Abstract
Bdellovibrio and like organisms (BALOs) form the group of predatory bacteria which require Gram-negative bacteria as prey. Genetic studies with Bdellovibrio bacteriovorus can be performed with vectors which are introduced into the predator via conjugation. The usefulness of the two vectors pSUP202 and pSUP404.2 as genetic tools were assessed. Both vectors were transferable into B. bacteriovorus by conjugative matings with an Escherichia coli K12 strain as donor. The transfer frequency was higher for vector pSUP404.2 (approx. 10−1–10−4) as for pSUP202 (approx. 10−5–10−6). Vector pSUP202 with a pMB1 origin is unstable in the predatory bacterium, whereas pSUP404.2 is stably maintained in the absence of selective antibiotics. pSUP404.2 harbors two plasmid replicons, the p15A ori and the RSF1010 replication region The copy number of pSUP404.2 was determined by quantitative PCR in B. bacteriovorus and averages seven copies per genome. pSUP404.2 harbors two resistance genes (chloramphenicol and kanamycin) which can be used for cloning either by selection for transconjugants or by insertional inactivation.
Introduction
Bdellovibrio bacteriovorus is the best characterized member of the Bdellovibrio and like organisms (BALOs [4]) which are predatory bacteria that attack and invade a number of Gram-negative bacteria. B. bacteriovorus depends on living bacteria for its growth and replication. The life cycle consists of a free-living attack phase and an intracellular replication phase. In the free living state predatory bacteria move with high velocity seeking prey bacteria. After attachment to prey bacteria B. bacteriovorus drills a pore through the outer membrane and invades the prey which is converted into a bdelloplast. In the following intracellular growth phase the invader consumes the cytoplasm of the prey and develops into a filament which septates into daughter cells. Subsequently the bdelloplast is lysed and newly differentiated attack phase cells are released [10, 13, 18].
In order to study this remarkable prokaryotic life cycle, genetic studies of B. bacteriovorus have been performed using a conjugation procedure which was first demonstrated by Cotter and Thomashow [2]. An E. coli K12 donor strain harboring the transfer function of the RP4 plasmid integrated in the chromosome transferred plasmids with a suitable mobilization region into predatory cells. Plasmids of the pMMB [8] and pSUP series [9] harboring an IncQ replicon were stably maintained in B. bacteriovorus, while plasmids with an IncP replicon were unstable. RP4 mobilizable plasmids with a pMB1/ColE1 replicon type which is present in most of the commonly used vectors (e.g., pBR vectors, pUC vectors, etc.) also do not replicate in B. bacteriovorus and have been used as suicide plasmids for insertional inactivation of chromosomal genes in the predator [5].
However, the number of genetic studies in B. bacteriovorus is limited and few vectors have been described more detailed in this host. For complementation studies in B. bacteriovorus only pMMB vectors have been used [2, 6, 16]. Because of the considerable importance for Bdellovibrio research to have functional plasmids we used pSUP202 and pSUP404.2 to analyze their properties for cloning purposes in B. bacteriovorus. These vectors have not been used in predatory bacteria before and especially vector pSUP404.2 offers some interesting features. pSUP404.2 possesses two replication regions, p15A origin and the IncQ replicon of RSF1010 [9] and has two antibiotic resistance genes (kanamycin resistance and chloramphenicol resistance) which can be used for selection or insertion of foreign genes. Vector pSUP202 contains a pMB1 ori and three antibiotic resistance genes which could be used for inserting genes of interest. We were interested in determination of the transfer frequency, stability and copy number of these vectors in B. bacteriovorus. We used the predatory streptomycin resistant mutant of B. bacteriovorus HD100-S1 as well as the prey independent derivatives M1 and M1.1 in this study (Table 1).
Table 1.
