradation, we entrapped single cells in gels containing the peptide Ac-FKGG-GPQGIAGF-ERCG-NH2 (28) which is not an MMP-cleavable substrate. As expected, over the course of the first 24 h in culture, cells remained completely trapped within stiff and intermediate MMP-insensitive gels (Fig. 2, C and D and Movies S2, S4, and S6). Interestingly, cellular activity in nondegradable gels remained intact, as was evident from protruding filopodia and pseudopodia and noticeable oscillations of cell bodies, presumably due to a mechanical deformation of the matrix.
To our surprise, cells embedded in the soft MMP-insensitive gels migrated to a similar degree as cells in the soft degradable gels (Fig. 2, A and B). These data thus indicate that three-dimensional cell migration behavior is dependent on both biochemical and biophysical matrix properties: stiffer matrices facilitated migration by proteolytic remodeling, whereas soft matrices appeared to support an MMP-insensitive migration mode.
MMP-independent cell migration behavior
To exclude that the differences in mechanical properties of MMP-sensitive and -insensitive gels (Fig. S2 A) were responsible for the MMP-independent migration in nondegradable gels, we performed migration experiments in the presence of the broad-range MMP-inhibitor GM 6001 (Fig. S5 and Fig. S6). Consistent with the previous data, we observed complete inhibition of three-dimensional migration in intermediate and stiff matrices but almost no change in migration within soft gels. These results indicate that the migratory machinery is not impaired by a synthetic MMP inhibitor, but, instead, that cells are not able to migrate above a certain matrix stiffness. This provides further confirmation that the inability to block migration in the MMP-insensitive hydrogel is due to the low stiffness of the hydrogel rather than the proteolytic degradation of negative control peptide.
Single cell migration parameters depend on biochemical and biophysical matrix properties
To substantiate the above observations, we quantified the three-dimensional migration rate and persistence time based on time-lapse imaging series of at least 24 consecutive time-frames by applying an unbiased random walk model (17). The percentage of migrating cells was based on the persistence length criterion of Lcrit = 1.2 μm. The histograms of the speed of migrating cells showed a rather broad and positively skewed distribution, except for soft gels (Fig. S7). The average speed in MMP-sensitive hydrogels decreased significantly with stiffness, from 1 μm/min (soft) to 0.5 μm/min (intermediate) and 0.2 μm/min (stiff) gels, respectively (Fig. 3 A and Fig. S7 A). The average migration speed within MMP-insensitive matrices remained as high as 0.5 μm/min in the soft gels and only decreased to 0.2 μm/min for the stiffer gels. These values may be due to the inaccuracy of the manual evaluation procedure as well as to the elasticity of the substrate, rather than from an effective movement of the cell body.
Figure 3.
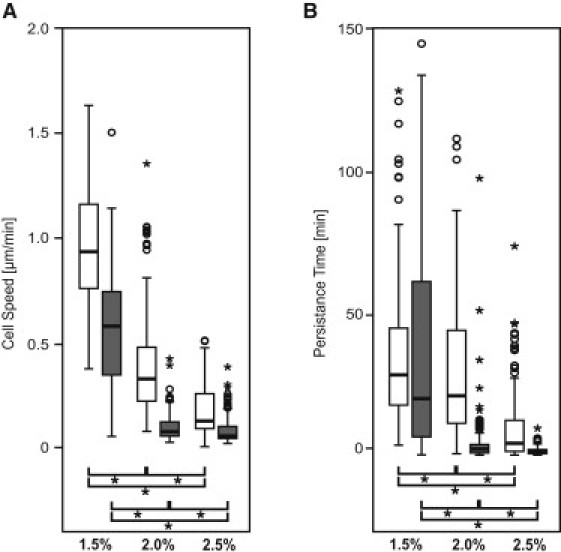
Three-dimensional single cell migration as a function of gel stiffness and MMP-sensitivity (degradable gels depicted in white, nondegradable gels in gray). (A) Mean cell speed and (B) persistence time of all cells of a group were analyzed from time-lapse images of at least 24 consecutive time points. Values are displayed as box plots ranging from 25th to 75th percentile including the whiskers from the 10th to the 90th percentile. (A) The average cell speed and (B) the average persistence time ((∗) p < 0.05) decreased significantly with increasing stiffness of the matrix ((∗) p < 0.05).
