Abstract
Although ubiquitous, the processes by which bacteria colonize surfaces remain poorly understood. Here we report results for the influence of the wall shear stress on the early-stage adhesion of Pseudomonas aeruginosa PA14 on glass and polydimethylsiloxane surfaces. We use image analysis to measure the residence time of each adhering bacterium under flow. Our main finding is that, on either surface, the characteristic residence time of bacteria increases approximately linearly as the shear stress increases (∼0–3.5 Pa). To investigate this phenomenon, we used mutant strains defective in surface organelles (type I pili, type IV pili, or the flagellum) or extracellular matrix production. Our results show that, although these bacterial surface features influence the frequency of adhesion events and the early-stage detachment probability, none of them is responsible for the trend in the shear-enhanced adhesion time. These observations bring what we believe are new insights into the mechanism of bacterial attachment in shear flows, and suggest a role for other intrinsic features of the cell surface, or a dynamic cell response to shear stress.
Introduction
The adhesion of bacteria to surfaces is a ubiquitous phenomenon. In addition to interacting with tissues where they can initiate infection, many bacterial cells also bind to abiotic surfaces. After adhesion, they can grow to form surface-attached, organized communities known as biofilms (1). These slimy aggregates, in which cells are connected by a self-excreted polymeric matrix, are thought to be the dominant form of microbial life (2,3). Biofilms are difficult to remove from surfaces, and bacteria within them exhibit an increased resistance to many biocides (4). Consequently, reducing the development and spread of biofilms is a challenge in many areas, such as clinical environments and water sanitation (5).
The adhesion of individual bacterial cells to a surface is the first step toward biofilm formation, and elucidating the mechanism of this process is of crucial importance. Bacterial adhesion, which involves fundamental electrostatic, van der Waals, and hydrophobic interactions, is thought to be mediated by micrometer-long surface organelles such as pili and flagella. It is a complex process that remains poorly understood, despite being the focus of numerous studies (6,7).
Because in many situations bacterial cells are dispersed in a flowing carrier fluid (water, urine, and blood are common examples) and colonize an immersed surface, studying the dynamics of bacterial adhesion under controlled conditions of both flow and chemical environment is particularly relevant. Utilizing flow cells (8), previous studies have focused on the influence on bacterial adhesion of parameters such as pH (9), ionic strength (10), or surface chemistry (11).
Although the effect of the shear has been addressed in the context of biofilm formation and biomass accumulation (12,13), few studies have focused on the effect of flow on the adhesion mechanism of individual cells. Meinders et al. (9) were the first to monitor simultaneously adsorption and desorption events, and showed that shear stress influences both the attachment and detachment rates of bacteria. Although their approach has been used to study the adhesion dynamics of microorganisms under different environmental conditions, the effect of shear stress itself on individual adhesion events remains only partially understood, and a systematic study has not yet been reported. To our knowledge, the detailed response of individual adhering bacteria to shear stress has only been elucidated for Escherichia coli on specially prepared surfaces: in their work, Thomas et al. (14) observed that the adhesion of E. coli to mannose-coated surfaces was enhanced by shear stress. The authors postulated that this phenomenon is the result of the formation of catch-bonds between bacteria type I pili adhesin and surface-attached mannose, whose strength is increased by shear stress (15). This result was reminiscent of previous observations of shear-induced adhesion for leukocytes resulting from specific P-selectin receptor bonds (16,17).
In this article, we address the question of whether shear-increased adhesion requires specific ligand-receptor interactions, or may be a more widespread phenomenon. For that purpose, we focus on the effect of shear stress on the initial adhesion of individual bacteria to an untreated, abiotic substrate. We use soft lithography technology to create straight microfluidic channels with dimensions in the range of tens of micrometers. The small dimensions of the channel enable conditions of very low to very high shear stress, while allowing for in situ monitoring of individual adhesion events. We report results from studying Pseudomonas aeruginosa, an opportunistic pathogen that can cause lethal infections in immuno-deficient patients (3), for which we investigate early adhesion dynamics on two different surfaces (glass and polydimethylsiloxane (PDMS)).
We find that the typical duration of adhesion events of P. aeruginosa strain PA14 increases when shear stress increases, for values up to ∼3.5 Pa. To understand the biological origin of this phenomenon, we investigate the adhesion behavior of various PA14 mutant strains. To our surprise, our results show that shear-enhanced adhesion is not regulated by the properties of the primary surface appendages. Moreover, our experiments seem to indicate that this behavior is not dependent on the nature of the substrate (hydrophilic glass or hydrophobic PDMS), but rather appears to be a general feature of the adhesive behavior of P. aeruginosa under many different environmental conditions. These results contrast with the specific interaction identified for E. coli, and suggest the existence of a previously unrecognized mechanism for shear-enhanced adhesion.
Methods
Bacterial strains and growth conditions
P. aeruginosa strain PA14 was used in all experiments and grown following a standard protocol: bacteria were taken from a freshly streaked Luria-Bertani agar plate (24–48 h old) to start a liquid culture in Tryptone Broth (TB, 10 g/liter tryptone). The culture was shaken at 37°C for 4–5 h, until it reached the early stationary phase. Optical density measured at 600 nm (OD600) was ∼1. The solution was diluted 1/10 in 10 mM phosphate-buffered saline (PBS). After dilution, the culture was further shaken at 37°C for 1 h, then infused in the channel, and flushed for ∼10 min before starting the experiment. The measured concentration right before the experiment ranged from OD600 = 0.1 to OD600 = 0.3. Alternatively, MSgg medium was used instead of TB, and prepared following the recipe described in Branda et al. (18).