Bacterial strains and plasmids used in this work
| Description | Source | |
|---|---|---|
| Strains | ||
| B. bacteriovorus HD100-S1 | Streptomycin resistant wild-type of HD100 (DSM50701) prey-dependent | BfR strain collectiona |
| B. bacteriovorus M1 | Saprophytic HD100 mutant strain | This study |
| B. bacteriovorus M1.1 | Axenic HD100 mutant strain | This study |
| E. coli K-12 | DSM423 | |
| E. coli JC3272 | K-12, streptomycin resistant | [7] |
| E. coli K-12 S17.1λpir | RecA thio, hsdR − M, Integrated plasmid RP4-2-Kn::Tn7/TC::M | DSM9526 |
| E. coli K-12 Gene Hogs® | F-mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (StrR) endA1 nupG fhuA::IS2 | Invitrogen, Darmstadt, Germany |
| Plasmids | ||
| pSUP202 | pMB1 replicon, ApR, TcR, CMR, mob | [15] |
| pSUP404.2 | p15A replicon, RSF1010 replicon, KnR, CmR, mob | [9] |
| pGFPuv | pMB1 replicon, ApR, gfpuv | Clontech (Mountain View, USA) |
| pBR329 | pMB1 replicon; ApR, TcR, CmR | [3] |
| pIV2 | p29807 replicon, KnR | [17] |
| pNR24 | Insertion of Plac and gfp uv into pSUP404.2 via the restriction sites NheI and HindIII | This study |
aSpontaneous mutant isolated by Dominik Schwudke (pers. communication)
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains used in this study are listed in Table 1. E. coli strains were grown in Luria–Bertani (LB) medium (liquid or agar) overnight at 37°C.
Bdellovibrio bacteriovorus HD100-S1 was cultivated either in liquid cultures with E. coli prey or on double layer agar plates. In the latter case prey bacteria were resuspended within the top layer [11]. Stationary phase prey bacteria (E. coli DSM 423 or E. coli JC3272) were harvested by centrifugation and dissolved in the same volume of Bdellovibrio buffer (3 mM ammonium acetate, 3 mM CaCl2, and 3 mM MgCl2, pH 7.5). For liquid cultures this suspension was directly inoculated with B. bacteriovorus HD100-S1 and shaken at 28°C until complete lysis of the prey bacteria (depending on the amount of predators 1–3 days).
Mutant B. bacteriovorus M1 is a spontaneous mutant which grows and replicates on heat killed prey bacteria. For this purpose, stationary phase prey bacteria were harvested by centrifugation, dissolved in the same volume of Bdellovibrio buffer and autoclaved for 20 min at 121°C.
The axenic B. bacteriovorus mutant M1.1 is a derivative of mutant M1 and was grown in standard bacteriological medium (PYE; liquid or agar [11]).
For synchronized cultivation of B. bacteriovorus, two cultivation steps were necessary. In the first step the prey suspension containing E. coli JC3272 was infected with B. bacteriovorus HD100-S1 from stock cultures and shaken at 28°C. After lysis cell debris and remaining prey bacteria were removed by centrifugation (10 min, 2,600×g at RT). 10 ml supernatant containing attack phase predators was added to 30 ml of freshly prepared prey bacteria to give a predator to prey ratio of approximately 1:3. Mixing was set as starting point of the synchronized cultivation, which was shaken at 28°C.
Bdellovibrio strains were stored at −80°C in either prey suspension or PYE supplemented with 30% glycerol.
Plasmid and Chromosomal DNA Preparations
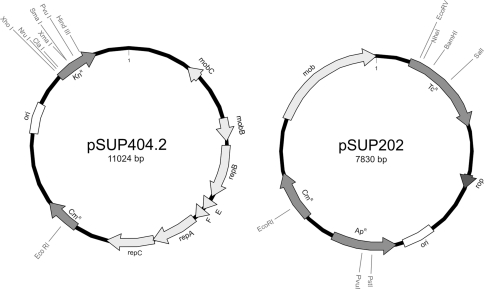
Plasmids used in this work are listed in Table 1. Vector pSUP202 (Fig. 1) contains single restriction sites for inactivation of ampicillin resistance (ApR) gene, tetracycline resistance (TcR) gene, and chloramphenicol resistance (CmR) gene. The replicon is derived from pMB1 and contains the RP4 mob region (Fig. 1). The complete sequence of pSUP202 is available in the NCBI database (accession AY428809).
Fig. 1.
Physical maps of the mobilizable vector plasmids pSUP404.2 and pSUP202. The map of pSUP202 is derived from the NCBI (accession AY428809). The map of pSUP404.2 was generated according to our sequence analyses (accession FN907923). Single restriction sites in antibiotic resistance genes are indicated, ori dedicates the p15A and pMB1 replicon regions of the plasmids pSUP404.2 and pSUP202, respectively
Plasmid preparations of B. bacteriovorus and E .coli were carried out using the Gene JetTM Plasmid Miniprep Kit (Fermentas, St. Leon-Rot, Germany).
