Abstract
Focal adhesions are critical to a number of cellular processes that involve mechanotransduction and mechanical interaction with the cellular environment. The growth and strengthening of these focal adhesions is dependent on the interaction between talin and vinculin. This study investigates said interaction and how vinculin activation influences it. Using molecular dynamics, the interaction between talin's vinculin binding site (VBS) and vinculin's domain 1 (D1) is simulated both before and after vinculin activation. The simulations of VBS binding to vinculin before activation suggest the proximity of the vinculin tail to D1 prevents helical movement in D1 and thus prevents binding of VBS. In contrast, interaction of VBS with activated vinculin shows the possibility of complete VBS insertion into D1. In the simulations of both activated and autoinhibited vinculin where VBS fails to fully bind, VBS demonstrates significant hydrophobic interaction with surface residues in D1. These interactions link VBS to D1 even without its proper insertion into the hydrophobic core. Together these simulations suggest VBS binds to vinculin with the following mechanism: 1), VBS links to D1 via surface hydrophobic interactions; 2), vinculin undergoes activation and D1 is moved away from the vinculin tail; 3), helices in D1 undergo conformational change to allow VBS binding; and 4), VBS inserts itself into the hydrophobic core of D1.
Introduction
Cellular survival, differentiation, migration, and other cellular processes are dependent upon the mechanical coupling of cells to their surroundings (1–9). The mechanical interaction of cells with the extracellular matrix (ECM) is typically mediated by membrane-bound integrin molecules and the focal adhesion complexes that form at sites of mechanical linkage (9–15). Focal adhesion complexes can best be described as a molecular-glue comprising an array of molecules binding to each other, to the ECM-bound integrins, and to the actin cytoskeleton. A number of studies have demonstrated that the formation of focal adhesions can be induced via an externally applied force (1,7,10,16–18). Furthermore, recruitment of several molecules, such as talin and vinculin, to focal adhesions is directly correlated with mechanical stimulus applied at the site of the focal adhesion formation (1,9,11,19–24). Taken together, these studies suggest mechanical sensation by talin and vinculin and motivate the study of the recruitment and activation of these mechanisms.
Talin binds integrin directly and is recruited at the formation of nascent adhesions that later mature into focal adhesions (21). Talin has a head domain that binds integrin, and a larger tail domain that can bind at least one actin filament directly and up to 11 additional actin filaments indirectly via binding to vinculin (21,23,25–27). Of the 11 vinculin binding sites (VBS), a number of them are known to be cryptic and require activation to bind vinculin (11,21,29,30). A number of both computational and experimental studies have demonstrated the force-induced activation of these cryptic VBSs (11,31–33).
Once activated, VBS will interact with vinculin. Computational simulations have suggested that VBS and vinculin interact via hydrophobic residues (34). Vinculin itself has five helical domains named: domain 1 (D1), domain 2 (D2), domain 3 (D3), domain 4 (D4), and the vinculin tail domain (Vt) (35) (Fig. 1 A). Talin's VBS binds to vinculin's D1 whereas actin filaments bind vinculin's Vt (27,35). Studies have also suggested a number of other binding partners for vinculin, including α-actinin, PIP2, and paxillin (1,35,36).
Figure 1.
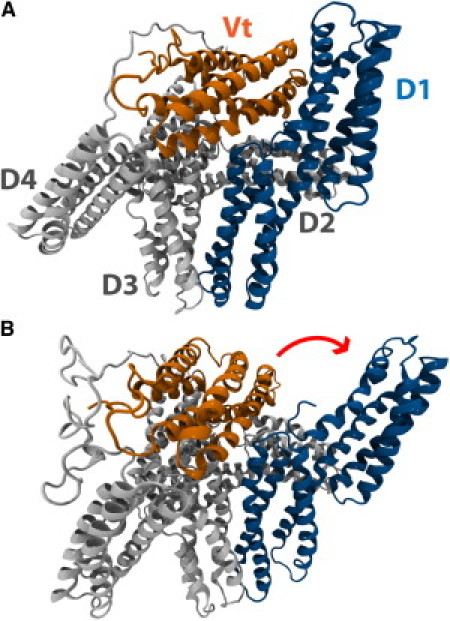
Vinculin can adopt an autoinhibited conformation or an activated conformation. (A) In its native state, vinculin is in an autoinhibited conformation. Vinculin has five helical domains: D1, D2, D3, D4, and Vt. Vt contains binding sites for F-actin whereas D1 contains binding sites for VBS. In its native conformation, the proximity of D1 to Vt prevents the binding of Vt to F-actin. The proximity of Vt to D1 could also impact the binding of D1 to VBS. (B) A suggested conformation of activated vinculin. Investigation of vinculin activation by a stretching force (46) using molecular dynamics has suggested that, during activation, D1 of vinculin undergoes a conformational change and rotates away from Vt. In this conformation, the proximity of Vt and D1 is reduced, potentially allowing for interaction of those domains with binding partners.
The binding of vinculin to talin and actin can function to multiply the number of actin filaments linked to focal adhesions. Similarly, the binding of vinculin to any of its numerous suggested binding partners can serve to multiply the number of focal adhesion forming molecules that have been recruited. However, vinculin's role as an agent to multiply and strengthen focal adhesions is critically dependent upon vinculin activation; in its native cytoplasmic state, vinculin adopts an autoinhibited conformation (37) in which the proximity of Vt to D1 can prevent binding to vinculin's binding partners (38,39) (Fig. 1 A). The actin binding site is at Vt and in its closed conformation, vinculin is unable to bind actin.
A number of studies have attempted to address vinculin activation and reveal how it is that vinculin transitions from an autoinhibited to an activated conformation (1,35,37,40–45). One study by Bois et al. (43) suggests that talin and α-actinin can bind D1 of autoinhibited vinculin and this binding can then lead directly to vinculin activation. In contrast, Chen et al. (42) suggest that vinculin activation occurs only after a simultaneous binding event involving the coincidence of both actin and talin with vinculin. More recently, molecular dynamic simulations of vinculin activation (46) suggest mechanical tension could play a role in activating vinculin, and even suggest a structure for activated vinculin (Fig. 1 B).