Persistence time showed a similar trend as cell speed (Fig. 3 B and Fig. S7 B). However, the distribution for all conditions was positively skewed. The highest persistence times were observed in the soft MMP-sensitive hydrogels, with average values of 30 min. This value is significantly larger than within intermediate (20 min) or stiff gels (5 min). The persistence time of cells in the soft MMP-sensitive and MMP-insensitive hydrogels are not significantly different, indicating that the predominant migration mode could be independent of proteolytic matrix remodeling.
These data show that three-dimensional cell migration can depend on biochemical and biophysical matrix properties (Fig. 4 and Fig. S8). Hydrogels which are loosely cross-linked, resulting in relatively high swelling ratios Q (Fig. 4 A) and low stiffness G′ (Fig. 4 B), might permit three-dimensional cell migration independent of the matrix's proteolytic sensitivity, for example via existing or newly formed macroscopic gel defects, whereas migration in relatively densely cross-linked gels strictly relies on proteolytic degradation of the elastically active macromolecules in close vicinity of the cell.
Figure 4.
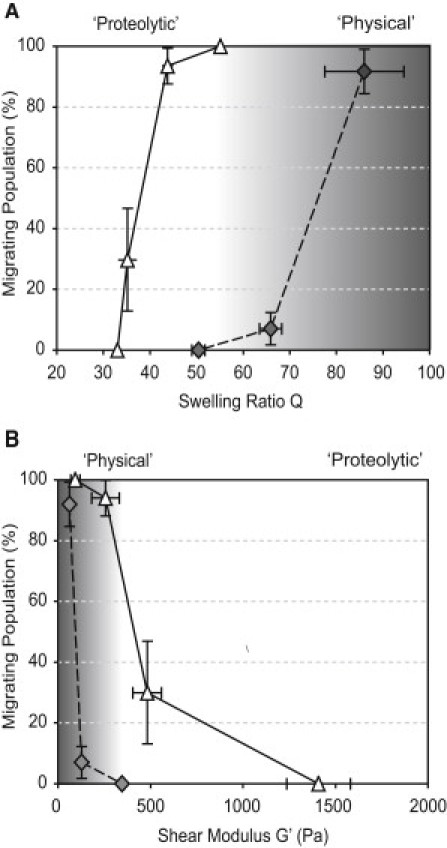
(A) The high swelling ratio Q results in a large population of migrating cells which, in MMP-sensitive gels (white triangles), is due to mostly proteolytic matrix remodeling and in MMP-insensitive gels (gray diamonds) due to physically controlled cell migration. With decreasing swelling ratios, the migrating population decreases. (B) In soft matrices, almost all the cells migrate in MMP-sensitive and MMP-insensitive gels. By increasing the stiffness, migration ceases and only cells in MMP-sensitive substrates can migrate.
Cell-mediated proteolytic remodeling leaves behind macroscopic tunnels of digested matrix
We next tried to assess, by live cell confocal microscopy, whether migration in PEG hydrogels is made possible by the formation of macroscopic channels, either generated by proteolytic digestion or by a physical mechanism (Fig. 5). For this purpose, single cells were fluorescently labeled with the membrane dye PKH26 and then encapsulated in FITC-labeled MMP-sensitive matrices formed at 1.5% precursor content, either in the absence (Fig. 5 A and Movie S7) or presence (Fig. 5 B and Movie S8) of the MMP inhibitor GM6001 to completely block proteolytic migration. Z-stacks of migrating cells were collected for a period of 8 h with a time-interval of 15 min. Three-dimensional-reconstruction of images was performed using either a maximum intensity projection method to highlight localized matrix compression around cells (Fig. 5, A and B, upper panels), or a minimum intensity projection method to highlight macroscopic matrix defects, i.e., putative cell tracks (Fig. 5, A and B, lower panels).
Figure 5.