Chromosomal DNA of B. bacteriovorus was prepared from purified predators cells by the Cetyltrimetylammoniumbromide (CTAB) extraction procedure [1]. Briefly, cells were harvested by centrifugation and inoculated with sodiumdodecylsulfate (0.5% final concentration), RNase (170 μg/ml), and Proteinase K (100 μg/ml) for 1 h. Sodium chloride to a concentration of 0.8 M and 1% CTAB were added and the solution was incubated for 10 min at 65°C. An equal volume of chloroform was added and the mixture was centrifuged at 14,900×g for 10 min at RT. The upper phase was transferred to a clean tube and an equal volume of phenol/chloroform/isoamylalcohol (25/24/1, v/v/v, premixed) was added. The mixture was centrifuged at RT for 10 min at 14,900×g. The supernatant was transferred to a clean tube and DNA was precipitated by addition of 0.6 volume of isopropanol. The precipitate was washed with 70% ethanol, dried in a vacuum concentrator and redissolved in distilled water.
Amplification of DNA by Polymerase Chain Reaction
For polymerase chain reaction (PCR), 0.5 μl Taq polymerase (5 U/μL), 5 μl dNTPs (2.5 mM each), 3 μl magnesium chloride (50 mM), 5 μl reaction buffer, 5 μl forward and reverse primer (5 μM each), and 1 μl Template DNA (0.1–1.0 ng) in 50 μl reaction volume were used. All chemicals were obtained from Bioline (Luckenwalde, Germany). The PCRs were performed with an initial denaturation step of 95°C for 5 min and 35 cycles (denaturation at 95°C for 30 s, annealing for 30 s and elongation at 72°C). Amplified PCR fragments were purified for either cloning or sequencing using the MSB®Spin PCRapace Kit from Invitek (Berlin, Germany). Oligonucleotide primers were designed using the Accelrys (DS-) gene program 2.5 (2006, Accelrys, Inc., San Diego, USA) (Table 2). Oligonucleotides were supplied by Metabion (Martinsried, Germany).
Table 2.
PCR primers and Taqman® probes for real-time PCR
| Target gene | Sequence 5′–3′ | Accession |
|---|---|---|
| Bd0108 (chromosome) | GACAGATGGGATTACTGTCTTCCA | NC_005363 |
| CGTGAGTGTATTGCCTACCGTAC | ||
| Fam-TTCTCTGGAAACGGTGTCGTCATGGC-BHQ-1 | ||
| cat gene (pSUP404.2) | TTTTATCCGGCCTTTATTCACATT | FN907923 |
| ACCGTCTTTCATTGCCATACG | ||
| Yakima Yellow-TTGCCCGCCTGATGAATGCTCA-BHQ-1 | ||
| gfp (pGFPuv) | GACGCTAGCCTAGCTAGAGAAGGTGATTAT | U62636 |
| ACCAAGCTTATTTGTAGAGCTCATCCATGC |
Mating Experiments
Matings were performed with the donor strain E. coli S17.1λpir harboring the RP4 transfer region chromosomally integrated. 20 ml of B. bacteriovorus recipients from liquid cultures containing about 108 Bdellovibro bacteria were concentrated tenfold by centrifugation and resuspended in 2 ml Bdellovibrio buffer. Donors were grown to stationary phase and concentrated tenfold in the same buffer. 100 μl of each suspension were mixed and transferred onto nitrocellulose filters (0.22 μm pore size) which were placed on top of LB agar plates. Matings were incubated overnight at 28°C. The next day, the bacteria were resuspended in 2 ml Bdellovibrio buffer and spread on double layer agar plates with E. coli JC3272 [pBR29] under selective conditions (25 μg/ml streptomycin, 12.5 μg/ml chloramphenicol) to determine the transfer frequency. The cells were incubated at 28°C until plaques became visible in the lawn of prey cells.