The aim of this procedure was to transfer bacteria in a nutrient-depleted medium, to prevent cell division, and work with cells that would be in as uniform a physiological state as possible. However, we decided to avoid centrifugation after we observed that this step could have a noticeable effect on the subsequent bacterial adhesion. Instead, we diluted the bacterial culture 1/10 in PBS and kept shaking at 37°C for 1 h. We checked that this was sufficient to reach a steady stage in which the bacterial concentration increased very slowly, probably due to nutrient depletion. In addition to wild-type, the following strains of PA14 were used in the experiments: cupA1::MrT7 (19), pilC::Tn5 (Tet), flgK::Tn5 (Tet) (20), and ΔpelA (21).
Experimental protocol and observations
Microfluidic devices were prepared using the standard soft lithography technique (22). Polydimethylsiloxane (PDMS, Sylgard 184 silicone elastomer kit, prepared with a ratio of 10 wt % cross-linker; Dow Corning, Midland, MI) stamps of a straight channel were plasma-sealed onto a glass surface (micro-cover glasses; VWR, Radnor, PA) 24 h before the experiment and kept at room temperature. The channels used were 3-cm-long, 200- or 400-μm-wide, with a height varying between 30 and 65 μm. Before starting bacterial adhesion measurements, the channel was rinsed for at least 15 min with PBS. The bacterial solution was then injected in the channel, and flushed at a constant flow rate Q using a syringe pump (PhD2000 nano; Harvard Apparatus, Holliston, MA). The wall shear stress created on the bottom and top walls of the channel is approximately
| (1) |
where w and h are the width and the height of the channel, and μ is the viscosity of the bacteria solution, which is close to the viscosity of water for the concentrations of bacteria we used. We applied flow rates between 0.1 and 80 μL/min, which correspond to values of the wall shear stress between ∼10−2 and 10 Pa (shear stress could be varied by changing both the imposed flow rate and the channel thickness).
The microfluidic device was placed on an inverted microscope (DM-IRB; Leica Microsystems, Bannockburn, IL), and the adhesion of bacteria on the glass surface was observed in situ with phase contrast microscopy (Fig. 1 A). To study adhesion on PDMS, we focused on the top surface of the channel. Test experiments were also performed using a 50 × 500-μm glass capillary (Hollow Rectangle Capillaries, fiber optic center; Vitrocom, Mountain Lakes, NJ) to form an all-glass channel. In all cases, the area of interest was taken far from the channel side walls, so that there was no spanwise velocity gradient and wall shear stress could be considered constant. To ensure similar environmental conditions, no more than three successive flow rates were studied in a given device.
Figure 1.
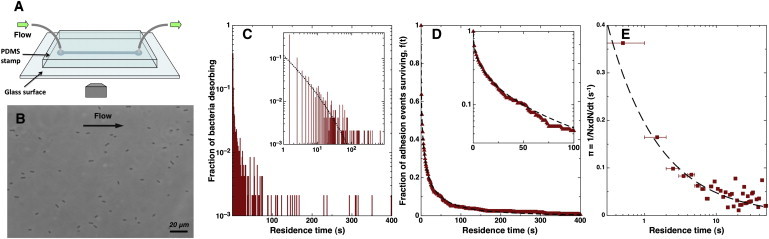
Principle of the experimental protocol and image analysis. (A) The straight channel is imprinted in a PDMS stamp, which is then sealed to a glass slide. Tubing is connected to the device and the flow rate is controlled by means of a syringe pump. (B) Typical field of view with a 40× phase contrast objective. For each measurement focused on the glass surface, 1 image/s was recorded for 25 min. (C–E) Analysis of the residence time distribution at a given flow rate: (C) Distribution of residence times for the population of bacteria. We note that with respect to the quantitative analysis in the Materials and Methods, the vertical axis in panel C is equivalent to P(t) = Ndes(t)/Ntot, where Ndes is the detachment rate of bacteria. (Inset) Log-log plot. (D) Fraction of adhesion events surviving longer than t (or renormalized number of cells on the surface f(t) = N(t)/Ntot with times renumbered so that all cells stick at t = 0). (Dashed line) Best fit with a stretched exponential function (Eq. 4). (Inset) Log-linear plot, which would be linear if the number of bacteria on the surface was decreasing exponentially. (E) Calculated detachment probability π as function of residence time. The data presented on the three graphs were obtained with PA14 wild-type flushed at 6 μL/min in a 400 × 35-μm cross-section channel (wall shear stress = 1.2 Pa). The total number of bacteria counted in this experiment was Ntot = 477. The best fit of f(t) with Eq. 4 corresponds to τ = 5.95 ± 0.11 s and α = 0.39 ± 0.003.These parameters were used to generate the curves superimposed to π(t) and P(t) using Eqs. 9 and 11, respectively.
At a given flow rate, successive images were recorded at 1 frame/s for 25 min, with an exposure time of 0.1 s. Image analysis was performed with a MATLAB (The MathWorks, Natick, MA) routine. For each image, the position and orientation of each bacterium were measured. Two successive images were compared to detect bacteria that had left the surface and new ones that had arrived, as well as to measure the displacement of adhered bacteria.
In a typical experiment, bacteria were flushed in the channel at a constant flow rate, and readily adhered onto the glass surface (Fig. 1 B); the majority of bacteria (>95%) typically stuck to the surface, attached for a limited time, and then detached. Only a small fraction of cells was not observed to leave the surface. To quantify the temporal dynamics of adhesion, we focused on the former population, i.e., cells that could be seen to leave the surface during the observation period.