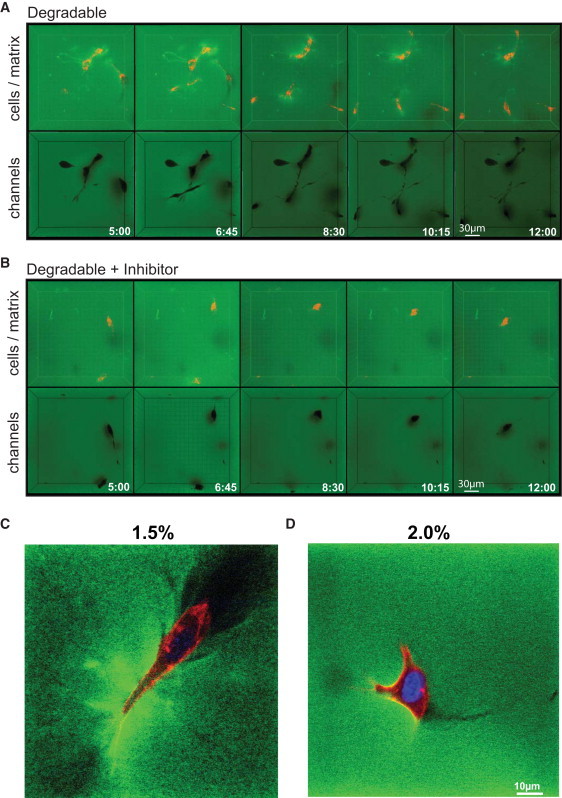
Visualization of cell-induced matrix deformation and macroscopic cavities. (A and B) Migrating PKH26-labeled cells were encapsulated in FITC-conjugated, soft (1.5%) matrices in the absence (A) or presence (B) of GM6001 and followed by four-dimensional time-lapse confocal microscopy for 8 h. In both cases, three-dimensional reconstruction of z stacks over time in maximum intensity projection mode (cells in red, matrix in green) revealed an increase in matrix intensity (green), indicating localized matrix deformation around migrating cells. Three-dimensional reconstruction of stack from the same time points performed in minimum intensity projection mode (matrix in green) revealed the presence of a network of interconnected macroscopic cavities and small cracks indicated the cell migrating paths. (C and D) High-resolution z section of encapsulated cells within soft and intermediate degradable gels after one day in culture (fixed samples).
As expected, zones of bright green color in close vicinity of the cells showing regions of higher FITC-concentration were seen, suggesting that migrating cells are exerting forces to locally deform their surrounding matrix. In contrast, in MMP-sensitive matrices in the absence of GM6001, a minimum intensity projection analysis of image stacks revealed a clearly visible network of interconnected macroscopic cavities, possibly harboring the encapsulated single cells initially, as well as micrometer-sized tracks generated by migrating cells (Fig. 5 A). These cavities and tracks appear to be permanent and were also seen in fixed samples at higher resolution (Fig. 5, C and D). The smaller cell tracks are presumably caused by MMP-mediated digestion of the hydrogel network, as is evident from an analysis of cells migrating in the presence of the MMP inhibitor (Fig. 5 B).
Although extensive migration under these conditions was observed and the larger cavities were still visible, smaller tracks caused by migrating cells had mostly disappeared. We assume that in the latter case cells migrate through de novo-created matrix defects most likely generated by a physical (e.g., rupture of the matrix) rather than a biochemical mechanism. Because the matrix is highly swellable and no material would be removed in that case, such defects could not be visualized by our method, because tracks would disappear after cell displacement.
Taken together, although we cannot fully exclude the existence of preexisting macroscopic defects (or pores) below the resolution by this microscopic evaluation, we believe that these defects are not preexisting but rather formed by cells that squeeze through the hydrogel network and induce the propagation of defects to generate cracks sufficient for cell migration.
Formation of three-dimensional cellular networks in vitro is dependent on matrix composition
Long-term cultures of single cells entrapped in matrices with different stiffness and MMP-sensitivity showed that matrix properties could influence the formation of three-dimensional cellular morphogenetic structures in vitro (Fig. 6). In line with the above migration results, highly interconnected cellular networks that permeate all gel areas were formed after three weeks in culture in soft MMP-degradable hydrogels.
Figure 6.
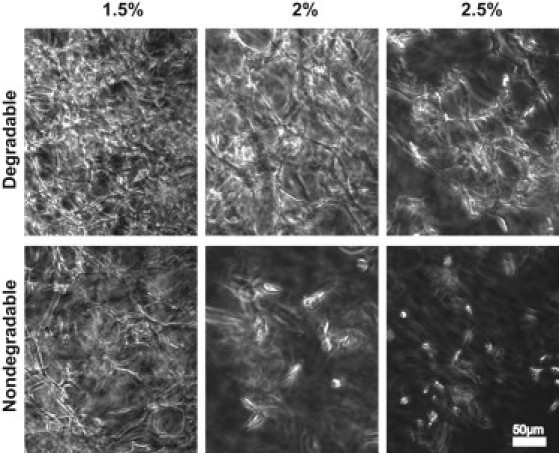
Matrix stiffness and MMP-sensitivity influence the formation of cellular networks. Starting from single cells, dense and evenly distributed cellular networks are formed in soft MMP-sensitive gels within three weeks. With increasing stiffness the cell distribution becomes less homogenous and less dense. The formation of cellular networks in MMP-insensitive soft hydrogels is largely reduced, and in intermediate or stiff hydrogels is almost absent.