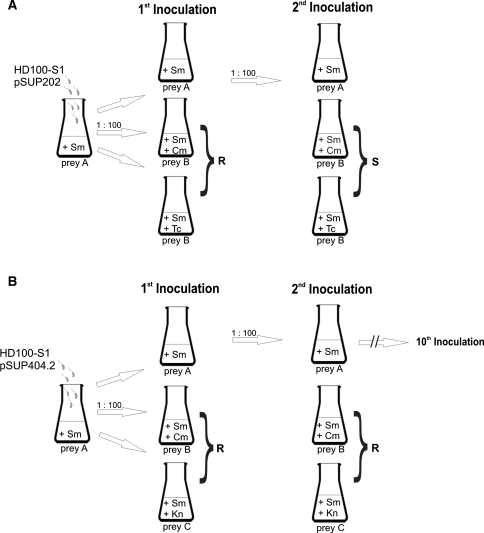
In case of stability experiments the resuspended bacteria were added to a freshly prepared prey suspension of E. coli JC3272 (see Fig. 2). After lysis of this first culture aliquots of the predatory bacteria were used for inoculation of the following cultures: cultures with E. coli JC3272, cultures with E. coli JC3272 [pBR29] under selective conditions (25 μg/ml streptomycin, 12.5 μg/ml chloramphenicol, 12.5 μg/ml tetracycline) or E. coli JC3272 [pIV2] (100 μg/ml kanamycin). Complete lysis of prey cells in the liquid cultures takes place within 2 days and is visible by clearance of the prey culture and microscopic control (Nikon-Optiphot-2, phase-contrast 1000×).
Fig. 2.
Stability of pSUP202 (a) and pSUP404.2 (b) in B. bacteriovorus HD100-S1. Following mating transconjugants were grown under nonselective conditions. From this culture three consecutive cultures were inoculated with a dilution of 1:100 (1st inoculation). For further inoculations only cultures grown with prey A were used. Prey A = streptomycin resistant E. coli JC3272, prey B = E. coli JC3272 [pBR329], prey C = E. coli JC3272 [pIV2]; Sm (streptomycin), Tc (tetracycline), Cm (chloramphenicol), Kn (kanamycin) show antibiotics added to media. R indicates resistance; S indicates sensitivity to antibiotics added to media
Determination of Plasmid Copy Number Using Real-Time PCR
To estimate the copy number of the plasmid pSUP404.2 within B. bacteriovorus, the number of plasmids was compared to the number of chromosomal copies using Real-Time PCR. Total DNA was isolated from the strain B. bacteriovorus HD100-S1 containing the plasmid pSUP404.2 and analyzed with the ABI-7500 Real-Time PCR System (Applied Biosystems, Foster City, USA). Primers and probes were designed (Table 2) using the Primer Express software version 3.0 (Applied Biosystems). The NanoDrop Spectrophotometer ND-1000 (peQLab, Erlangen; Germany) was used to determine the DNA concentrations. Serial dilutions from 1.66 × 10−1 to 1.66 × 10−8 ng DNA (plasmid pSUP404.2) and 9.882 ng to 9.882 × 10−7 ng DNA (chromosome) were prepared for calibration. The number of plasmid molecules (size of 11,026 bp) contained in 1 μg was calculated to be 2.25 × 1010. In comparison 1 μg chromosomal DNA (3,782,950 bp) consists of approximately 2.41 × 108 molecules ([10], accession NC_005363).
Plasmid copy number was calculated by dividing the number of plasmids by the number of chromosomal molecules. All data were analyzed using the 7500 Software version 2.0.1 (2008, Applied Biosystems).
DNA Sequencing and Computer Analysis
DNA sequencing was done by primer walking. Sequencing of either the pSUP404.2 plasmid or the newly generated gfp-construct (pNR24) was done by QIAGEN sequencing services (Hilden, Germany). The obtained sequences were analyzed using the Lasergene programme “SeqMan” (DNASTAR, Inc., Madison, USA). The nucleotide sequence of pSUP404.2 was deposited at EMBL database (accession FN907923).
Fluorescence Microscopy
Fluorescent bacteria were diluted 1:2 with 0.8% agarose and spotted on glass slides. Bacteria were visualized with a Zeiss Axioskop microscope at a magnification of 1000× using a mercury lamp with a filter for blue excitation.
Results and Discussion
Sequence Analysis of Plasmid pSUP404.2
Vector pSUP404.2 was constructed as part of a series of cloning vectors [9] and only incomplete sequence information was available. To accomplish its detailed physical map including all restriction sites we sequenced the plasmid prior to use (accession FN907923). Figure 1 shows the map of the vector which has a size of 11,024 bp. The largest part (nucleotides 482–6135) of pSUP404.2 originates from the plasmid RSF1010 and contains its mob and broad host range replication functions. The other parts of pSUP404.2 are derived from different cloning vectors. The main functional genetic elements are the origin of replication (ori) and the kanamycin resistance (KnR) gene from pACYC177 and the chloramphenicol resistance gene (CmR). Table 3 shows singular restriction sites that could be used for cloning purposes.