Under our experimental conditions, all bacteria appeared to lie flat on the substrate (as opposed to attached by one pole and rotating, as sometimes described (23)). Although most tended to align with the flow, it was common to observe bacteria permanently misaligned or even perpendicular to the flow, suggesting that attachment occurred at several points of the bacterial surface. All cells exhibited very little motility while they were on the surface, as shown by the systematic measurement of the position and orientation of individual cells as a function of time (see Fig. S1 in the Supporting Material). The analysis shows that, whereas adsorbed bacteria preferentially move in the direction of the flow, typically the total average displacement between attachment and detachment barely exceeds the length of one bacterium (∼2 μm). This adhesive behavior is therefore intrinsically different from the stick-and-roll response previously reported for E. coli (24), which is displaced over large distances by the flow (typically tens of micrometers). In contrast, P. aeruginosa seems to stick-and-slide over short distances. In this case, a change in position corresponds to an undetermined number of bonds breaking and rearranging, so that trying to measure the lifetime of single bonds appeared impossible. Instead, we chose to focus on the total time each bacterium spent adhered to the surface, which we henceforth refer to as residence time.
Data analysis
For a given flow rate, image analysis allowed us to measure the residence time of each bacterium that adhered on and left the surface during the acquisition period. Bacteria that were already present at the beginning or still adhered at the end of the observation period, which were never >4–5% of the total count, were not taken into account. This analysis resulted in a distribution of residence times, which can be renormalized as a probability distribution of residence times (Fig. 1 C):
| (2) |
Here, Ndes(ti) is the detachment rate of bacteria that have spent time ti on the surface (i.e., the number of bacteria detected on exactly i successive images, and desorbing between ti and ti+1), and Ntot is the total number of bacteria counted during the 25-min acquisition period (i.e., i = 1–1500). We can transform this data to present results in a manner classical for adhesion events or bond formation, by defining
| (3) |
which is the fraction of adhesion events that last time tj or longer (Fig. 1 D). This way of presenting results has the advantage of highlighting long-time differences between different series of data of P(t). In all assays, the majority of bacteria left the surface within a few seconds. Nevertheless, significant variations were observed in the fraction of bacteria that had longer residence times, and constitute the right tail of the distribution.
As clearly shown by the log-linear plot in the inset of Fig. 1 D, this response is significantly different from a simple exponential response, which would be expected if the probability of detachment was independent of time. We found that f(t) can be well fitted with a stretched exponential function, given by
| (4) |
as shown in Fig. 1 D. Stretched-exponential distributions are commonly used to describe relaxation processes in complex systems (e.g., kinetics of polymer adsorption and desorption (25,26)) and are characterized by two adjustable parameters: a shape parameter α, and a characteristic time τ.
To further understand the dynamics of bacterial adhesion, it is useful to focus on the time-dependence of the bacterial detachment rate coefficient π(t). A priori, π depends on the time spent on the surface, so that the detachment rate of bacteria at a given time is
| (5) |
where Nτ(t) is the number of bacteria that have been residing on the surface for a duration τ at the time t, and π(t) is their detachment rate coefficient. Two simplifying cases can be considered: the case when the detachment probability does not depend on the residence time, in which case Ndes is simply proportional to the number of bacteria on the surface N(t); and the case when all bacteria have adsorbed on the surface at the same time t = 0. In the latter case, the detachment rate of bacteria is given by
| (6) |
with N(t) the number of bacteria left on the surface at time t.
In all of our experiments, although bacteria adhere at any moment during the acquisition period, our protocol (low bacterial and nutrient concentrations) makes it reasonable to assume that conditions remain similar throughout the experiment, i.e., that the chemical environment and the densities of bacterial populations on the surface and in solution do not vary significantly. Under this assumption we can represent data as if all bacteria adsorbed at the same time t = 0. In this case, f(t) can also be interpreted as the fraction of bacteria still remaining on the surface at time t,
| (7) |
with N(t) the number of bacteria left on the surface at time t. Using Eq. 6, the detachment rate coefficient can be defined as
| (8) |
and is easily calculated from f(t).
The function π(t) represents the likelihood of detachment of a bacterium as a function of its residence time on the surface (Fig. 1 E). We observe that π(t) decreases with the residence time, which indicates that bacteria become less likely to detach as they spend time on the surface. Similar qualitative observations have sometimes been attributed to bond aging (27), although the exact origin of this phenomenon has not yet been elucidated.
Using Eq. 6, it is straightforward to show that the stretched exponential model used for f(t) (Eq. 4) leads to fitting π(t) with a power-law function,
| (9) |
shown as the dashed curve in Fig. 1 E. Finally, combining Eqs. 2 and 6 yields the relation
| (10) |
which yields an expression for the probability distribution function of residence times:
| (11) |
In order to avoid artifacts from bacteria detected without truly adhering, to study the evolution of the characteristic residence time τ as a function of shear stress (in Figs. 3–5 below) we renormalized data so that the residence times were counted starting from the second image (i.e., t = 0 and f(t) = 1 corresponds to the number of bacteria detected on two successive images). Only data sets for which >50 adhesion events were detected were fitted. Fits were performed using the MATLAB curve-fitting tool, and error bars are given for a 99% confidence interval.
Figure 3.