Recently, a number of experimental data have emerged that vinculin is important in cellular force generation and transduction, for example experiments by Grashoff et al. (47) using a FRET mechanosensor report mechanical tension across activated vinculin, further supporting the suggestion born from the molecular dynamics simulations (46) that vinculin can be activated by a stretching force. Furthermore, other studies have suggested a phosphorylation event would be necessary to achieve vinculin activation (48–50).
The hypothesis of vinculin activation by phosphorylation is not necessarily a competing hypothesis to vinculin activation by a stretching force. It is conceivable that phosphorylation enhances the ability of vinculin to be activated by a stretching force, or perhaps a stretching force enhances the ability of vinculin to be activated by phosphorylation. It is likely that each event enhances the probability of activation and together lead to vinculin activation. Considering the controversy surrounding the mechanism for vinculin activation, it is important to evaluate these hypotheses and refine our understanding of the process of focal adhesion growth via vinculin activation and recruitment.
In this study, we evaluate the influence of talin binding to D1 on the activation of vinculin using molecular dynamics simulation. The binding of talin VBS to D1 involves the insertion of VBS into the hydrophobic patch formed by the four helices of D1 (34). Upon binding of talin, D1 is suggested to undergo helical bundle conversion (51). Using molecular dynamics, Lee et al. (34) have simulated the binding of VBS to D1 alone and elaborated a three-step binding process:
-
1.
Insertion of the VBS helix between helices 1 and 2 of D1.
-
2.
Movement of helices 1 and 2 away from VBS.
-
3.
Rotation of hydrophobic residues on VBS into the hydrophobic patch of D1.
In this study, we expand on the previous simulations and evaluate the influence of the full-length vinculin structure on the VBS binding event.
In our approach, we first simulate the binding of VBS to full-length vinculin in its autoinhibited conformation. We then simulate binding of VBS to full-length vinculin in an activated conformation. Simulation of binding to autoinhibited vinculin will evaluate whether:
-
1.
Helical bundle conversion and insertion of VBS into D1 is possible with the close proximity of Vt to D1.
-
2.
Binding of VBS to D1 can subsequently cause conformational changes in vinculin leading to activation.
For simulation of VBS binding to activated vinculin, we use the suggested structure of activated vinculin from the previous molecular dynamics study (46), and simulate the interaction of VBS with D1 of vinculin in this conformation. Simulation of VBS binding to an activated vinculin will evaluate the validity of that conformation being activated. An activated vinculin would bind each of its binding partners, including Talin.
Methods
Simulation of VBS near full-length vinculin
To explore the interaction between talin and vinculin in its autoinhibited conformation (Fig. 1 A) molecular dynamics simulations are utilized. The crystal structure of full-length vinculin (PDB ID: 1TR2) (39) is used as the autoinhibited vinculin structure and the VBS structure from the crystal structure of D1 bound to talin VBS (PDB ID: 1T01) (52) is used after alignment. Swiss-PDB (53) is used to structurally align D1 bound to VBS with D1 from full-length vinculin, effectively establishing the correct VBS orientation needed for binding. Once aligned, VBS is translated 12 Å away from D1 while remaining in the orientation needed for binding. Structural rotation and translations are done with VMD (54).
The system is minimized with 3000 steps of the steepest descent method followed by the adopted basis Newton-Raphson method and then heated to 310 K over 80 ps. Once heated, harmonic constraints are placed on the center of mass of vinculin—residues E186, W258, A490, and E569—to prevent its stochastic translation. Initially no other constraints are applied and the two molecules are simulated in the NPT ensemble for 30 ns in each trial. The SHAKE algorithm (55) is used to constrain bond lengths between heavy atoms and hydrogen atoms, hence allowing the use of a 2-fs timestep. The CHARMM19 force fields (56) are used in conjunction with an implicit solvent model, the effective energy function (EEF1) (57).
An implicit solvent model is an appropriate approximation of solvent effects for this simulation because the driving interaction between VBS and D1 of vinculin is hydrophobic. Implicit solvent models, including EEF1, have been extensively validated by experimental results (58) and have been used numerous times in similar simulations (59). Implicit solvent models lack electrostatic shielding effects, but these shielding effects should have minimal impact on our results considering the hydrophobic nature of the VBS and D1 interaction. The implicit solvent model also lacks accurate viscous solvent effects, which in these simulations allows for binding events to occur within our 30-ns simulations window. To track the binding event we monitor both the distance between the center of the VBS helix and the center of the fourth helix in D1 as a reporter of the distance between D1 and VBS, and we monitor the distance between helix 1 and helix 2 as a reporter of the helical bundle conversion in D1. All postsimulation calculations, visualizations, and analyses are carried out using VMD (54).
To increase the possibility of binding events between VBS and D1 of autoinhibited vinculin the molecular dynamics are repeated using an initial nudging force on VBS. After minimization and heating, a nudging force of 15 pN is applied to VBS at residues Q610, L615, G617, and E621 for a duration of 1 ns in the direction of D1. The short application of the nudging force gives VBS an initial velocity toward D1, increasing the possibility of a binding event. After the nudge, the VBS and autoinhibited vinculin system is simulated for an additional 30 ns with the absence of any externally applied force. As before, the center of mass of vinculin is harmonically constrained to prevent its stochastic translation. Both simulation of the brief nudging of VBS and the subsequent simulation of the binding of VBS to D1 are carried out as before, with the EEF1 implicit solvent model (57), the CHARMM19 force fields (56), and the SHAKE algorithm (55). All simulations are run with the CHARMM software package (60).
Simulation of VBS near an activated vinculin
To simulate the interaction of talin's VBS with vinculin in an activated conformation (Fig. 1 B), the suggested structure for activated vinculin from our previous molecular dynamics investigation (46) was used along with the structure of VBS from the crystal structure of VBS bound to D1 (PDB ID: 1T01) (52). VBS bound to D1 was aligned with D1 of the activated vinculin structure using Swiss-PDB (53). VBS in its aligned orientation is then used for simulation along with the activated vinculin structure. The aligned VBS is translated 12 Å away from D1, using VMD (54), before the start of the molecular dynamics simulations. Before the start of simulation, the activated vinculin structure is minimized with adopted basis Newton-Raphson for 3000 steps and then equilibrated for 2 ns. The additional equilibration is to allow the activated vinculin structure to reach an equilibrium in its new conformation.