Table 3.
Selected singular restriction sites in the plasmid pSUP404.2 useful for cloning
| Restriction enzyme | Restriction site | Gene/target |
|---|---|---|
| AatII | 7824 | Plasmid backbone |
| AsiSI | 10053 | KnR |
| BanII | 9708 | KnR |
| BmtI | 9292 | Plasmid backbone |
| Bsu36I | 7400 | Plasmid backbone |
| ClaI | 9744 | KnR |
| EcoRI | 6745 | CmR |
| HincII | 254 | Plasmid backbone |
| HindIII | 10171 | KnR |
| MluI | 7744 | Plasmid backbone |
| NaeI | 399 | Plasmid backbone |
| NheI | 9288 | Plasmid backbone |
| NruI | 9710 | KnR |
| PvuI | 10053 | KnR |
| SmaI | 9927 | KnR |
| XhoI | 9651 | KnR |
| XmaI | 9925 | KnR |
Plasmid RSF1010 is a broad host range plasmid and its replicon belongs to the IncQ-type family that was shown to replicate autonomously in B. bacteriovorus [2]. The pACYC177 ori stems from the p15A plasmid [14]. The p15A replicon is closely related to the pMB1/ColE1 replicon which does not replicate in Bdellovibrio, as enzymes of the bacterial host are required for its replication [12]. The presence of the p15A ori increases the copy number of pSUP404.2 in E. coli and facilitates its isolation from this standard E. coli K-12 host strains.
The two vectors pSUP202 (“Materials and Methods” section) and pSUP404.2 carry mobilization regions that enable their conjugative transfer by the RP4 transfer system.
Conjugative Transfer of the pSUP202 and pSUP404 into B. bacterivorus
Mobilization of the two plasmids into the streptomycin resistant B. bacteriovorus strain HD100-S1 was carried out with E. coli S17.1λpir as donor, which provides the RP4 transfer functions chromosomally integrated.
The matings were carried out with the donor strain E. coli S17.1λpir and B. bacteriovorus HD100-S1 as recipient. To determine the number of transconjugant bdellovibrios, serial dilutions of mating experiments were plated after overnight incubation on double layer agar plates supplemented with streptomycin and chloramphenicol containing the prey bacterium E. coli JC3272 [pBR329] in the top layer. After 2 days of incubation plaques became visible. The transfer frequency was calculated from the ratio of donors to transconjugants, whereby the number of transconjugants was determined by counting the PFU (plaque forming units). In case of pSUP404.2 transfer frequencies between 10−1 and 10−4 were obtained (Table 4). Despite these variations conjugative transfer was easily reproducible. Conjugative transfer was also detected for pSUP202 (10−5–10−6) which, however, is quickly lost after prolonged incubation (see below). Cotter and Thomashow [2] found transfer frequencies in the range of 10−3 for IncQ plasmids, while ColE1/pMB1 plasmids were not successfully conjugated.
Table 4.
Transfer frequency and stability of pSUP202 and pSUP404.2
| pSUP202 | pSUP404.2 | |
|---|---|---|
| Transfer frequencya | 4.8 × 10−5 | 3.5 × 10−3 |
| 4.8 × 10−6 | 3.4 × 10−1 | |
| 4.9 × 10−4 | ||
| Stabilityb | ||
| After 1st inoculation | + | + |
| After 2nd inoculation | − | + |
| After 10th inoculation | − | + |
aTransfer frequency determined in independent experiments: number of CmR SmR B. bacteriovorus HD100-S1 transconjugants per E. coli S-17.1 donor; number of transconjugants were determined on double layer plates containing streptomycin and chloramphenicol with prey E. coli K-12 JC3272 [pBR329]
bStability determined in liquid cultures. + growth in liquid cultures containing streptomycin and antibiotic selective for the corresponding plasmid. − No growth because of loss of plasmid mediated antibiotic resistance
To test if plasmid transfer was really a conjugative process, matings with an E. coli DH5α [pSUP404.2] strain as donor were conducted which does not provide RP4 transfer function. As was found by Cotter and Thomashow [2] we did not find plaque formation under selective conditions on double layer plates in these experiments, thus confirming that plasmid transfer was conjugative in nature.