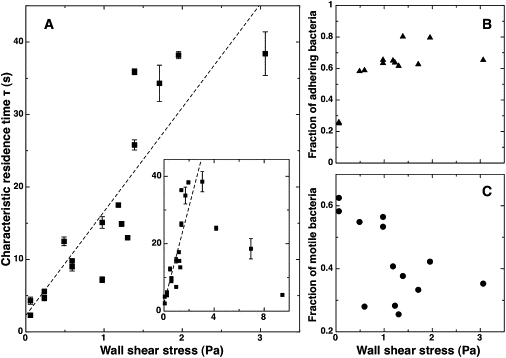
Adhesion dynamics on glass of wild-type PA14 as a function of shear stress. (A) Characteristic residence time τ, obtained by taking into account only bacteria detected on two or more successive images, as a function of wall shear stress. The value τ increases approximately linearly up to a critical value of the shear rate, then decreases (inset). Each point corresponds to an independent experiment, for which the shear was varied by changing the flow rate or the thickness of the channel. Error bars result from the fit of f(t) with 99% confidence bounds. (B) Fraction of strongly adhering bacteria, i.e., bacteria detected on two or more successive images, as a function of shear stress. (C) Fraction of visibly motile bacteria among adhering bacteria as a function of shear stress. Data was collected from 11 different sets of experiments, with a maximum of 2–3 measurements carried out in a given microfluidic device.
Figure 4.
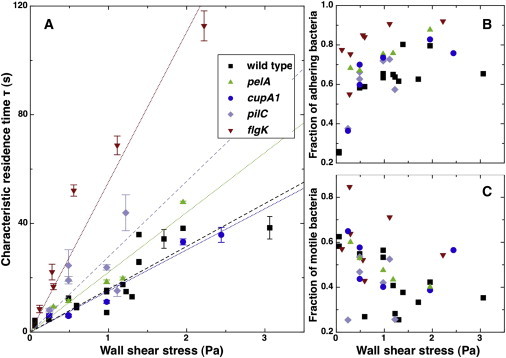
Adhesive behavior of matrix (pelA), type I pili (cupA1), type IV pili (pilC), and flagellum-defective (flgK) mutants on glass. (A) Characteristic residence time τ as a function of wall shear stress. Each point corresponds to an independent experiment. Error bars result from the fit of f(t) with 99% confidence bounds. Lines are linear fits to the experimental data, imposing τ(0) = 0. (B) Fraction of strongly adhering bacteria as a function of shear stress. (C) Fraction of motile bacteria among adhering bacteria as a function of shear stress.
Figure 5.
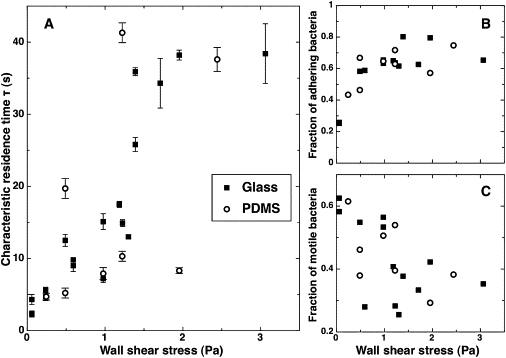
Adhesive behavior of wild-type PA14 on PDMS. (A) Characteristic residence time τ as a function of wall shear stress. Each point corresponds to an independent experiment. Error bars result from the fit of f(t) with 99% confidence bounds. (B) Fraction of strongly adhering bacteria as a function of wall shear stress. (C) Fraction of motile bacteria among adhering bacteria as a function of wall shear stress.
In summary, we are able to carefully characterize the adhesion dynamics of P. aeruginosa under flow. Next, we focus on the effect of shear stress on adhesion, i.e., how the typical set of data shown in Fig. 1 is modified when the shear stress varies. We will then use mutant strains to investigate the importance of surface appendages and matrix production on early time adhesion.
Results
Influence of shear stress
To study how shear stress influences bacterial attachment, we analyzed the adhesion of P. aeruginosa strain PA14 over a wide range of shear rates, 50–10,000 s–1, equivalent to a range of wall shear stresses of ∼0.05–10 Pa, with all other experimental conditions remaining identical. Our first observation was that the number of binding events tended to decrease as the shear increased (Fig. 2 A). In the range of wall shear stress 0–4 Pa, the number of adhering bacteria remained high enough for us to quantify residence time distributions. Normalized distributions f(t) for three different flow rates are shown in Fig. 2 B: on glass, detected bacteria typically adhered for a longer time as the shear stress increased, which is reflected by an increased fraction of long-time adhesion events. Examination of the detachment rate coefficient showed that this response was the consequence of a strong decrease of the detachment probability π(t) during the first seconds after adsorption (t < 10 s) (Fig. 2 C).
Figure 2.
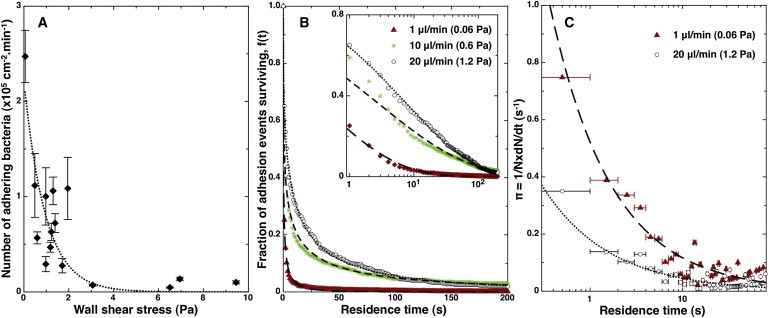
Influence of the shear stress on the adhesion of wild-type PA14 on glass. (A) Frequency of adhesion events recorded as a function of wall shear stress. (Dotted line) Best fit with an exponential function. OD600 were renormalized to 0.2. Because small concentration variations could occur during the experiment, error bars were calculated assuming that OD600 was known ±0.03. (B) Fraction of adhesion events surviving as a function of time, for three values of the shear stress (lin-log plot in inset). Fitting with a stretched exponential function (Eq. 4) gives the characteristic residence time τ. (C) Associated detachment rate coefficient as a function of residence time. Curves are obtained with a power-law model (Eq. 9) and the same fitting parameters as in plot B.