With increasing stiffness, the density of these cellular networks decreased, as cells were increasingly hindered from proliferating and penetrating the matrix. Remarkably, cells in soft, nondegradable gels and cells in intermediate, degradable gels formed similar morphogenetic structures, indicating that although cells efficiently penetrate the gel while migrating, this might be restricted to preferentially used gel areas. The network formation in nondegradable gels of higher cross-link density, or in gels incubated with the MMP inhibitor GM6001 (Fig. S9), was almost absent, in line with the above three-dimensional migration data.
In vivo remodeling and tissue regeneration is dependent on matrix composition
We next tested how the reported in vitro behaviors would translate to a more complex in vivo cell migration context. We chose a previously established rat bone regeneration model (25). 8-mm calvarial defects were allowed to heal in the presence of MMP-sensitive or -insensitive gel implants with different stiffness which also contained 1 μg of BMP-2. These conditions had previously been shown to induce bone healing within ∼5 weeks (25). We reasoned that this experimental model could be explored to assess in vivo invasion of endogenous cells at early time-points (i.e., 1–3 weeks) preceding de novo bone formation.
Gels were explanted after only two weeks and assessed by histological analysis and μCT. Similarly to the previously characterized in vitro behavior, the in vivo recruitment of a cellular invasion front was largely dependent on the hydrogel properties (Fig. 7 A). Matrix stiffness appeared to be an important determinant of cell invasion in vivo. Soft gels were completely penetrated by endogenous cells and only minor quantities of detectable gel remained. With increasing stiffness, the distribution of cells within the matrices became less regular and preferential paths of multicellular assemblies separated by intact gel mass were often seen. At the highest stiffness, cells did not invade the hydrogel and the formation of new tissue was entirely restricted to the cell-matrix interface.
Figure 7.
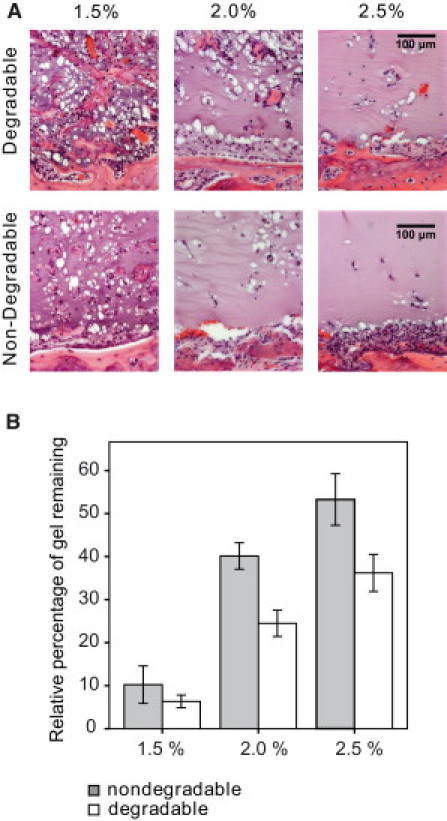
In vivo matrix remodeling is controlled by gel properties. (A) Critical-sized calvarial defects in rats were treated with soft, intermediate, and stiff MMP-sensitive and -insensitive hydrogels containing 1 μg BMP-2. Within two weeks, endogenous cells invade, degrade, and remodel provisional MMP-sensitive matrices in a stiffness-dependent manner. Cell invasion is homogenous in soft gels and becomes less regular in more dense gels. MMP-insensitive gels are less efficiently invaded by cells (n = 5). (B) The stiffness and MMP-dependent degradation rate of the aECM was confirmed by histomorphometrical quantification of the gel leftovers (mean ± SD, n = 5).