To prevent stochastic translation, harmonic constraints are placed on the center of mass of vinculin. To prevent a conformational switch back to the autoinhibited conformation, weak harmonic constraints (K = 1.0 kcal/mol/Å2) are placed along helix 4 of domain 1. Previous results from binding of VBS to domain 1 alone (34) showed helix 4 of domain 1 to act as a scaffold for VBS binding with no movement during the binding event, thus application of these weak harmonic constraints on helix 4 should not affect results from our binding simulations using the activated vinculin conformation. VBS should interact with D1 hydrophobically, allowing the use of an implicit solvent treatment without significant artifacts (34). The EEF1 (57) implicit solvent model is used for solvent effects, the SHAKE algorithm (55) is used to constrain hydrogen atoms to heavy atoms, and CHARMM19 force fields (56) are used for the physics definitions. Two-femtosecond timesteps are used for each step of the molecular dynamics. After alignment of VBS, and preparation of activated vinculin, the two molecules are minimized together and then heated to 310 K over 80 ps. For simulation of the binding event, the two molecules are simulated together for 30 ns. Results from all simulations were visualized using VMD; trajectory data was also recorded using VMD (54).
As with binding of VBS to the autoinhibited vinculin conformation, simulation of VBS binding to the activated vinculin conformation are further stimulated after the application of initial brief nudging forces of 5 pN, 12.5 pN, or 15 pN are applied to VBS, at residues Q610, L615, G617, and E621, for a duration of 1 ns. The brief nudging force is applied to accelerate VBS toward domain 1 and to increase the possibility of binding. For both simulation of the initial brief nudging of VBS and the subsequent binding of VBS to D1, the same simulation parameters as above are used: weak harmonic constraints are applied to helix 4 of D1 to prevent conformational switch back to the autoinhibited vinculin conformation, harmonic constraints are applied to the center of mass of vinculin to prevent stochastic translation, the SHAKE algorithm (55) is used to allow 2fs timesteps, and the EEF1 implicit solvent model (57) is used along with CHARMM19 (56) force fields for efficient simulation.
Results
VBS fails to bind autoinhibited vinculin
Simulation of VBS binding to D1 isolated from other parts of vinculin showed binding of VBS to D1 after movement of helices 1 and 2 of D1 and hydrophobic insertion of VBS (34). In extension of that study, binding of VBS to D1 is simulated with inclusion of all domains of vinculin in the autoinhibited conformation. To allow for binding within a reasonable simulation time, VBS is aligned with D1 of full-length vinculin in the orientation predicted by the crystal structure of VBS bound to D1 (44). While maintaining this correct orientation, VBS is then translated away from D1 to be 12 Å away from D1, ensuring a layer of solvation between vinculin and VBS. Twelve simulations, 30 ns each, are produced with this setup.
These simulations produce two possible outcomes:
-
1.
VBS demonstrates hydrophobic contact with regions of D1 but fails to undergo hydrophobic insertion (Fig. 2).
-
2.
VBS fails to initiate any hydrophobic contact with D1 and drifts away from D1.
Figure 2.
VBS does not bind autoinhibited vinculin. Simulation of the binding of VBS to D1 of vinculin in its autoinhibited conformation suggests that VBS can interact with hydrophobic residues on the surface of D1 between helix 1 and helix 2, but it cannot insert into the hydrophobic core of D1. For VBS to insert into D1, helix 1 and helix 2 of D1 need to separate. VBS is linked to D1 by the surface interactions, but without helical separation it fails to fully insert into D1.
It has been predicted that talin binds vinculin by insertion of its hydrophobic residues into the hydrophobic core of D1 (34), therefore we define a complete binding event as any event resulting in complete insertion of VBS into the hydrophobic core of D1. None of the 12 trials show complete binding and hydrophobic insertion of VBS to D1 during the 30-ns simulation window.
In eight out of 12 trials (66%) VBS initiates hydrophobic contact with regions of D1. On average, VBS migrates toward D1 and forms contact with D1 within 5 ns of simulation. These initial intermolecular forces link VBS of talin to D1 of vinculin. Initially, P607 and L608 of VBS form hydrophobic interactions with V62, V57, I12, and L54 of D1 (Fig. 3 A). After these interactions, I615 and A616 proceed to interact with P15 and A50 of D1. Together these interactions prevent the drift of VBS away from D1 for the remaining 25 ns of simulation. Despite this prolonged link of VBS to D1 at the surface of helix 1 and helix 2, VBS fails to insert into the hydrophobic core of D1. For full binding, the distance between helix 1 and helix 2 needs to increase; yet in each of these trials the distance between these helices remains relatively unchanged (see Fig. S1 in the Supporting Material).
Figure 3.
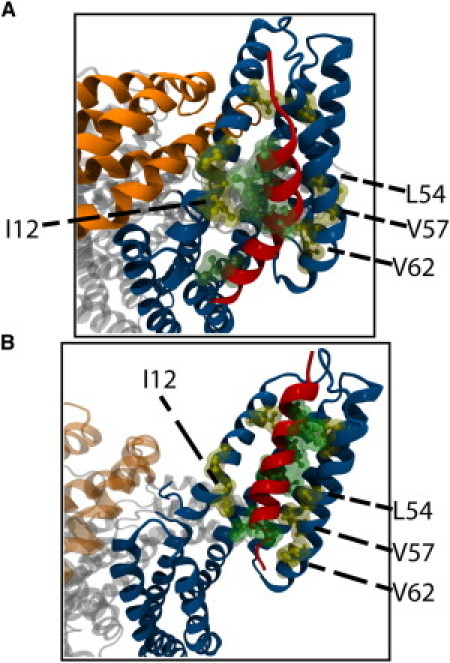
Hydrophobic interactions link VBS to D1. (A) Interaction among VBS residues P607, L608, I615, and A616 with D1 residues I12, P15, A50, L54, V57, and V62 link lower-VBS to D1. Lower-VBS links to D1 by these interactions in simulation with autoinhibited vinculin (shown here) and with activated vinculin. (B) In addition to the lower-VBS interactions, a second set of surface hydrophobic interactions link upper-VBS to D1 in simulations with activated vinculin. Residues V619, L622, and L623 of VBS interact with residues L23 and P43 of D1. D1 Helical separation near lower-VBS allows interaction of these residues near upper-VBS.