To quantify these observations, we fitted our data with the stretched exponential of Eq. 4. The characteristic residence time τ increased with increasing shear stress, and did so in an approximately linear fashion over a wide range of shear stresses (0.05–3.5 Pa) (Fig. 3 A). Although high shear rate measurements were difficult due to the low number of adhering bacteria, we reproducibly observed a decrease in τ at higher values of the shear stress, >4 Pa (Fig. 3 A, inset). Focusing on lower shear rates, we made two further observations:
-
1.
The fraction of bacteria that form long-lived adhesion events on the surface (i.e., bacteria that stick for two or more successive images) increased as the shear stress increased (Fig. 3 B).
-
2.
The fraction of visibly motile bacteria on the surface, defined as bacteria whose position varied significantly between attachment and detachment (by more than half the minor axis length), decreased as the shear stress increased (Fig. 3 C). Together, these trends led us to conclude that the adhesion of P. aeruginosa to glass was enhanced by shear stress.
Experiments with mutant strains
To determine the origin of the shear-enhanced adhesion of P. aeruginosa, we performed experiments with several types of mutant strains: bacteria defective in the synthesis of type I pili (cupA1), type IV pili (pilC), or flagellum (flgK), and a strain that is not able to produce the main polysaccharide of the extracellular matrix of strain PA14 (pelA). P. aeruginosa possesses two types of pili. Short type I pili are distributed all over the bacterial surface and play an important role in adhesion (28). Type IV pili, which are only a few and measure up to several micrometers in length, are necessary for twitching motility on surfaces and are involved in the early stages of biofilm development (20). Another surface appendage of P. aeruginosa is a single flagellum, which the bacteria use not only to swim but also to anchor to surfaces (29). All of these surface organelles are important for biofilm development, and, in the case of type IV pili and flagellum, have been shown to influence the initial steps of surface colonization by P. aeruginosa (21,20).
For all of the mutant strains, we observed a similar decrease in the total number of adhering cells as shear stress increased (see Fig. S2). However, more significant was the observation that the absolute number of adhering bacteria varied significantly between strains. Bacteria lacking the flagellum or type IV pili adhered less frequently to the substrate (total count was approximately two orders-of-magnitude lower than wild-type for the same acquisition time). This result is consistent with previous observations made in the absence of shear (20). Nevertheless, bacteria for which type I pili were missing showed an adhesion frequency close to the wild-type. For matrix mutants the number of adhesion events was even slightly higher than for the wild-type strain.
The residence times τ as a function of the wall shear rate for the different mutant strains (as well as for wild-type PA14) are shown in Fig. 4 A. We first asked if P. aeruginosa was similar to E. coli regarding surface adhesion, in that shear-enhanced adhesion in E. coli has been shown to be mediated by type I pili. To test the role of pili, we measured the residence time of P. aeruginosa PA14 mutants lacking type I (CupA1) or type IV (PilC) pili. As shown in Fig. 4, the results are qualitatively similar to what we observed with the wild-type strain. For both mutants the characteristic residence time increased as shear stress increased. However, whereas type I pili-deficient bacteria were indistinguishable from the wild-type, for type IV pili mutants the values of τ increased noticeably. Thus, our observations demonstrate that shear-enhanced adhesion of P. aeruginosa does not require the presence of either type I or type IV pili. This result highlights the fundamentally different nature of adhesion enhancement by shear stress in E. coli and P. aeruginosa.
Next we measured the residence time under shear of nonmotile mutants, which do not possess a flagellum. Here again, shear stress increased the fraction of long-term adhesion events (Fig. 4 A). However, we noticed an important difference: the characteristic residence times τ were much longer than with all other strains, and approximately three times longer than the ones obtained with wild-type bacteria. This result indicates that 1), bacteria could in fact anchor to the surface without a flagellum; and 2), the presence of a functional flagellum influences the rate of detachment. This latter point is further supported by the observation that the fraction of bacteria sticking for two images or more is significantly higher for flgK mutants than for other strains (Fig. 4 B): the probability of detaching during the first second after adsorption is reduced for bacteria lacking a flagellum.
All together, our results suggest that shear-enhanced adhesion is not related to surface appendages of P. aeruginosa. Instead, this behavior could be linked to receptors and structures located directly on the cell surface. This hypothesis is in agreement with the observation that bacteria seem to establish a close contact with the surface, along the entire body length. The bacterial surface is covered by a multitude of membrane proteins, polysaccharides, and the lipopolysaccharides (30,31). In addition, P. aeruginosa secretes polymeric substances to form the biofilm matrix. After adhesion of individual cells, the next step toward biofilm formation is the development of microcolonies where bacteria are connected by extracellular polymeric substances, but the timescale for the onset of matrix production is not well known.
Could bond aging and shear-enhanced adhesion be related to matrix production? To test whether extracellular polymeric substances are responsible for adhesion strengthening, we analyzed results obtained with a mutant unable to produce the main matrix exopolysaccharide (Pel) of P. aeruginosa PA14 (pelA) (21). As shown in Fig. 4, the trend observed for the characteristic residence time τ was not significantly modified when bacteria were unable to produce matrix. In the low shear stress region, the characteristic residence time increased with shear stress in a manner similar to the wild-type strain, and values of τ are of the same order of magnitude. This result suggests that the Pel exopolysaccharide, the main matrix building block, plays no significant role in the early bacterial adhesion process.