The overall remodeling kinetics of the implants was assessed by histomorphometric determination of remaining gel mass (Fig. 7 B). The slow remodeling of the MMP-insensitive compared to the MMP-sensitive hydrogels indicated that MMPs significantly contribute to the remodeling process. However, the degradation by protease-independent mechanisms and/or by proteases other than MMPs cannot be excluded.
Discussion
The understanding and optimization of three-dimensional cell migration within and into biomaterials scaffolds is critically important for the field of tissue engineering. Here we have utilized TG-PEG gels as a model system to shed light on the role of matrix stiffness and proteolytic remodeling on three-dimensional cell migration in vitro and in vivo.
Structural components of the native ECM such as fibrillar collagen provide an interconnected microporous network intercalated with other matrix components such as glycosaminoglycans. Cell migration within this complex milieu is the net result of proteolysis-dependent and -independent mechanisms (5), a distinction that often cannot be made easily. Indeed, interpretations of cell migration data from in vivo ECMs, or in vitro models that are derived from natural ECM components, can be confounded by the difficulty to engineer ECM characteristics and/or to parse the various cell-ECM interactions that influence three-dimensional cell migration in these systems. For example, Sabeh et al. (29) showed that cancer cells migrate within noncovalently cross-linked collagen gels independently of proteases, while in covalently cross-linked gels, MT1-MMP was demonstrated to be indispensable.
In contrast, engineering approaches now allow us to build aECMs with very good control over the materials microstructure, in some cases to form scaffolds that are essentially amorphous and pore-free. Indeed, by using synthetic and natural protease inhibitors, some of us had previously shown that three-dimensional migration in dense chemically crosslinked PEG hydrogels is strictly dependent on proteolysis (17,27). Here, using enzymatically cross-linked PEG-based matrices, we expected a similar protease-dependency, across the entire stiffness range tested. We calculated molecular mesh sizes from swelling ratios of all gel conditions to be approximately tens of nanometers, and thus far below the threshold that a cell could breach to penetrate the surrounding physical matrix barrier.
Indeed, cells encapsulated within relatively stiff gels, formed at 2 or 2.5% solid content, were unable to migrate in the absence of proteolytic activity, as was shown by blocking proteolysis or assessing cell migration in stable gels. However, just a slight decrease in cross-linking density, induced by changing the solid content to 1.5%, resulted in matrices through which cells were able to efficiently migrate. Although the activity of proteases other than MMPs cannot be fully excluded, our data strongly suggest protease-independent migration mechanisms under these conditions.
In addition to the restriction of migration in the absence of a microscopic porosity, the viscoelastic matrix properties might also directly impact the migratory behavior of cells in three dimensions (4,30–32). Studies have demonstrated that certain cell types can adopt an amoeboid mode of migration that is independent of matrix remodeling and is instead characterized by localized ECM deformation and cell shape changes (33). Such a mechanism might be strongly dependent on the biophysical properties of the microenvironment and could possibly explain our data. However, we think that this is rather unlikely.
First, fibroblastlike preosteoblastic cells would be expected to migrate by a mesenchymal-type mechanism (34).
Second, ameboid migration has thus far only been observed in physically cross-linked matrices that allow material displacement by cellular forces, while the PEG-based gels used here are cross-linked by much stronger covalent bonds.
Alternatively, one could think of a mechanism in which cells exploit existing, macroscopic gel defects or even exert sufficiently large forces to produce local propagating cracks within the hydrogels; unfortunately this is a hypothesis that is not easily testable. Interestingly, it is well documented that single-component hydrogels, and in particular those formed from synthetic polymers such as PEG, are mechanically very fragile—a problem that can be overcome by choosing more sophisticated multicomponent design strategies (35). Such cracks might be accessible for migrating cells and could result in a migration speed that exceeds that of cells that rely exclusively on proteolytic migration in higher density hydrogels.
The hydrogel matrices used in vitro under well-defined culture conditions were subjected to a considerably more complex in vivo situation to assess the migratory events during early bone regeneration. Similarly to the in vitro situation, more densely cross-linked matrices led to marginal migration to the inside of the gel and, consequently, bone formation limited on the outside. Although differences were significant, the sensitivity of the implanted materials to MMPs seemed to play a less important role during in vivo remodeling compared to the in vitro migration model.