In one particular trial, helix 2 of D1 begins to separate from helix 1 (Fig. S1) as VBS forces its way into the hydrophobic core. The separation between helix 1 and helix 2 continues for 10 ns. In the end, VBS fails to insert and is eventually forced out of the gap between the two helices. Contact between helix 1 of D1 and Vt prevents the necessary separation between the helices and prevents the hydrophobic insertion of VBS into D1 (Fig. 4).
Figure 4.
Contact between D1 and Vt prevents VBS binding to autoinhibited vinculin. VBS insertion into D1 requires separation of helix 1 and helix 2. Helix 1 is near Vt and can sterically contact Vt residues during separation. The proximity of Vt to D1 limits helix 1 movement; residues R7, E10, Q18, I20, S21, V24, and I25 clash with residues G940, S941, T943, R945, A946, P989, T993, K996, and I997. Separation of Vt and D1 by vinculin activation removes these clashes, allowing for helix 1 movement and VBS insertion.
In four out of 12 trials (33%), VBS forms no contact with D1 and drifts away from its binding site on vinculin. Interaction with and binding to D1 requires 1), VBS maintain the correct orientation necessary for binding; 2), that it stochastically moves toward vinculin; 3), that it forms interactions with D1 that stabilize it and reduces its movement away from D1; and 4), that it proceeds to force its way into the hydrophobic patch in D1. In the trials where VBS drifts away from D1, VBS fails at achieving step 2, and stochastically moves in other directions. In the trials where VBS interacts with D1 yet fails to bind, VBS fails to achieve step 4. In none of the trials does VBS achieve all the steps and fully bind vinculin.
VBS binds the activated vinculin conformation
Previous molecular dynamics simulation of vinculin activation (46) has produced a suggested conformation for activated vinculin. In this suggested activated conformation, D1 has been moved away from Vt, removing the contact between D1 and Vt (Fig. 1 B). To further test the impact of the Vt contact with D1, the binding of VBS to vinculin in this activated conformation is simulated. These simulations also serve to evaluate whether this suggested conformation will be consistent with the expected binding behavior of activated vinculin; vinculin should bind its binding partners, namely talin and actin, in its activated conformation. Although it would be more informative to simulate vinculin activation and VBS binding simultaneously, limitations in computing power limit us to simulation of VBS binding before and after vinculin activation.
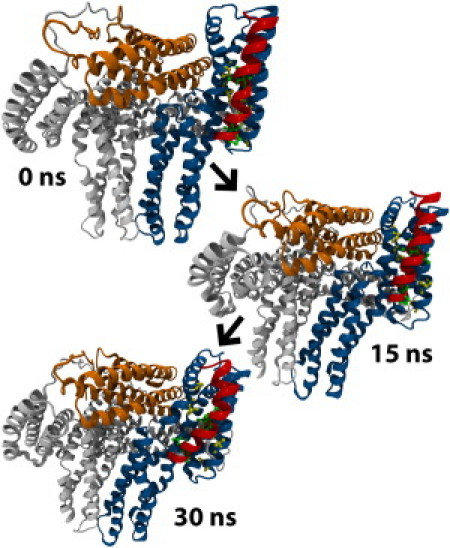
Simulation of activation and binding could report the impact of VBS binding on the rate of vinculin activation, whereas our present simulations will contrast the binding of VBS to vinculin in its inactive, and its suggested active, conformation. VBS is simulated interacting with D1 of this activated conformation using only one harmonic constraint to prevent translation of vinculin, and a second weak harmonic constraint to hold vinculin in the suggested activated conformation. Out of 15 trials of 30-ns simulations, two simulations show binding of VBS with insertion of VBS into the hydrophobic core of D1 (Fig. 5) and 13 simulations show partial binding of VBS to D1 with incomplete insertion (Fig. 5). These results are comparable to simulation results from a previous study of VBS binding to an isolated D1 (34), where VBS inserted into an isolated D1 in only one simulation when no nudging force was used.
Figure 5.
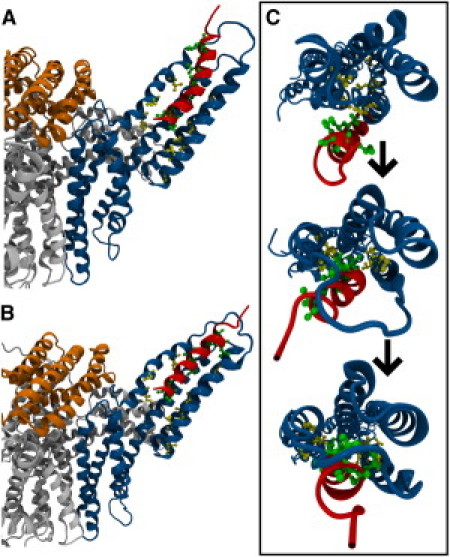
VBS can bind activated vinculin. (A) Interaction between hydrophobic residues in VBS and D1, links lower-VBS to D1. Separation of Vt from D1 in activated vinculin allows for helix 1 movement and insertion of lower-VBS after linking to D1. (B) Upper-VBS can link D1 after lower-VBS insertion. After surface interactions between upper-VBS and D1, if D1 helices continue to separate, upper-VBS can also insert into D1. (C) VBS inserts into activated vinculin, first, by inserting lower-VBS, followed by insertion of upper-VBS, and finally, rotation of VBS hydrophobic residues into the hydrophobic core of D1. D1 is shown with a view looking down its helices.
The use of an initial nudging force reduces the possible translational movements of VBS to those that are toward D1, thereby reducing the sampling necessary to evaluate binding. In both the previous study of binding to an isolated D1 and in our simulations with an activated vinculin conformation, a nudging force greatly increases the number of binding events (see below).