Discussion
The effect of shear stress on the adhesion of bacteria, cells, or colloidal particles to a substrate has been addressed by multiple studies. A decrease of the total number of adhesion events as shear increases has often been reported, e.g., for functionalized microspheres (32) as well as for bacteria (33,34), in agreement with our observations. Although an increased flow rate results in an increased mass transport, the decrease of the sticking efficiency suggests the existence of an energy barrier to form a reversible binding state. Focusing on the duration of adhesion events, or arrests in the case of particles rolling on a surface, the usual behavior is that an increased shear rate leads to shorter binding events. For example, in a study of streptavidin-coated micron-size spheres adhering to a biotinylated surface, an increase of the shear rate led to a higher detachment rate at short times (32). The same result was obtained for other ligand-receptor pairs (35). The opposite trend we report here for P. aeruginosa is thus counterintuitive.
What is the origin of the shear-increased adhesion time of P. aeruginosa? A longer residence time could result from an increased number of bonds forming between the cell and the surface, or may be due to the bonds themselves becoming stronger. Such bonds, whose lifetime increases under load, have been referred to as catch-bonds, as opposed to the more usual slip-bonds (36). In the case of E. coli, shear-enhanced adhesion is a consequence of specific catch-bonds formed between FimH, an adhesion protein present on E. coli type I pili, and mannose adsorbed on the substrate. Enhancement of adhesion is triggered by a conformational change of FimH under load (37).
By analogy with E. coli, could shear-enhanced adhesion in P. aeruginosa also be explained by the formation of catch-bonds? Let us assume that we are in the presence of catch-bonds formed between bacterial receptors and ligands on the surface. Our experiments demonstrate that in the case of P. aeruginosa, surface appendages (flagellum, pili) are not involved in this process. Specific receptors would thus have to be located directly on the cell surface. Our results also show that the Pel exopolysaccharide is not involved in catch-bond formation. Possibly, the shear-increased residence time could involve catch-bonds between cell-surface molecules and the substrate. However, an important feature of our experiments is that we used clean hydrophilic glass surfaces, which did not receive any specific treatment. This important point suggests that shear-enhanced adhesion on abiotic surfaces could be nonspecific. To further evaluate this hypothesis, we studied the adhesion of P. aeruginosa on a different substrate, PDMS, which is a hydrophobic polymer: as shown in Fig. 5, we observe a similar trend for the residence time, as well as for the fractions of adhering and mobile bacteria as shear stress increases. These results support the idea that adhesion could be nonsurface-specific.
Nevertheless, although they are initially bare, surfaces are undoubtedly rapidly conditioned by macromolecules present in the flowing medium, which could then act as specific ligands. Investigating this point, we first have been able to observe shear-enhanced adhesion both in high-ionic strength 90% PBS (10% TB), and in pure TB medium without salt (Fig. S3). To test the possible impact of surface conditioning by specific contaminants coming from the growth medium, we tried to grow bacteria in the minimal medium MSgg (18), which does not have any common component with TB. In addition, to rule out the potential influence of PDMS oligomers released from the channel walls, we also carried out experiments using a device made out of a rectangular glass capillary instead of molded PDMS. In both cases, we still observed shear-enhanced adhesion (Fig. S4). Hence the shear-increased residence time does not appear to depend on the medium used, nor the substrate or wall material.
However, surfaces could also be conditioned by molecules excreted by the bacteria themselves. To test this hypothesis, we tried to resuspend bacteria in totally clean medium immediately before measurements. Unfortunately, in this process we found out that centrifugation significantly modified the adhesive behavior of cells, even when resuspended in the original medium. Centrifugation might remove surface appendages, trigger matrix production, or more generally modify bacterial surface properties. As a consequence we were unable to draw conclusions from these experiments; however, this observation shows that P. aeruginosa bacteria are particularly sensitive to mechanical stress, and opens the way for future investigations in that domain.
In summary, our experiments indicate that if shear-enhanced adhesion of P. aeruginosa results from the formation of specific interactions, these have to take place between receptors located directly on the cell body, and bacteria-excreted molecules able to adsorb on different types of abiotic surfaces.
Let us now consider the possibility that our observations are not due to catch-bonds, but instead involve other mechanisms. For instance, our observations could result from a selectivity process, if the bacterial population was inhomogeneous and contained cells intrinsically stickier than others. Two different sets of experiments seem to rule out this possibility.
First, cells were left to adhere at very low shear rate (<100 s–1) for a couple of minutes, following which the flow rate was progressively increased by steps every 5 min. No bacteria that were originally adhered to the surface were observed to leave when the shear increased, which seems to disagree with the idea that bacteria have different adhering strengths due to varying cell surface properties.
In a second set of experiments, we recorded the detachment of recently adhered bacteria after a 30-fold decrease of the shear stress, from 0.732 to 0.024 Pa. Results show a significant increase in the detachment probability of bacteria, from to 29% to 46%, when compared to a control where the shear rate was kept constant (Fig. 6). This result is crucial because it proves that shear stress is indeed directly influencing the residence time of individual bacteria, even though they adhered under identical conditions.
Figure 6.