Under in vivo conditions, many more factors influencing the gel performance come into play. For example, the amount and range of proteases could be considerably different between cell culture conditions and in a healing environment due to the interaction of the implant with a higher number of cells specialized in rapid remodeling in the in vivo condition (e.g., inflammatory cell types). Nevertheless, the good correlation between in vivo and in vitro behaviors suggests that our in vitro migration model can be successfully applied to predict the response of modulated material characteristics in specific tissue regeneration applications.
Indeed, a good control over in vivo cell fates via engineered ECMs would be an extremely useful capability (36). Our results suggest that through variation of network stiffness or proteolytic susceptibility and specificity, recruited cells could be restricted to the tissue-material interface or directed to migrate into the material. In comparison to healing that relies on surface erosion, the generation of new tissue from the inside out of an implant might be considerably faster. In agreement with this notion, we generally found that the most efficient bone formation in long-term experiments occurs with gel compositions that allowed efficient cell migration. It can also be noted that soft gels which led to very fast gel degradation failed to support complete healing, presumably because the implant stability did not support bone tissue formation (unpublished data).
Acknowledgments
We thank Flora Nicholls for help with animal trials and Esther Kleiner for help with histology.
This work has been supported by grants from the Swiss National Science Foundation (Nos. CR3213-125426/1 and 310000-116240), the European Community's Angioscaff Program contract No. 214402, and a European Young Investigators award to M.P.L.
Contributor Information
M. Ehrbar, Email: martin.ehrbar@usz.ch.
M.P. Lutolf, Email: matthias.lutolf@epfl.ch.
Supporting Material
References
- 1.Lutolf M.P., Hubbell J.A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Phelps E.A., García A.J. Engineering more than a cell: vascularization strategies in tissue engineering. Curr. Opin. Biotechnol. 2010;21:704–709. doi: 10.1016/j.copbio.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mooney D.J., Vandenburgh H. Cell delivery mechanisms for tissue repair. Cell Stem Cell. 2008;2:205–213. doi: 10.1016/j.stem.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Zaman M.H., Matsudaira P., Lauffenburger D.A. Understanding effects of matrix protease and matrix organization on directional persistence and translational speed in three-dimensional cell migration. Ann. Biomed. Eng. 2007;35:91–100. doi: 10.1007/s10439-006-9205-6. [DOI] [PubMed] [Google Scholar]
- 5.Ilina O., Friedl P. Mechanisms of collective cell migration at a glance. J. Cell Sci. 2009;122:3203–3208. doi: 10.1242/jcs.036525. [DOI] [PubMed] [Google Scholar]
- 6.Friedl P., Wolf K. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moroni L., de Wijn J.R., van Blitterswijk C.A. Integrating novel technologies to fabricate smart scaffolds. J. Biomater. Sci. Polym. Ed. 2008;19:543–572. doi: 10.1163/156856208784089571. [DOI] [PubMed] [Google Scholar]
- 8.Lutolf M.P. Integration column: artificial ECM: expanding the cell biology toolbox in 3D. Integr. Biol. (Camb) 2009;1:235–241. doi: 10.1039/b902243k. [DOI] [PubMed] [Google Scholar]
- 9.Tibbitt M.W., Anseth K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrbar M., Rizzi S.C., Lutolf M.P. Enzymatic formation of modular cell-instructive fibrin analogs for tissue engineering. Biomaterials. 2007;28:3856–3866. doi: 10.1016/j.biomaterials.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 11.Ehrbar M., Rizzi S.C., Lutolf M.P. Biomolecular hydrogels formed and degraded via site-specific enzymatic reactions. Biomacromolecules. 2007;8:3000–3007. doi: 10.1021/bm070228f. [DOI] [PubMed] [Google Scholar]
- 12.Gobin A.S., West J.L. Cell migration through defined, synthetic ECM analogs. FASEB J. 2002;16:751–753. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- 13.Halstenberg S., Panitch A., Hubbell J.A. Biologically engineered protein-graft-poly(ethylene glycol) hydrogels: a cell adhesive and plasmin-degradable biosynthetic material for tissue repair. Biomacromolecules. 2002;3:710–723. doi: 10.1021/bm015629o. [DOI] [PubMed] [Google Scholar]