In the 13 simulations lacking complete VBS insertion, residues closer to the N-terminus of VBS (lower-VBS) show partial insertion of VBS in between helix 1 and helix 2 of D1 (Fig. 5). At these D1 regions with VBS insertion, helix 1 and helix 2 of D1 are forced apart whereas D1 regions' interaction with VBS residues near the C-terminus of VBS (upper-VBS) show no separation between the helices (Fig. S2). Separation of helix 1 and helix 2 by at least 20.5 Å at all regions of D1 interacting with lower-VBS and upper-VBS is necessary for complete insertion of VBS (34). After complete insertion, VBS should be within 13 Å of helix 4 (34). Insertion of lower-VBS into D1 helices is stabilized by hydrophobic interactions. VBS residues P607, L608, I615, and A616 interact with D1 residues V57, V62, P15, I12, A50, and L54 (Fig. 3 B); VBS interacts with autoinhibited vinculin at the same residues (see above).
Simulations resulting in the complete insertion of VBS into the hydrophobic core of D1 initially show the same interaction between residues in lower-VBS and D1. The partial insertion of lower-VBS anchors VBS to vinculin, allowing upper-VBS to force itself in between helix 1 and helix 2 of D1 (Fig. 5). The results show that regions of D1 nearest lower-VBS show helical separation before regions of D1 nearest upper-VBS (Fig. S2). Insertion of upper-VBS into D1 within the 30-ns simulation window occurs in only two of the trials. In these trials, VBS insertion occurs, after at least 20 ns of simulation, after interaction of VBS residues V619, L622, and L623 with D1 residues L23 and P43 (Fig. 3 B). Removal of the contact between helix 1 and Vt allows for insertion of both lower-VBS and upper-VBS into D1.
Acceleration of VBS toward D1 only enhances VBS complete insertion in the activated vinculin conformation
To enhance the binding and full insertion of VBS into D1, the 30-ns binding simulations are repeated with the additional application of an initial small nudging force on VBS, applied over 1 ns. The nudging force is applied to VBS because of its smaller size as compared to vinculin. The nudging force accelerates VBS toward its binding groove between helix 1 and helix 2 in D1. Twelve trials simulate the binding of VBS to vinculin in the autoinhibited conformation after a 15-pN nudge, and 12 trials simulate the binding of VBS to the activated conformation after an initial nudge—four trials with a 15-pN nudge, four trials with a 12.5-pN nudge, and four trials with a 5-pN nudge. Application of the nudging force accelerates VBS toward D1, yet no binding interactions occur within the 1-ns window during which the nudge is applied.
Application of the initial accelerating nudge force enhances binding of VBS to the activated vinculin conformation, but none of the simulations of VBS binding to the autoinhibited vinculin conformation after a nudging force show complete binding (Table S1). VBS inserts into D1 in 25% of the trials with a 5-pN initial nudging force, in 25% of the trials with a 12.5-pN initial nudging force, and in 75% of the trials with a 15-pN initial nudging force. The insertion of VBS after an initial nudge follows the same trajectory as insertion of VBS in the previous sections; first, lower-VBS inserts between helix 1 and helix 2, then, upper-VBS inserts between helix 1 and helix 2, and finally, VBS rotates into the hydrophobic core in D1. The initial force increases the rate of binding of VBS to the activated vinculin structure, within the 30-ns simulation window, yet, the mechanism of binding is unchanged. The contact between Vt and D1 prevents VBS binding to autoinhibited vinculin, even after the initial nudge.
Discussion
Simulation of the interaction between talin and vinculin in each of its two conformations—autoinhibited and activated—suggests that a prior vinculin activation event is necessary to allow for full binding of talin to D1 of vinculin. Talin interacts with D1 via the insertion of VBS into the hydrophobic core of D1 (34,35,40,45,62). The insertion of VBS into D1 while vinculin is in its autoinhibited conformation (Fig. 1 A) fails to occur in simulation, even after an initial nudge of VBS toward D1 (Table S1 and Fig. 2) because D1 cannot undergo the necessary conformational change to accommodate VBS insertion while in close proximity to Vt (Fig. 4). In contrast, the insertion of VBS into D1 of vinculin in its activated conformation (Fig. 1 B) occurs in simulation after first, separation of D1 helices near lower-VBS; second, separation of D1 helices near upper-VBS; and third, rotation of VBS into the hydrophobic core (Fig. 5). This binding to the activated vinculin conformation is enhanced by an initial nudge of VBS toward D1 (Table S1). Vinculin activation before talin binding could reduce the proximity of D1 and Vt (46) allowing for helix 1 and helix 2 to separate and for VBS to follow these steps to activation.
The trajectory suggested here for the binding of VBS to D1 can be compared to the trajectory for VBS binding to D1 in that absence of other vinculin domains (34). Simulation of VBS binding to D1 alone described the binding event as comprising of three essential steps:
-
1.
Insertion of VBS into the groove between helix 1 and helix 2.
-
2.
Separation of helix 1 from helix 2.
-
3.
Rotation of VBS into the hydrophobic core of D1.
The results presented here expand these steps to account for effects of the other vinculin domains; VBS binding to D1 can now be described with the following steps:
-
1.
Insertion of lower-VBS into the groove between helix 1 and helix 2 in D1.
-
2.
Separation of helix 1 and helix 2 at regions near lower-VBS.
-
3.
Insertion of upper-VBS into the groove between helix 1 and helix 2 in D1.
-
4.
Separation of helix 1 and 2 at regions near upper-VBS.
-
5.
Rotation of VBS hydrophobic residues into the hydrophobic core of D1.
The steps suggested here are similar to and confirm the steps suggested by Lee et al. (34). In the previous simulations, VBS from α-actinin and other talin VBSs all bound vinculin D1 with the same mechanism. Simulation of VBS from α-actinin in Lee et al. (34), however, suggested α-VBS binds with an inverted orientation. The results presented here demonstrate a significant difference between binding to lower-VBS and upper-VBS, therefore it is unclear whether α-VBS would bind D1 of full-length vinculin as it did D1 in the previous simulations. The nature of α-actinin binding to full-length vinculin should be a topic for future investigations.