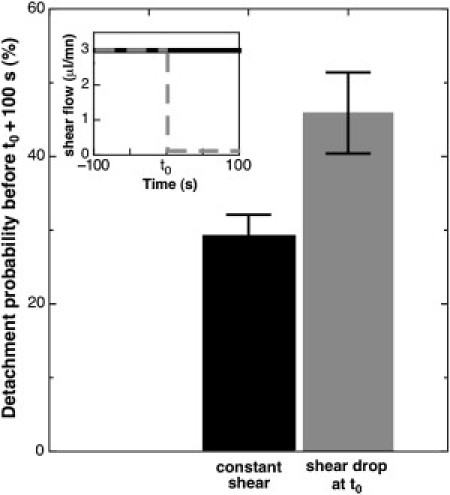
Influence of an abrupt decrease of the shear rate on the residence time of recently adhered bacteria. Bacteria were left to adhere for 100 s at a high flow rate value (3 μL/min), following which at t0 the flow rate was decreased to 0.1 μL/min (equivalent to changing shear stress from 0.73 to 0.024 Pa), or kept constant. Desorption events were recorded for 100 s after t0. Data represent the fraction of adhered bacteria detaching at constant shear stress (solid) or after a decrease of the shear stress (shading). (Inset) Flow rate as a function of time for both cases. Data are the average of 17 and 23 measurements, represented with standard errors.
Our observations are thus not the result of a selection process, but instead reflect a shear-sensitive mechanism at the cell level. This result would be consistent with the formation of catch-bonds, but does not rule out nonspecific behaviors. For example, mechanical stress could trigger a physical (e.g., deformation) or a biological (e.g., chemical surface modification, higher density of binding sites) response of bacteria, resulting in an increased number of bonds created between the cell and the surface. In that case, one would expect shear-enhanced adhesion to take place under a wide variety of environmental conditions, as seen in our experiments. Ongoing studies should hopefully allow us to determine the precise origin of shear-enhanced adhesion in P. aeruginosa.
Finally, although they do not elucidate the exact origin of the shear-enhanced adhesion, experiments with mutant strains provide a thorough investigation of the bacterial adhesion process. We observed that the flagellum and type IV pili are important for the establishment of initial adhesion, and strongly influence the frequency of adhesion events. However, paradoxically, once bacteria have adhered, the presence of these appendages also increases the likelihood of surface detachment at short times.
These observations suggest that bacterial desorption could be an active process involving the pili and/or the flagellum. One can imagine that bacteria that twitch or swim are more likely to be removed by the flow. As shear increases, we also observe that attached bacteria become less motile, which is consistent with this hypothesis and can account for longer residence times. Shear-enhanced adhesion also seems to involve the effect of shear itself on the adhesion strength. It is known that there is a delay in transforming the initial attachment into a close-contact with the substrate, during which the detachment probability is high. In the presence of shear stress this delay could be significantly shortened, resulting in a lower detachment probability. In the absence of long surface appendages, the dependence of the residence time on shear stress is more pronounced, possibly because the establishment of a close contact is easier.
The observations presented in this article outline a different adhesion mechanism under shear flow. They could also possibly impact the usual developmental model for the formation of a P. aeruginosa biofilm (38), which has been proposed from observations made either in the absence of flow or experiments using low-shear flow cells.
In conclusion, in this study we measured simultaneously the frequency of adhesion events and their duration for varying shear stresses. We discovered that P. aeruginosa cells create more long-lived adhesion events to immersed surfaces under shear stress, even though their probability of sticking is reduced. Furthermore, bacteria attaching under identical flow conditions are more likely to detach when shear is suddenly decreased, which shows that individual cells dynamically respond to shear rate variations, modifying their adhesion state. This phenomenon is not due to specific properties of surface organelles (pili, flagellum) or to exopolysaccharide production, but instead takes place under a wide variety of environmental conditions and on different surfaces. Although we were unable to identify a specific cell-surface interaction so far, their existence cannot be discarded and future studies are needed to clarify this point. On the other hand, a nonspecific mechanism would qualify shear-enhanced adhesion as a potentially widespread response, also observed for other microorganisms. Other recent works also seem to hint at this possibility (39).
Our results add to the understanding of the way bacteria colonize surfaces under flow. Such insights are important because controlling cell adhesion is of broad interest, whether it is to fight bacterial contamination and biofilm formation, or to promote surface colonization by desirable organisms.
Acknowledgments
We thank R. Losick for valuable discussions, and Y. Chai for technical advice. We are grateful to the Ausubel lab for the cupA1 strain. We thank U. Hees, J. Rieger, M. Rueckel, B. Sobotka, and the BASF-Harvard working group for useful feedback, and an anonymous referee for the suggestion to investigate a sudden reduction in shear rate.
This work was supported by the BASF Advanced Research Initiative at Harvard University, a grant from the Fulbright foundation to S.L., and a grant from the National Institutes of Health to R.K. (No. GM58213).