Furthermore, electroporation of pSUP404.2 into the mutant strain B. bacteriovorus M1.1, which is able to grow on complex bacteriological medium (PYE), was tested. Two different voltages were applied (2.5 and 3 kV), but this method was not successful. So far there is only one report on successful electroporation of bdellovibrios [19].
Stability of Vectors in B. bacteriovorus
To investigate the stability of the plasmids pSUP202 and pSUP404.2 B. bacteriovorus strain HD100-S1 was used as recipient. After mating the transconjugant HD100-S1 bacteria were cultivated in liquid cultures containing streptomycin with prey bacteria (E. coli JC3272) without addition of any of the plasmid specific antibiotics. After prey lysis aliquots of these primary cultures were taken for inoculation of freshly prepared prey suspensions containing (i) JC3272 with only streptomycin, (ii) JC3272 [pBR329] with streptomycin and chloramphenicol, or streptomycin and tetracycline, and (iii) JC3272 [pIV2] with streptomycin and kanamycin (see Fig. 2).
After 2 days of cultivation these cultures were controlled microscopically for presence of predatory bacteria. If transconjugants were present under selective conditions for pSUP404.2 (streptomycin and chloramphenicol; streptomycin and kanamycin) and for pSUP202 (streptomycin and chloramphenicol; streptomycin and tetracycline), from the culture that contained only streptomycin an aliquot was taken for inoculation of new liquid cultures with the above described prey bacteria and antibiotic combinations. The procedure was repeated in case of pSUP404.2 ten times, while in case of pSUP202 after the second inoculation no growth was obtained in media containing the antibiotics tetracycline and chloramphenicol.
It can be concluded that pSUP202 is rapidly lost in B. bacteriovorus, whereas plasmid pSUP404.2 is stably maintained. It was possible to isolate pSUP404.2 after 10 successive inoculations and growth under non-selective conditions from predatory bacteria. Transformation of the isolated plasmid into E. coli Gene Hogs® and growth of E. coli transformants on LB medium containing either kanamycin or chloramphenicol, as well as restriction digests with different enzymes verified that the isolated plasmid was the cloning vector pSUP404.2.
As B. bacteriovorus replicates by septation of a filament that releases varying numbers of daughter cells it is not possible to express the stability of plasmids in generation times. An active Bdellovibrio culture has a concentration of approximately 2 × 108 cells/ml and for the inoculation of a new liquid culture an aliquot was chosen giving a 1:100 dilution.
Vector pSUP404.2 harbors the minimal region of plasmid RSF1010 needed for replication and mobilization by RP4 transfer with a size of 6 kbp [9]. RSF1010 is known to replicate autonomously in a wide variety of gram-negative bacteria, while the pMB1 ori of pSUP202 is dependent on replication enzymes of the bacterial host [12]. The host range of ColE1 ori is limited to Enterobacteriaceae and pSUP202 sequences can be maintained in B. bacteriovorus only by homologous recombination with chromosomal sequences resulting in integration. This can be achieved by insertion of chromosomal B. bacteriovorus DNA into the vector.
Copy Number of pSUP404.2 in B. bacteriovorus
The determination of the pSUP404.2 copy number was carried out with synchronized cultures of B. bacteriovorus HD100-S1 using E. coli JC3272 [pIV2] as prey bacteria. For a period of 5 h samples were taken hourly, plus one sample taken after an incubation of 21.5 h.
To identify the copy number of the plasmid in comparison to the bacterial chromosome, quantitative real time PCR was applied and two different Taqman® probes were designed. The first probe was specific to the pSUP404.2 encoded chloramphenicol resistance gene (CmR) and the second one detected the B. bacteriovorus specific gene Bd0108 (hit—host interaction locus) which is located in the chromosome.
Table 5 shows that the plasmid pSUP404.2 exists in a ratio of approximately seven plasmids per one copy of bacterial chromosome. This means that the cloning vector is maintained as a low-copy number plasmid in B. bacteriovorus. The direct isolation of pSUP404.2 from B. bacteriovorus is tedious because of the limited amount of plasmid DNA which can be recovered. However, by transformation of crude B. bacteriovorus plasmid preparations into standard E. coli K12 host strains (e.g. Gene Hogs®), the p15A origin increases the copy number in these transformants allowing easy recovery of recombinant plasmids from these secondary hosts.