The suggestion that vinculin activation is required for full talin binding extends the current understanding of vinculin autoinhibition. Vinculin in its native conformation is defined as being autoinhibited mainly because the interaction of Vt with actin is not possible in this conformation (38). It has been suggested that actin binding to Vt is inhibited by the proximity of D1 to Vt; D1 would sterically clash with any nearby actin filaments that would otherwise bind Vt. The results from these simulations suggest that vinculin is also autoinhibited in its native conformation because this proximity between Vt and D1 also prevents the complete binding of talin to D1.
Although talin can form a weak hydrophobic link to D1 via interaction at the lower-VBS residues, it is important to recognize that this differs from the mechanisms of talin binding to vinculin previously described (35,43,45). The conformation of vinculin can regulate the strength of the talin-vinculin linkage; talin and vinculin would likely link with the greatest strength only after a transition of vinculin to an activated conformation. This observation can lead us to two specific assertions concerning vinculin recruitment to focal adhesions:
-
1.
It is possible that talin is not the only mechanosensor at focal adhesions but vinculin might also be dependent on a mechanical environment for its activation.
-
2.
Vinculin reinforcement at focal adhesions and mechanosensitive focal adhesion growth is a holistic event that depends on a coordinated set of events between numerous molecules.
The cooperative nature of focal adhesion growth accounts for the abundance of a number of different molecules at focal complexes.
In the simulations that failed to show full VBS insertion into D1, hydrophobic residues in lower-VBS interact with hydrophobic residues in D1 (Fig. 3). Although these interactions are a weaker intermolecular bond than the full insertion of VBS into the hydrophobic core would have been, they link talin to vinculin and prevent the drifting of VBS away from D1. Considering that the suggested mechanism for vinculin activation is that actin and talin cooperatively interact with vinculin to cause its activation (42,46), this proximity of talin to vinculin by the weaker hydrophobic interaction at lower-VBS could allow for vinculin activation. Furthermore, phosphorylation of vinculin could also contribute to vinculin activation by enhancing the cooperative binding of actin and talin to vinculin.
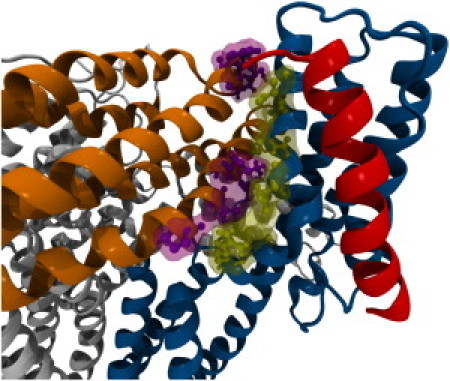
With D1 weakly interacting with lower-VBS, an electrostatic interaction of Vt with actin (38) could then stretch vinculin and cause its activation (46). The stretching of vinculin can lead to a conformational change in which D1 moves away from Vt and vinculin becomes activated (Fig. 1 B). Then, after vinculin activation by movement of D1 away from Vt, talin's VBS would be able to fully insert into D1 to solidify and strengthen the talin-vinculin link (Fig. 6). In this way vinculin can serve as an intermediary for actin binding to talin. The strengthening of the interaction between vinculin and talin after vinculin activation allows for vinculin to take a larger mechanical load at focal adhesions, reflecting its role as a reinforcing agent. With vinculin activation by this mechanism, it is possible that the number of actin filaments linked to each talin rod can be multiplied via vinculin, although there is currently no experimental evidence supporting this hypothesis. The expansion of actin filaments linked to integrin-bound talin molecules is essential to focal adhesion growth and maturation. In force-induced focal adhesion formation, as the load on the developing focal complex increases we would expect more vinculin activation and recruitment of activated vinculin to crosslink talin and actin filaments.
Figure 6.
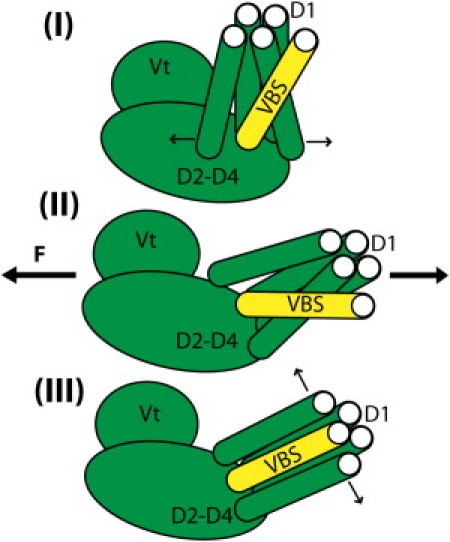
A mechanism for vinculin activation: VBS linking followed by binding. Simulation of VBS interaction with autoinhibited vinculin suggests that VBS can link to D1 but cannot insert into D1 with Vt in close proximity. Simulation of VBS interaction with activated vinculin suggests that VBS can link to and insert into D1 after the proximity of Vt is removed. Together, these results suggest the following mechanism for vinculin activation: (I) lower-VBS links to D1; (II) vinculin is stretched (by simultaneous interaction with talin and actin perhaps); and (III) VBS fully inserts into D1.
That vinculin becomes activated by a stretching force, and furthermore, that after its stretch and activation vinculin strengthens its link to at least one of its binding partners, talin, is entirely consistent with vinculin's role as a linker molecule. Vinculin recruitment to focal adhesions serves to strengthen and reinforce the cytoskeletal link to the ECM (1,11,27,31); vinculin is a reinforcing agent. The ability of vinculin to strengthen its binding upon stretch is a feature that enables its reinforcement. Recent in vivo experiments using a FRET probe to measure mechanical tension across single molecules reports that vinculin is under 2.5 pN of tension in vivo when reinforcing focal adhesions (47). It is likely that this tension stretches vinculin to a conformation similar to the one tested here (46) (Fig. 1 B), and also that it strengthens the link of vinculin to talin. Mechanosensation at focal adhesions is not limited to talin and vinculin. Other critical components of focal adhesions, such as membrane-bound integrins, are also involved in mechanosensation by focal adhesions: the process of integrin clustering at focal adhesions can be force-induced, and the binding of integrin to ECM can be mechanosensitive (63).