Supporting Material
References
- 1.Costerton J.W., Lewandowski Z., Lappin-Scott H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Dunne W.M., Jr. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Toole G., Kaplan H.B., Kolter R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 4.Stewart P.S., Costerton J.W. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 5.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 6.An Y.H., Friedman R.J. Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J. Biomed. Mater. Res. 1998;43:338–348. doi: 10.1002/(sici)1097-4636(199823)43:3<338::aid-jbm16>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 7.Neu T.R. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 1996;60:151–166. doi: 10.1128/mr.60.1.151-166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busscher H.J., van der Mei H.C. Microbial adhesion in flow displacement systems. Clin. Microbiol. Rev. 2006;19:127–141. doi: 10.1128/CMR.19.1.127-141.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meinders J.M., van der Mei H.C., Busscher H.J. Physicochemical aspects of deposition of Streptococcus thermophilus-b to hydrophobic and hydrophilic substrata in a parallel-plate flow chamber. J. Colloid Interface Sci. 1994;164:355–363. [Google Scholar]
- 10.Busscher H.J., van de Belt-Gritter B., van der Mei H.C. Streptococcus mutans and Streptococcus intermedius adhesion to fibronectin films are oppositely influenced by ionic strength. Langmuir. 2008;24:10968–10973. doi: 10.1021/la8016968. [DOI] [PubMed] [Google Scholar]
- 11.Katsikogianni M.G., Missirlis Y.F. Bacterial adhesion onto materials with specific surface chemistries under flow conditions. J. Mater. Sci. Mater. Med. 2010;21:963–968. doi: 10.1007/s10856-009-3975-y. [DOI] [PubMed] [Google Scholar]
- 12.Paris T., Skali-Lami S., Block J.C. Effect of wall shear rate on biofilm deposition and grazing in drinking water flow chambers. Biotechnol. Bioeng. 2007;97:1550–1561. doi: 10.1002/bit.21321. [DOI] [PubMed] [Google Scholar]
- 13.Purevdorj B., Costerton J.W., Stoodley P. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2002;68:4457–4464. doi: 10.1128/AEM.68.9.4457-4464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas W.E., Trintchina E., Sokurenko E.V. Bacterial adhesion to target cells enhanced by shear force. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 15.Thomas W. Catch bonds in adhesion. Annu. Rev. Biomed. Eng. 2008;10:39–57. doi: 10.1146/annurev.bioeng.10.061807.160427. [DOI] [PubMed] [Google Scholar]
- 16.Marshall B.T., Long M., Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- 17.Zhu C., Yago T., McEver R.P. Mechanisms for flow-enhanced cell adhesion. Ann. Biomed. Eng. 2008;36:604–621. doi: 10.1007/s10439-008-9464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Branda S.S., González-Pastor J.E., Kolter R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberati N.T., Urbach J.M., Ausubel F.M. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 21.Friedman L., Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 22.Xia Y.N., Whitesides G.M. Soft lithography. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- 23.Sjoblad R.D., Doetsch R.N. Adsorption of polarly flagellated bacteria to surfaces. Curr. Microbiol. 1982;7:191–194. [Google Scholar]
- 24.Thomas W.E., Nilsson L.M., Vogel V. Shear-dependent ‘stick-and-roll’ adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol. 2004;53:1545–1557. doi: 10.1111/j.1365-2958.2004.04226.x. [DOI] [PubMed] [Google Scholar]
- 25.Douglas J.F., Johnson H.E., Granick S. A simple kinetic model of polymer adsorption and desorption. Science. 1993;262:2010–2012. doi: 10.1126/science.262.5142.2010. [DOI] [PubMed] [Google Scholar]
- 26.Laherrere J., Sornette D. Stretched exponential distributions in nature and economy: “fat tails” with characteristic scales. Eur. Phys. J. B. 1998;2:525–539. [Google Scholar]
- 27.Vadillo-Rodríguez V., Busscher H.J., Van Der Mei H.C. Comparison of atomic force microscopy interaction forces between bacteria and silicon nitride substrata for three commonly used immobilization methods. Appl. Environ. Microbiol. 2004;70:5441–5446. doi: 10.1128/AEM.70.9.5441-5446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallet I., Olson J.W., Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lillehoj E.P., Kim B.T., Kim K.C. Identification of Pseudomonas aeruginosa flagellin as an adhesin for Muc1 mucin. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;282:L751–L756. doi: 10.1152/ajplung.00383.2001. [DOI] [PubMed] [Google Scholar]
- 30.Rocchetta H.L., Burrows L.L., Lam J.S. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ubbink J., Schar-Zammaretti P. Colloidal properties and specific interactions of bacterial surfaces. Curr. Opin. Colloid Interface Sci. 2007;12:263–270. [Google Scholar]
- 32.Pierres A., Touchard D., Bongrand P. Dissecting streptavidin-biotin interaction with a laminar flow chamber. Biophys. J. 2002;82:3214–3223. doi: 10.1016/S0006-3495(02)75664-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickinson R.B., Cooper S.L. Analysis of shear-dependent bacterial adhesion kinetics to biomaterial surfaces. AIChE J. 1995;41:2160–2174. [Google Scholar]
- 34.McClaine J.W., Ford R.M. Characterizing the adhesion of motile and nonmotile Escherichia coli to a glass surface using a parallel-plate flow chamber. Biotechnol. Bioeng. 2002;78:179–189. doi: 10.1002/bit.10192. [DOI] [PubMed] [Google Scholar]
- 35.Pierres A., Benoliel A.M., van der Merwe P.A. Determination of the lifetime and force dependence of interactions of single bonds between surface-attached CD2 and CD48 adhesion molecules. Proc. Natl. Acad. Sci. USA. 1996;93:15114–15118. doi: 10.1073/pnas.93.26.15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans E. Probing the relation between force lifetime and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 37.Forero M., Yakovenko O., Vogel V. Uncoiling mechanics of Escherichia coli type I fimbriae are optimized for catch bonds. PLoS Biol. 2006;4:e298. doi: 10.1371/journal.pbio.0040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monds R.D., O'Toole G.A. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Ding A.M., Palmer R.J., Jr., Kolenbrander P.E. Shear-enhanced oral microbial adhesion. Appl. Environ. Microbiol. 2010;76:1294–1297. doi: 10.1128/AEM.02083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