Table 5.
Copy number of vector pSUP404.2 in B. bacteriovorus HD100-S1
| Time | Probea | ∅ Ct-valueb | DNA molecules | Mean of DNA molecules | Ratio chromosome:plasmid |
|---|---|---|---|---|---|
| 1 h | Chlor-I | 18.81 | 2.90 × 106 | 2.96 × 106 | |
| Chlor-II | 19.83 | 3.05 × 106 | 1:6.96 | ||
| Hit-I | 18.36 | 4.15 × 105 | 4.25 × 105 | ||
| Hit-II | 19.19 | 4.34 × 105 | |||
| 2 h | Chlor-I | 18.74 | 2.89 × 106 | 3.01 × 106 | |
| Chlor-II | 20.3 | 3.13 × 106 | 1:6.95 | ||
| Hit-I | 18.65 | 4.21 × 105 | 4.33 × 105 | ||
| Hit-II | 19.71 | 4.45 × 105 | |||
| 3 h | Chlor-I | 19.30 | 2.93 × 106 | 3.06 × 106 | |
| Chlor-II | 20.63 | 3.18 × 106 | 1:7.08 | ||
| Hit-I | 18.54 | 4.19 × 105 | 4.32 × 105 | ||
| Hit-II | 19.67 | 4.45 × 105 | |||
| 4 h | Chlor-I | 18.65 | 2.87 × 106 | 3.01 × 106 | |
| Chlor-II | 20.39 | 3.14 × 106 | 1:7 | ||
| Hit-I | 18.16 | 4.10 × 105 | 4.30 × 105 | ||
| Hit-II | 19.88 | 4.49 × 105 | |||
| 5 h | Chlor-I | 17.25 | 2.66 × 106 | 2.76 × 106 | |
| Chlor-II | 18.48 | 2.85 × 106 | 1:6.78 | ||
| Hit-I | 17.46 | 3.95 × 105 | 4.07 × 105 | ||
| Hit-II | 18.53 | 4.19 × 105 | |||
| 21.5 h | Chlor-I | 13.56 | 2.09 × 106 | 2.08 × 106 | |
| Hit-I | 13.68 | 2.07 × 106 | 1:6.75 | ||
| Chlor-II | 13.43 | 3.09 × 105 | 3.08 × 105 | ||
| Hit-II | 13.55 | 3.06 × 105 | |||
| ∅ | Mean value | 1:6.9 | |||
aTwo independent sets of experiments were performed, indicated by Roman numerals I and II
bEach Ct value was determined in quadruplicate
gfp Expression in B. bacteriovorus
To demonstrate the applicability of plasmid pSUP404.2 for cloning, the gene of the green fluorescence protein (gfp) was chosen for a test cloning. For this purpose, the whole gene plus its promoter sequence (Plac) was amplified from the vector pGFPuv and cloned into the plasmid pSUP404.2, using the restriction sites NheI and BspI. The recombinant plasmid was designated pNR24. Matings, as described above, were carried out between E. coli S17.1λpir [pNR24] and either B. bacteriovorus HD100-S1 or the saprophytic growing mutant 100-M1.
The matings were examined by fluorescence microscopy. Fluorescent cells could be seen in an intense green color (data not shown). Expression of green fluorescence protein in wild type B. bacteriovorus has been also reported by Flannagan et al. [6]. The observation of fluorescent B. bacteriovorus cells demonstrates that pSUP404.2 was functional either in wild type bacteria or saprophytic mutants.
Conclusion
As the number of vectors for genetic manipulation of predatory bacteria is limited, we studied two vectors of the pSUP series in detail [9]. pSUP202 is a vector with the pMB1 replicon that does not replicate in B. bacteriovorus, but can efficiently be mobilized into the predator and therefore could be used as a suicide vector. Vector pSUP404.2, in contrast, is stably maintained in B. bacteriovorus with a copy number of approximately seven per genome even in the absence of selective conditions. This vector can be applied for expression of foreign genes and complementation experiments in B. bacteriovorus. The kanamycin and chloramphenicol resistance genes can be used for selection of transconjugants, or can be used for insertional inactivation as both genes have singular restriction sites.