The results reported here are limited to the structures used in simulation. Just as including other regions of vinculin aside from D1 in these simulations demonstrated an impact of those regions on the conformational changes D1 is able to make during binding to VBS, we expect that including other regions of talin near VBS in simulation would help us investigate the impact of those nearby regions on the ability of VBS to bind to and interact with D1. To expand on the results presented here and further investigate the mechanisms of this critical interaction between talin and vinculin, it would be important explore the impact of the talin rod on the binding of talin to vinculin. In doing so, one challenge that would need to be addressed is the consistency of conformation of the talin rod with VBS activation.
Some studies have reported a talin conformation in which VBS is rotated out of its hydrophobic grove in the rod domain during VBS activation (31), whereas other studies have suggested an unraveling of the rod domain during talin-VBS activation (32,33). Of course both conformations assume talin VBS activation by exposure to an external stress, whereas it is entirely possible that talin mechanosensation is a result of external strain: movement of the integrin binding head domain from each talin monomer would cause a talin dimer to shift in its arrangement of rod domains from parallel to elongated, effectively doubling the number of actin filaments that could be cross-linked by vinculin.
Here we have investigated the interaction of activated vinculin with talin and contrasted it with the interaction of autoinhibited vinculin with talin. The other significant binding partner of vinculin is the actin filament. It is essential to understand the mechanisms of actin-vinculin binding. It has been suggested that electrostatic forces might play a significant role in driving the binding of vinculin to actin (38). The interactions between Vt and actin filaments that drive this binding event should be studied and characterized.
Is the suggested conformation of activated vinculin (46)—which removes the proximity of D1 to Vt via movement of D1—sufficient to allow Vt to interact with actin?
Will there be a clash between an actin filament aiming to bind vinculin and a talin rod domain aiming to bind D1? And if so, what are the additional conformational changes in vinculin that would prevent this clash?
The results presented in this article evaluated the activated vinculin conformation for binding to talin, but numerous significant questions remain.
Acknowledgments
The authors are thankful to Yousef Jamali and other members of Molecular Cell Biomechanics Laboratory for their invaluable input.
Financial support by National Science Foundation (CAREER-0955291) is gratefully acknowledged.
Supporting Material
References
- 1.Mierke C.T., Kollmannsberger P., Goldmann W.H. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys. J. 2008;94:661–670. doi: 10.1529/biophysj.107.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarz U.S., Erdmann T., Bischofs I.B. Focal adhesions as mechanosensors: the two-spring model. Biosystems. 2006;83:225–232. doi: 10.1016/j.biosystems.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 3.Critchley D.R. Focal adhesions—the cytoskeletal connection. Curr. Opin. Cell Biol. 2000;12:133–139. doi: 10.1016/s0955-0674(99)00067-8. [DOI] [PubMed] [Google Scholar]
- 4.Bershadsky A.D., Ballestrem C., Kozlov M.M. Assembly and mechanosensory function of focal adhesions: experiments and models. Eur. J. Cell Biol. 2006;85:165–173. doi: 10.1016/j.ejcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Shen T.L., Park A.Y., Guan J.L. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J. Cell Biol. 2005;169:941–952. doi: 10.1083/jcb.200411155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu M.H. Endothelial focal adhesions and barrier function. J. Physiol. 2005;569:359–366. doi: 10.1113/jphysiol.2005.096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamir E., Geiger B. Components of cell-matrix adhesions. J. Cell Sci. 2001;114:3577–3579. doi: 10.1242/jcs.114.20.3577. [DOI] [PubMed] [Google Scholar]
- 8.Ingber D.E. Mechanobiology and diseases of mechanotransduction. Ann. Med. 2003;35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 9.Critchley D.R. Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem. Soc. Trans. 2004;32:831–836. doi: 10.1042/BST0320831. [DOI] [PubMed] [Google Scholar]
- 10.Geiger B., Spatz J.P., Bershadsky A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 11.Ziegler W.H., Gingras A.R., Emsley J. Integrin connections to the cytoskeleton through talin and vinculin. Biochem. Soc. Trans. 2008;36:235–239. doi: 10.1042/BST0360235. [DOI] [PubMed] [Google Scholar]
- 12.Xiong J.P., Stehle T., Arnaout M.A. New insights into the structural basis of integrin activation. Blood. 2003;102:1155–1159. doi: 10.1182/blood-2003-01-0334. [DOI] [PubMed] [Google Scholar]
- 13.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 14.Huveneers S., Danen E.H. Adhesion signaling—crosstalk between integrins, Src and Rho. J. Cell Sci. 2009;122:1059–1069. doi: 10.1242/jcs.039446. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y.L., McNamara L.M., Weinbaum S. A model for the role of integrins in flow induced mechanotransduction in osteocytes. Proc. Natl. Acad. Sci. USA. 2007;104:15941–15946. doi: 10.1073/pnas.0707246104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galbraith C.G., Yamada K.M., Sheetz M.P. The relationship between force and focal complex development. J. Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamm R.D., Kaazempur-Mofrad M.R. On the molecular basis for mechanotransduction. Mech. Chem. Biosyst. 2004;1:201–209. [PubMed] [Google Scholar]
- 18.Mofrad M.R., Golji J., Kamm R.D. Force-induced unfolding of the focal adhesion targeting domain and the influence of paxillin binding. Mech. Chem. Biosyst. 2004;1:253–265. [PubMed] [Google Scholar]
- 19.Zhang X., Jiang G., Sheetz M.P. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 2008;10:1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Clainche C., Carlier M.F. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 21.Critchley D.R., Gingras A.R. Talin at a glance. J. Cell Sci. 2008;121:1345–1347. doi: 10.1242/jcs.018085. [DOI] [PubMed] [Google Scholar]
- 22.Moes M., Rodius S., Kieffer N. The integrin binding site 2 (IBS2) in the talin rod domain is essential for linking integrin β-subunits to the cytoskeleton. J. Biol. Chem. 2007;282:17280–17288. doi: 10.1074/jbc.M611846200. [DOI] [PubMed] [Google Scholar]
- 23.Gingras A.R., Bate N., Critchley D.R. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H., Cohen D.M., Craig S.W. Spatial distribution and functional significance of activated vinculin in living cells. J. Cell Biol. 2005;169:459–470. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brett T.J., Legendre-Guillemin V., Fremont D.H. Structural definition of the F-actin-binding THATCH domain from HIP1R. Nat. Struct. Mol. Biol. 2006;13:121–130. doi: 10.1038/nsmb1043. [DOI] [PubMed] [Google Scholar]
- 26.Hemmings L., Rees D.J., Critchley D.R. Talin contains three actin-binding sites, each of which is adjacent to a vinculin-binding site. J. Cell Sci. 1996;109:2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- 27.Humphries J.D., Wang P., Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reference deleted in proof.