- 14.Bryant S.J., Anseth K.S. Hydrogel properties influence ECM production by chondrocytes photoencapsulated in poly(ethylene glycol) hydrogels. J. Biomed. Mater. Res. 2002;59:63–72. doi: 10.1002/jbm.1217. [DOI] [PubMed] [Google Scholar]
- 15.Shu X.Z., Liu Y., Prestwich G.D. Disulfide-crosslinked hyaluronan-gelatin hydrogel films: a covalent mimic of the extracellular matrix for in vitro cell growth. Biomaterials. 2003;24:3825–3834. doi: 10.1016/s0142-9612(03)00267-9. [DOI] [PubMed] [Google Scholar]
- 16.Kim S., Chung E.H., Healy K.E. Synthetic MMP-13 degradable ECMs based on poly(n-isopropylacrylamide-co-acrylic acid) semi-interpenetrating polymer networks. I. Degradation and cell migration. J. Biomed. Mater. Res. A. 2005;75:73–88. doi: 10.1002/jbm.a.30375. [DOI] [PubMed] [Google Scholar]
- 17.Raeber G.P., Lutolf M.P., Hubbell J.A. Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration. Biophys. J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almany L., Seliktar D. Biosynthetic hydrogel scaffolds made from fibrinogen and polyethylene glycol for 3D cell cultures. Biomaterials. 2005;26:2467–2477. doi: 10.1016/j.biomaterials.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 19.Peyton S.R., Raub C.B., Putnam A.J. The use of poly(ethylene glycol) hydrogels to investigate the impact of ECM chemistry and mechanics on smooth muscle cells. Biomaterials. 2006;27:4881–4893. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Lutolf M.P., Hubbell J.A. Synthesis and physicochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromolecules. 2003;4:713–722. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 21.Schense J.C., Hubbell J.A. Cross-linking exogenous bifunctional peptides into fibrin gels with factor XIIIa. Bioconjug. Chem. 1999;10:75–81. doi: 10.1021/bc9800769. [DOI] [PubMed] [Google Scholar]
- 22.Hu B.H., Messersmith P.B. Rational design of transglutaminase substrate peptides for rapid enzymatic formation of hydrogels. J. Am. Chem. Soc. 2003;125:14298–14299. doi: 10.1021/ja038593b. [DOI] [PubMed] [Google Scholar]
- 23.Lutolf M.P., Tirelli N., Hubbell J.A. Systematic modulation of Michael-type reactivity of thiols through the use of charged amino acids. Bioconjug. Chem. 2001;12:1051–1056. doi: 10.1021/bc015519e. [DOI] [PubMed] [Google Scholar]
- 24.Sala A., Ehrbar M., Weber F.E. Enzyme mediated site-specific surface modification. Langmuir. 2010;26:11127–11134. doi: 10.1021/la1008895. [DOI] [PubMed] [Google Scholar]
- 25.Lutolf M.P., Weber F.E., Hubbell J.A. Repair of bone defects using synthetic mimetics of collagenous extracellular matrices. Nat. Biotechnol. 2003;21:513–518. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- 26.Flory P.J. University Press; Ithaca, NY: 1953. Principles of Polymer Chemistry. [Google Scholar]
- 27.Raeber G.P., Lutolf M.P., Hubbell J.A. Mechanisms of 3-D migration and matrix remodeling of fibroblasts within artificial ECMs. Acta Biomater. 2007;3:615–629. doi: 10.1016/j.actbio.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Lutolf M.P., Lauer-Fields J.L., Hubbell J.A. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc. Natl. Acad. Sci. USA. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabeh F., Shimizu-Hirota R., Weiss S.J. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen J.A., Swartz M.A. Mechanobiology in the third dimension. Ann. Biomed. Eng. 2005;33:1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- 31.Zaman M.H., Trapani L.M., Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dikovsky D., Bianco-Peled H., Seliktar D. Defining the role of matrix compliance and proteolysis in three-dimensional cell spreading and remodeling. Biophys. J. 2008;94:2914–2925. doi: 10.1529/biophysj.107.105841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf K., Mazo I., Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedl P., Bröcker E.B. The biology of cell locomotion within three-dimensional extracellular matrix. Cell. Mol. Life Sci. 2000;57:41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong J.P., Katsuyama Y., Osada Y. Double-network hydrogels with extremely high mechanical strength. Adv. Mater. 2003;15:1155–1158. [Google Scholar]
- 36.Chan G., Mooney D.J. New materials for tissue engineering: towards greater control over the biological response. Trends Biotechnol. 2008;26:382–392. doi: 10.1016/j.tibtech.2008.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