Acknowledgments
We would like to acknowledge Dr. Jochen Reetz (BfR) for help with the fluorescence microscopy. Furthermore, we would like to thank Andreas Schlueter (Bielefeld University) for providing the plasmids pSUP404.2 and pSUP202. N.R. receives a Grant from DFG (STR 1034/1-1).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidmon JG, Smith JA, et al. Current protocols in molecular biology. New York: Greene Publishing Associates, Wiley Interscience; 1987. [Google Scholar]
- 2.Cotter TW, Thomashow MF. A conjugation procedure for Bdellovibrio bacteriovorus and its use to identify DNA sequences that enhance the plaque-forming ability of a spontaneous host-independent mutant. J Bacteriol. 1992;19:6011–6017. doi: 10.1128/jb.174.19.6011-6017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Covarrubias L, Bolivar F. Construction and characterisation of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 428-base-pair inverted duplication. Gene. 1982;17:79–89. doi: 10.1016/0378-1119(82)90103-2. [DOI] [PubMed] [Google Scholar]
- 4.Davidov Y, Jurkevitch E. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovoraxstarrii as Peredibacterstarrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int J Syst Evol Microbiol. 2004;54:1439–1452. doi: 10.1099/ijs.0.02978-0. [DOI] [PubMed] [Google Scholar]
- 5.Fenton AK, Lambert C, Wagstaff PC, Sockett RE. Manipulating each mreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J Bacteriol. 2010;192:1299–1311. doi: 10.1128/JB.01157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flannagan RS, Valvano MA, Koval SF. Downregulation of the motA gene delays the escape of the obligate predator Bdellovibrio bacteriovorus 109 J from bdelloplasts of bacterial prey cells. Microbiology. 2004;150:649–656. doi: 10.1099/mic.0.26761-0. [DOI] [PubMed] [Google Scholar]
- 7.Maidhof H, Guerra B, Abbas S, Elsheikha HM, Whittam TS, Beutin L. A multiresistant clone of Shiga toxin producing Escherichia coli O118:[H16] is spread in cattle and humans over different European countries. Appl Environ Microbiol. 2002;68:5834–5842. doi: 10.1128/AEM.68.12.5834-5842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales VM, Bäckmann A, Bagdasarian M. A series of wide-host-range lo-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-X. [DOI] [PubMed] [Google Scholar]
- 9.Priefer UB, Simon R, Pühler A. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J Bacteriol. 1985;163:324–330. doi: 10.1128/jb.163.1.324-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 11.Schwudke D, Strauch E, Krüger M, Appel B. Taxonomic studies of predatory bdellovibrios based on 16 s rRNA analysis, ribotyping and the hit locus and characterisation of isolates from the gut of animals. Syst Appl Microbiol. 2001;24:385–394. doi: 10.1078/0723-2020-00042. [DOI] [PubMed] [Google Scholar]
- 12.Scott JR. Regulation of plasmid replication. Microbiol Rev. 1984;48:1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seidler RJ, Starr MP. Factors affecting the intracellular parasitic growth of Bdellovibrio bacteriovorus developing within Escherichia coli. J Bacteriol. 1969;97:912–923. doi: 10.1128/jb.97.2.912-923.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selzer G, Som T, Itoh T, Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983;32:119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- 15.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gramnegative bacteria. Bio/Technology. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 16.Steyert SR, Pineiro SA. Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus. Appl Environm Microbiol. 2007;73:4717–4724. doi: 10.1128/AEM.00640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strauch E, Voigt I, Broll H, Appel B. Use of a plasmid of a Yersinia enterocolitica biogroup 1 A strain for the construction of cloning vectors. J Biotechnol. 2000;72:63–72. doi: 10.1016/S0168-1656(00)00216-9. [DOI] [PubMed] [Google Scholar]
- 18.Strauch E, Schwudke D, Linscheid M. Predatory mechanisms of Bdellovibrio and like organisms. Future Microbiol. 2007;2:63–73. doi: 10.2217/17460913.2.1.63. [DOI] [PubMed] [Google Scholar]
- 19.Tudor JJ, Davis JJ, Panichella M, Zwolak A. Isolation of predation-deficient mutants of Bdellovibrio bacteriovorus by using transposon mutagenesis. Appl Environ Microbiol. 2008;74:5436–5443. doi: 10.1128/AEM.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]