- 29.Critchley D.R. Genetic, biochemical and structural approaches to talin function. Biochem. Soc. Trans. 2005;33:1308–1312. doi: 10.1042/BST0331308. [DOI] [PubMed] [Google Scholar]
- 30.Gingras A.R., Ziegler W.H., Emsley J. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J. Biol. Chem. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- 31.Lee S.E., Kamm R.D., Mofrad M.R. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J. Biomech. 2007;40:2096–2106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Hytönen V.P., Vogel V. How force might activate talin's vinculin binding sites: SMD reveals a structural mechanism. PLOS Comput. Biol. 2008;4:e24. doi: 10.1371/journal.pcbi.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Rio A., Perez-Jimenez R., Sheetz M.P. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S.E., Chunsrivirot S., Mofrad M.R. Molecular dynamics study of talin-vinculin binding. Biophys. J. 2008;95:2027–2036. doi: 10.1529/biophysj.107.124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegler W.H., Liddington R.C., Critchley D.R. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Diez G., List F., Goldmann W.H. Direct evidence of vinculin tail-lipid membrane interaction in β-sheet conformation. Biochem. Biophys. Res. Commun. 2008;373:69–73. doi: 10.1016/j.bbrc.2008.05.182. [DOI] [PubMed] [Google Scholar]
- 37.Cohen D.M., Chen H., Craig S.W. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- 38.Janssen M.E., Kim E., Hanein D. Three-dimensional structure of vinculin bound to actin filaments. Mol. Cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 39.Borgon R.A., Vonrhein C., Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Cohen D.M., Kutscher B., Craig S.W. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J. Biol. Chem. 2006;281:16006–16015. doi: 10.1074/jbc.M600738200. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y., Dokholyan N.V. Insights into allosteric control of vinculin function from its large scale conformational dynamics. J. Biol. Chem. 2006;281:29148–29154. doi: 10.1074/jbc.M605512200. [DOI] [PubMed] [Google Scholar]
- 42.Chen H., Choudhury D.M., Craig S.W. Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 2006;281:40389–40398. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- 43.Bois P.R., O'Hara B.P., Izard T. The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J. Biol. Chem. 2006;281:7228–7236. doi: 10.1074/jbc.M510397200. [DOI] [PubMed] [Google Scholar]
- 44.Izard T., Vonrhein C. Structural basis for amplifying vinculin activation by talin. J. Biol. Chem. 2004;279:27667–27678. doi: 10.1074/jbc.M403076200. [DOI] [PubMed] [Google Scholar]
- 45.Nhieu G.T., Izard T. Vinculin binding in its closed conformation by a helix addition mechanism. EMBO J. 2007;26:4588–4596. doi: 10.1038/sj.emboj.7601863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golji J., Mofrad M.R. A molecular dynamics investigation of vinculin activation. Biophys. J. 2010;99:1073–1081. doi: 10.1016/j.bpj.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grashoff C., Hoffman B.D., Schwartz M.A. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Küpper K., Lang N., Hoffmann B. Tyrosine phosphorylation of vinculin at position 1065 modifies focal adhesion dynamics and cell tractions. Biochem. Biophys. Res. Commun. 2010;399:560–564. doi: 10.1016/j.bbrc.2010.07.110. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z., Izaguirre G., Haimovich B. The phosphorylation of vinculin on tyrosine residues 100 and 1065, mediated by SRC kinases, affects cell spreading. Mol. Biol. Cell. 2004;15:4234–4247. doi: 10.1091/mbc.E04-03-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziegler W.H., Tigges U., Jockusch B.M. A lipid-regulated docking site on vinculin for protein kinase C. J. Biol. Chem. 2002;277:7396–7404. doi: 10.1074/jbc.M110008200. [DOI] [PubMed] [Google Scholar]
- 51.Izard T., Evans G., Bois P.R. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427:171–175. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- 52.Papagrigoriou E., Gingras A.R., Emsley J. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 2004;23:2942–2951. doi: 10.1038/sj.emboj.7600285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arnold K., Bordoli L., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 54.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 55.Kräutler V., Gunsteren W.F.v., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. [Google Scholar]
- 56.Neria E., Fischer S., Karplus M. Simulation of activation free energies in molecular systems. J. Chem. Phys. 1996;105:1902–1921. [Google Scholar]
- 57.Lazaridis T. Effective energy function for proteins in lipid membranes. Proteins. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 58.Best R.B., Clarke J., Karplus M. What contributions to protein side-chain dynamics are probed by NMR experiments? A molecular dynamics simulation analysis. J. Mol. Biol. 2005;349:185–203. doi: 10.1016/j.jmb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Feig M., Brooks C.L., 3rd Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr. Opin. Struct. Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 60.Brooks B.R., Bruccoleri R.E., Karplus M. CHARMM—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 61.Reference deleted in proof.
- 62.Patel B., Gingras A.R., Critchley D.R. The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J. Biol. Chem. 2006;281:7458–7467. doi: 10.1074/jbc.M508058200. [DOI] [PubMed] [Google Scholar]
- 63.Friedland J.C., Lee M.H., Boettiger D. Mechanically activated integrin switch controls α5β1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


